Abstract
Type I interferons (IFNs) play an essential role in the host response to viral infection through the induction of numerous IFN-stimulated genes (ISGs), including important antiviral molecules such as PKR, RNase L, Mx, and iNOS. Yet, additional antiviral ISGs likely exist. IFN-stimulated gene 15 (ISG15) is a ubiquitin homolog that is rapidly up-regulated after viral infection, and it conjugates to a wide array of host proteins. Although it has been hypothesized that ISG15 functions as an antiviral molecule, the initial evaluation of ISG15-deficient mice revealed no defects in their responses to vesicular stomatitis virus or lymphocytic choriomeningitis virus, leaving open the important question of whether ISG15 is an antiviral molecule in vivo. Here we demonstrate that ISG15 is critical for the host response to viral infection. ISG15−/− mice are more susceptible to influenza A/WSN/33 and influenza B/Lee/40 virus infections. ISG15−/− mice also exhibited increased susceptibility to both herpes simplex virus type 1 and murine gammaherpesvirus 68 infection and to Sindbis virus infection. The increased susceptibility of ISG15−/− mice to Sindbis virus infection was rescued by expressing wild-type ISG15, but not a mutant form of ISG15 that cannot form conjugates, from the Sindbis virus genome. The demonstration of ISG15 as a novel antiviral molecule with activity against both RNA and DNA viruses provides a target for the development of therapies against important human pathogens.
Keywords: innate immunity, ubiquitin, viral pathogenesis, RNA viruses, DNA viruses
Type I interferons (IFN-α and -β) play a critical role in the innate immune response to viral infection. They exert their biological effects through the induction of hundreds of IFN-stimulated genes (ISGs). Although a subset of these genes have known antiviral activity, the function of many of them remains unknown (1–3).
ISG15 is an IFN-stimulated gene that is rapidly up-regulated after stimulation with either IFNs or viral infection. It contains two domains with structural homology close to ubiquitin, connected by a proline-containing linker (4). ISG15 can be released from cells and function as an extracellular cytokine; or similar to ubiquitin, it can conjugate to target proteins through its C-terminal LRLRGG motif (5–9). ISG15 utilizes an IFN-inducible conjugation cascade that includes an E1 (UBE1L), E2 (UbcM8), and to date two identified E3 ligases (estrogen-responsive finger protein and Herc5) as well as a deconjugating enzyme, UBP43 (10–14). Recent studies have identified >100 target proteins of ISG15, which encompass diverse cellular pathways (15–17). However, the consequence of ISGylation of target proteins and thus the cellular function(s) of ISG15 remain unknown (18, 19).
Given the dramatic up-regulation of ISG15 after viral infection, there has been speculation that ISG15 functions as an antiviral molecule. Recent studies have shown that the influenza B virus targets the disruption of the ISG15 pathway through the activity of its NS1 protein (10). NS1-B binds to ISG15, inhibiting its interaction with its E1 enzyme, UBE1L, and disrupting the formation of ISG15 conjugates (10). ISG15 expression has also been shown to inhibit the release of HIV-1 virions in vitro (20). Finally, more direct evidence comes from the observation that the overexpression of ISG15 by a recombinant Sindbis virus attenuates infection in IFN-αβ receptor-deficient (IFNαβR−/−) mice (21). Yet, the initial evaluation of ISG15-deficient mice revealed normal IFN signaling and ISG induction as well as normal resistance to vesicular stomatitis virus (VSV) and lymphocytic choriomeningitis virus (LCMV) infection (18). In this work, we provide evidence that ISG15−/− mice are deficient in their ability to respond to viral infection. We show that mice lacking ISG15 have increased susceptibility to influenza, herpes virus type 1 (HSV-1), and Sindbis virus infection. This work clearly establishes ISG15 as an important antiviral molecule in vivo.
Results
ISG15 Is Induced in Vivo After Influenza Infection.
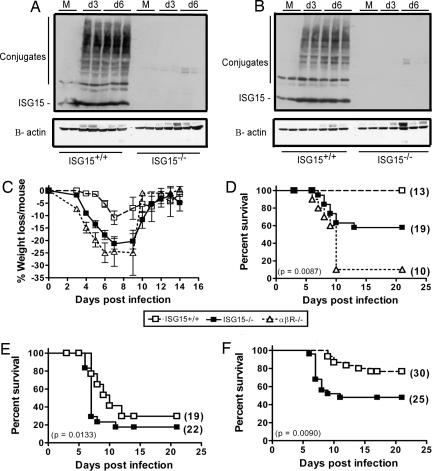
Previous studies have shown that ISG15 is rapidly up-regulated after infection with multiple viruses both in vitro and in vivo (21). Yet, infection of cultured cells with influenza A virus inhibited ISG15 expression, and after infection with influenza B virus, conjugation of ISG15 to target proteins was specifically blocked (10). We therefore evaluated whether ISG15 was up-regulated in vivo after influenza A or B viral infection. As early as 3 days after intranasal (i.n.) infection with either influenza B/Lee virus or influenza A rWSN virus, ISG15+/+ mice expressed large amounts of both free ISG15 and ISG15 conjugates in the lung. As expected, ISG15−/− mice did not express ISG15 (Fig. 1A and B). Therefore, in contrast to the in vitro results, ISG15 and ISG15 conjugates are dramatically up-regulated in influenza virus-infected tissue in vivo.
Fig. 1.
ISG15−/− mice demonstrate increased susceptibility to both influenza A and influenza B viral infection. (A and B) Lung homogenates from either ISG15+/+ or ISG15−/− mice that were mock-infected (M) or infected with influenza B/Lee (A) or influenza A rWSN (B) virus were analyzed for expression of ISG15 by Western blot analysis. (C and D) ISG15−/− (filled squares), ISG15+/+ (open squares), and IFNαβR−/− (open triangles) mice were infected with influenza B/Lee virus at 1 × 106 pfu i.n. and followed daily for weight loss (C) or lethality (D). (E and F) ISG15−/− and ISG15+/+ mice were infected with influenza A rWSN virus at either 1 × 105 pfu (E) or 1 × 104 pfu (F) i.n. and followed for lethality. The number of mice per group is indicated in parentheses and is equal for C and D.
ISG15-Deficient Mice Demonstrate Increased Susceptibility to Influenza Virus Infection.
We next evaluated the susceptibility of ISG15−/− mice to influenza virus infection. We challenged wild-type (WT) and ISG15−/− mice with a sublethal dose of influenza B/Lee virus. ISG15−/− mice displayed clinical signs of disease (ruffled fur and increased weight loss) beginning at 4 days postinfection (Fig. 1C), whereas WT mice displayed minimal signs of disease. Although none of the WT mice succumbed to infection, 43% of the ISG15−/− mice died by 21 days postinfection (P = 0.0087; Fig. 1D). This finding proves that ISG15 can have antiviral effects in vivo.
To determine whether ISG15 accounted for all of the antiviral effects mediated by IFN, we compared the susceptibility of ISG15−/− mice with that of mice lacking the IFNαβ receptor. The degree of weight loss seen in the ISG15−/− mice was similar to that seen in IFNαβR−/− mice infected with the same viral dose (Fig. 1C). Yet, when the mice were followed for lethality, the IFNαβR−/− mice displayed 90% lethality at a dose of virus that killed only 43% of ISG15−/− mice (P < 0.0001; Fig. 1D). Therefore, additional ISGs contribute to the IFN-mediated resistance to influenza B virus infection.
ISG15−/− mice were also more susceptible to influenza A virus infection. Inoculation with 1 × 105 pfu of influenza A rWSN virus resulted in more rapid onset of death in ISG15−/− mice compared with WT mice. Seventy percent of the ISG15−/− mice succumbed to infection by 7 days postinfection compared with only 23% of control mice (P = 0.0133; Fig. 1E). When the dose of influenza A rWSN virus was lowered to 1 × 104 pfu, the ISG15−/− mice displayed increased lethality (52% lethality) compared with the ISG15+/+ mice (23% lethality) (P = 0.009; Fig. 1F). Once again, ISG15 did not account for all of the IFN-induced antiviral effects during influenza A rWSN infection because it has been reported previously that the IFNαβR−/− and STAT1−/− mice have an LD50 2 logs lower than their WT counterparts (22). It should be noted that in our experiments there was no contribution to antiviral immunity of the IFN-inducible protein Mx because all inbred strains of mice commercially available lack a functional Mx1 gene. Thus, ISG15 is an important IFN-induced host protein that protects against both influenza A and B virus infection.
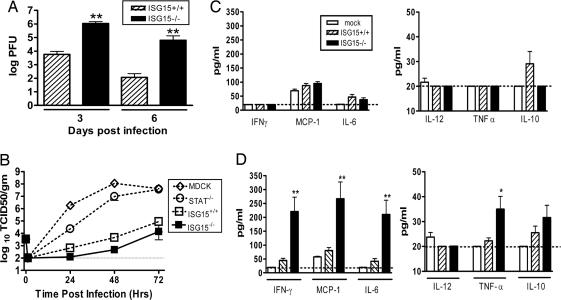
STAT1−/− and IFNαβR−/− mice infected with influenza A develop a fulminate local lung infection followed by systemic spread of the virus (22). In contrast, WT mice limit viral replication and spread of the virus to the respiratory tract (22). Three days after inoculation with influenza B/Lee virus, ISG15−/− mice had viral titers in their lungs that were >100-fold higher than in WT mice (P < 0.0001; Fig. 2A). By day 6, when WT mice were clearing the infection, the lungs of ISG15−/− mice still contained viral titers >105 pfu (P < 0.0001; Fig. 2A). A similar increase in viral lung titers was also seen in ISG15−/− mice after influenza A rWSN infection at 1 × 104 pfu (data not shown). Thus, similar to mice lacking either STAT1 or the IFNαβR, a deficiency of ISG15 resulted in enhanced local influenza virus replication. However, we did not detect dissemination of either influenza A rWSN or B/Lee viruses to the brain, spleen, or liver in ISG15−/− mice (data not shown). Thus, ISG15 may significantly contribute to the capacity of IFNs to control local influenza virus infection in the lung, but it is not required for IFN-mediated control of dissemination of influenza viruses.
Fig. 2.
Characterization of influenza pathogenesis in ISG15−/− mice. (A) Lung homogenates from ISG15+/+ (hatched bars) or ISG15−/− (filled bars) mice infected with influenza B/Lee virus at 1 × 106 pfu i.n were titered by plaque assay. Error bars represent SEM. Fifteen to 17 animals were harvested for each time point. (B) Growth of influenza A rWSN was determined on Madin–Darby canine kidney (MDCK) cells (open diamonds), STAT1−/− (open circles), ISG15+/+ (open squares), or ISG15−/− (filled squares) murine embryonic fibroblasts (MEFs). Data are representative of two independent experiments. Error bars represent SEM. (C and D) Serum cytokine levels were analyzed at 3 (C) or 6 (D) days postinfection in mock-infected (open bars), ISG15+/+ (hatched bars), or ISG15−/− (filled bars) mice infected with 1 × 106 pfu influenza B/Lee virus i.n. ∗, P < 0.001; ∗∗, P < 0.0001.
Previous work has also shown that influenza viral growth is limited in WT MEFs by an IFN- and STAT1-dependent protein (22). We therefore examined whether ISG15 mediated the blockade of influenza replication by evaluating viral growth in MEFs isolated from ISG15−/−, STAT−/−, and WT mice. As seen previously, influenza A rWSN virus displayed significant growth in STAT1−/− MEFs (Fig. 2B). In contrast, there was little evidence of viral growth seen in either WT or ISG15−/− MEFs (Fig. 2B). Thus, we could obtain no evidence that ISG15 is a STAT1-dependent molecule required for the control of influenza viral replication in MEFs.
To evaluate further the response of ISG15−/− mice to influenza B/Lee virus infection, we evaluated serum cytokine levels during the course of infection. Three days after i.n. infection, there was little evidence of systemic cytokine production in either WT or ISG15−/− mice (Fig. 2C), despite significant viral titers in the lungs (Fig. 2A). Even at 6 days postinfection, WT mice demonstrated no significant increase in serum cytokine levels compared with mock-infected mice (Fig. 2D). In contrast, at 6 days postinfection, ISG15−/− mice had 4-fold increased serum levels of IFN-γ, monocyte chemoattractant protein 1, and IL-6 compared with WT mice (P < 0.0001) as well as modestly increased levels of TNF-α (P < 0.001; Fig. 2D). No detectable levels of IL-2, IL-4, IL-5, or IFN-α or -β were found in either WT or ISG15−/− mice at these time points (data not shown). We also evaluated both WT and ISG15−/− mice for the presence of type I IFNs as early as 18 h postinfection, and we were unable to detect the presence of either cytokine in their serum (data not shown), despite the clear importance of type I IFNs for control of influenza virus infection (Fig. 1 C and D) (22). Thus, in addition to increased susceptibility to influenza infection, ISG15 deficiency resulted in a systemic inflammatory cytokine response during infection.
ISG15−/− Mice Display Increased Susceptibility to Herpes Virus Infection.
Microarray analysis had previously shown that HSV-1 strongly induces ISG15 expression (23). Therefore, we evaluated whether the antiviral activity of ISG15 extended beyond influenza virus to HSV-1. ISG15−/− mice displayed increased lethality compared with ISG15+/+ mice after infection with either 20 pfu (P = 0.0349; Fig. 3A) or 2.0 pfu (P = 0.0112; Fig. 3B) of HSV-1 intracranially (i.c.). To evaluate a more physiological route of infection, we also infected mice by corneal infection. ISG15−/− mice displayed more rapid onset of death, and they were more susceptible to infection, with 46% of the mice dying at 2 × 104 pfu/eye (P = 0.013; Fig. 3C) and 70% dying at 2 × 105 pfu/eye (P = 0.0037; Fig. 3D) compared with only 10–20% lethality in ISG15+/+ mice at either dose. To determine whether ISG15 is important for resistance across herpesviruses, we also evaluated the response of ISG15−/− mice to gammaherpesvirus 68 (γHV68) infection. There was no lethality observed in either ISG15+/+ or ISG15−/− mice after i.p. infection with γHV68 (data not shown). ISG15+/+ and ISG15−/− mice displayed similar viral titers in spleen (data not shown), liver, and lungs at 1 and 2 days postinfection (Fig. 3 E and F). However, by both 3 and 4 days postinfection, ISG15−/− mice displayed increased viral titers in both the liver (P < 0.01; Fig. 3E) and lung (P < 0.001; Fig. 3F). By 6 days postinfection, both the ISG15+/+ and ISG15−/− mice were able to clear the infection from the liver and control the lung infection to equivalent titers. Thus, the antiviral activity of ISG15 is not restricted to influenza viruses, suggesting a broadly important antiviral role for ISG15.
Fig. 3.
ISG15−/− mice display increased susceptibility to herpes virus infection. (A and B) ISG15+/+ (open squares) or ISG15−/− (filled squares) mice were infected with HSV-1 by intracranial (i.c.) infection at either 20 pfu i.c. (A) or 2 pfu i.c. (B) and were followed for lethality. (C and D) ISG15+/+ (open squares), ISG15−/− (filled squares), or ISG15+/− (open triangles) mice were infected with HSV-1 by corneal scarification at either 2 × 104 pfu per eye (C) or 2 × 105 pfu per eye (D) and followed for lethality. (E and F) ISG15−/− (filled bars) or ISG15+/+ (open bars) mice were infected with murine γHV68 at a dose of 1 × 106 pfu i.p. At the indicated times postinfection, liver (E) or lungs (F) were isolated and titered by standard plaque assay. ∗, P < 0.01; ∗∗, P < 0.001. The number of mice per group is indicated in parentheses. All experiments were performed in 129OLA/C57BL/6 chimeric mice except for those in (B), which was confirmed in animals backcrossed to C57BL/6 for 10 generations.
ISG15−/− Mice Are Susceptible to Sindbis Virus Infection.
We have previously shown that when overexpressed by a recombinant double subgenomic Sindbis virus, ISG15 functions as an antiviral molecule and protects IFNαβR−/− mice from lethality (21). We therefore infected 4-day-old pups with recombinant chimeric Sindbis virus, dsTE12Q, and observed both a more rapid time to death and increased lethality in ISG15−/− mice compared with WT mice (P < 0.0001; Fig. 4A). The use of a recombinant chimeric virus provided an opportunity to define the amino acids within ISG15 required for its antiviral effects against Sindbis virus. We therefore determined whether expression of ISG15 from the dsTE12Q genome was able to complement the ISG15−/− phenotype. As seen in Fig. 4B, infection of 4-day-old ISG15−/− pups with dsTE12Q expressing WT ISG15 (ISG15-LRLRGG) protected ISG15−/− mice from lethality, with 42% of the mice surviving compared with no survival among ISG15−/− mice infected with dsTE12Q (P < 0.0001). Previous studies show that mutation of the C-terminal LRLRGG motif to LRLRAA prevents ISG15 from forming conjugates in vitro, and it abrogates the protection from lethality seen in IFNαβR−/− mice (21). We therefore determined whether ISG15 with the LRLRGG motif mutated to LRLRAA could protect ISG15−/− mice from Sindbis virus infection. We infected 4-day-old ISG15−/− pups with the LRLRAA virus, and we found that unlike mice infected with the LRLRGG virus (42% survival, n = 14), pups infected with the LRLRAA virus all succumbed to infection (0% survival, n = 13, P < 0.0001; Fig. 4B). These results demonstrate that an intact LRLRGG motif is required for ISG15 antiviral activity and is consistent with protein conjugation playing a role in its antiviral function.
Fig. 4.
Increased susceptibility to Sindbis virus infection in ISG15−/− mice can be complemented by Sindbis viruses expressing ISG15-LRLRGG but not ISG15-LRLRAA. (A) ISG15−/− (filled squares) or ISG15+/+ (open squares) pups were infected with dsTE12Q at 1,000 pfu i.c., and they were followed for lethality. (B) ISG15−/− pups were infected with dsTE12Q expressing either WT ISG15-LRLRGG (filled triangles) or mutant ISG15-LRLRAA (filled circles) at 1,000 pfu i.c., and they were followed for lethality. The number of mice per group is indicated in parentheses.
Discussion
IFNs are the most important early defense against acute virus infection. This work defines ISG15 as an important host IFN-induced antiviral protein that functions in vivo against several important human pathogens. The loss of ISG15 results in increased susceptibility to both RNA and DNA viruses, including influenza A and B viruses, the herpes viruses HSV-1 and γHV68, and Sindbis virus. Although it is clear that ISG15 has a broad importance in antiviral responses, it is not clear that ISG15 will protect against all viruses because previous analysis of the ISG15−/− mice found no alterations in their response to either VSV or LCMV (18). The lack of a phenotype of ISG15−/− mice infected with these viruses could be the result of the assays used. For example, in our studies of γHV68, the ISG15−/− mice did not demonstrate increased lethality at the dose examined, but they did show alterations in viral titer on specific days after infection. A more detailed analysis may reveal a role for ISG15 in control of LCMV or VSV.
Alternatively, there may be no role for ISG15 in resistance to either VSV or LCMV. As is the case with many antiviral molecules, ISG15 may display activity against only a subset of viruses, which may reflect either the mechanism by which ISG15 is exerting its activity, or it may be secondary to viruses (e.g., VSV or LCMV) having evolved strategies to counteract ISG15 action.
Many of the previously defined host antiviral molecules function by inhibiting viral replication. Although increased viral titers were found in vivo after influenza (Fig. 2A) and γHV68 (Fig. 3) virus infections, we were unable to demonstrate a difference in viral growth in vitro with any of the viruses in MEFs lacking ISG15: influenza A (Fig. 2B); or HSV-1, γHV68, or Sindbis virus [see supporting information (SI) Fig. 5]. This finding does not eliminate the possibility that ISG15 blocks viral replication because the activity of ISG15 may be limited to a specific cell type or to primary cells that have yet to be tested. Alternatively, ISG15 could function in a cell-extrinsic manner either by altering the function of cells responding to virally infected cells or through its cytokine activity (6–9). In either case, ISG15 antiviral activity would only be detected during in vivo analysis. These results underline the need for screens to be performed in vivo when trying to dissect apart the complexities of the actions of interferons.
The antiviral activity of ISG15 may be the result of its cytokine activity or its ability to conjugate to target proteins or both. Human ISG15 is released from cells after stimulation with IFNs (5), and it is found in the serum of persons treated with IFN (5). It is reported to stimulate natural killer cell proliferation, IFN-γ production, neutrophil recruitment, and the maturation of dendritic cells (6–9). We have detected ISG15 in the sera of mice after treatment with LPS, pI:pC, and after viral infection (SI Fig. 6), indicating that it could function as a cytokine. However, extensive data now indicate that ISG15 couples to a very broad array of proteins in either infected or IFN-treated cells (15, 16), raising the possibility that an important antiviral role for ISG15 may be altering the function of host cell or viral proteins through conjugation.
To date, the antiviral action of ISG15 has been restricted to in vivo assays such as infection with viruses that overexpress ISG15 or infection of ISG15−/− mice, as reported here. To begin to evaluate the mechanism of action of ISG15, we used a recombinant chimeric Sindbis virus system to identify amino acids required for the antiviral action of ISG15. We found that mutation of the C-terminal LRLRGG motif, present on mature processed ISG15, to LRLRAA abrogated protection. This motif is required for conjugation of ISG15 to intracellular targets (21). These data, along with the previous observation that influenza B virus specifically disrupts ISG15 conjugate formation, demonstrate the importance of ISG15 conjugation for its antiviral activity in these two viral systems. Recent studies have identified >100 proteins targeted for ISG15 conjugation that encompass multiple cellular pathways such as protein translation, cell cycle regulation, carbohydrate metabolism, cell structure and motility, and signal transduction (15, 16). Interestingly, included in these target proteins are several known IFN-induced antiviral molecules such as PKR, Mx, RIGI, and GBP-1. ISGylation of these antiviral molecules may regulate their activity during viral infection. Alternatively, ISG15 may conjugate to viral proteins within infected cells and alter their localization or function. Interestingly, it has been reported that the cytokine activity of ISG15 also requires the terminal diglycine (7), raising the possibility that conjugated proteins serve as “warning signals” that alert the body to damaged cells. Indeed, we find that a high molecular mass form of ISG15 is present in the serum of infected mice (SI Fig. 6), raising the possibility that the conjugation and cytokine activity of ISG15 are linked.
Identifying which target proteins are altered by ISGylation during viral infection, the fate of proteins after conjugation to ISG15, and the role of conjugation in its putative cytokine activity will be required to determine how ISG15 functions in vivo as an antiviral molecule. Defining the molecular mechanisms of ISG15 action against specific viruses may ultimately allow for the development of potential therapies against these important human pathogens.
Methods
Mice.
IFNαβR−/− mice on the 129/SV/Pas background were initially obtained from M. Aguet, Swiss Intstitute of Experimental Cancer Research (Epalinges, Switzerland) (24, 25). ISG15−/− mice were described in ref. 18. ISG15−/−, ISG15+/−, and ISG15+/+ mice used in the HSV-1 experiments were 129OLA-C57BL/6 mixed background unless indicated otherwise. For all influenza and Sindbis virus experiments, ISG15−/− mice were backcrossed to C57BL/6 mice 10 generations. WT mice were either ISG15+/+ mice backcrossed 10 generations to C57BL/6, or they were C57BL/6 obtained from Jackson Laboratory (Bar Harbor, ME) and bred in our mouse facility. Results from these WT mice were indistinguishable. STAT−/− mice were obtained from D. Levy (New York University School of Medicine, New York, NY) (26). All mice were bred and maintained at Washington University School of Medicine in accordance with all federal and university guidelines.
Viruses.
Influenza A virus.
The recombinant influenza A/WSN/33 (rWSN) virus was generated from cDNA and has been described previously in refs. 27 and 28. The virus was grown on MDCK cells using DMEM containing 1 μg/ml N-acetyltrypsin (Sigma Chemicals, St. Louis, MO), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). The cells were infected at a multiplicity of infection of 0.01 pfu per cell, harvested 48 h postinfection, and titered by plaque assay in MDCK cells.
Influenza B virus.
Recombinant WT influenza B/Lee/40 (B/Lee) virus was grown in 10-day-old embryonated chicken eggs and titered by plaque assay in MDCK cells (29).
HSV-1.
HSV-1 strain 17 was propagated and titered on African green monkey kidney (Vero) cells as described in ref. 30.
HV68.
γHV68 was propagated and titered on 3T12 cells as described in ref. 31.
Sindbis viruses.
Recombinant double-subgenomic Sindbis viruses were produced as described in ref. 21. Briefly, dsTE12Q was produced from a viral cDNA clone by in vitro transcription and RNA transfection of baby hamster kidney-21 cells. The ISG15-expressing Sindbis viruses were generated with the ISG15-LRLRGG virus containing nucleotides 1–465, the 3′ primer containing the GGT GGG TAA sequence, and the ISG15 LRLRAA virus containing nucleotides 1–465 with the 3′ primer containing the GCT TCT TAA sequence. These viruses were generated and characterized as described in ref. 21.
Viral Studies.
For influenza B virus experiments 6- to 8-week-old male mice were infected with 1 × 106 pfu of influenza B/Lee virus (in 40 μl of PBS) i.n. Viral titers were determined by plaque assay on MDCK cells. After absorption, cells were overlayed with MEM containing a 0.6% oxoid agar and 1 μg/ml l-1-tosylamide-2-phenylmethyl chloromethyl ketone-treated trypsin. Plaques were visualized by crystal violet. For influenza A virus experiments, 6- to 8-week-old female mice were infected with either 1 × 104 or 1 × 105 pfu of influenza A rWSN virus (in 40 μl of PBS) i.n. Titers were determined by 50% tissue culture infectious dose (TCID50) assay on a 10% (wt/vol) suspension as described in ref. 32. For HSV experiments, i.c. inoculations were performed on 8- to 10-week-old male mice that were anesthetized with xylazine and ketamine and then inoculated i.c. with 20 μl of medium containing either 20 or 2 pfu of HSV-1 strain 17 (30). Corneal infections were performed as described previously (30). Corneas of 8- to 10-week-old male mice were scarified with 10 interlocking strokes with a 27-gauge needle, and 5 μl of medium containing either 2 × 104 or 2 × 105 pfu of virus was added per eye. For γHV68 infections, 6- to 10-week-old mice were injected i.p. with 1 × 106 pfu, and viral titers were determined by plaque assay on 3T12 cells. For Sindbis virus experiments, 4-day-old pups were infected with the indicated virus at a dose of 1,000 pfu in 10 μl of HBSS into the right cerebral hemisphere.
Viral Growth Curves.
The indicated cells were infected with either influenza A rWSN or influenza B/Lee viruses at a multiplicity of infection of ≈0.1 pfu/cell for 1 h at 37°C. The inoculum was removed, and cells were washed. DMEM containing 0.5% FBS was added to the infected cells. At the indicated times postinfection, the infected cell supernatants were harvested, clarified by centrifugation, and stored at −70°C. Infectious virus was quantified by TCID50 assay.
Cytokine Analysis.
Serum cytokine levels were analyzed on sera isolated from mice infected with influenza B/Lee at 1 × 106 pfu i.n. on the indicated days. Samples were analyzed for IL-6, IL-10, monocyte chemoattractant protein 1, IFN-γ, TNF, and IL-12 p70 by using the cytometric bead array mouse inflammation kit as directed by the manufacturer (BD Biosciences PharMingen, San Diego, CA). Samples were analyzed for IFN-α or -β by ELISA as directed by the manufacturer (PBL Biomedical Laboratories, Piscataway, NJ).
In Vivo Induction of ISG15.
ISG15−/− or ISG15+/+ mice were infected with influenza B/Lee (1 × 106 pfu i.n.) or influenza A rWSN (1 × 105 pfu i.n.) viruses. Lung samples were homogenized and mixed with SDS loading buffer and boiled for 30 min before Western blot analysis. ISG15 expression was detected as previously described with anti-ISG15 mAb (3C2) (21) and then developed with goat anti-Armenian hamster horseradish peroxidase secondary antibody (Jackson ImmunoResearch, West Grove, PA). For loading controls, parallel blots were probed with anti-β-actin mAb (clone AC-74; Sigma) and then developed with a goat anti-mouse horseradish peroxidase secondary antibody (Jackson ImmunoResearch). All blots were developed with ECL Plus chemiluminescent reagent (Amersham, Piscataway, NJ).
Statistical Analyses.
All data were analyzed with Prizm software (GraphPad, San Diego, CA). Survival data were analyzed by the Mantel–Haenzsel test, with death as the primary variable. Acute titer data were analyzed by using the Mann–Whitney test. Error bars in figures represent the SEM.
Supplementary Material
Acknowledgments
We thank Darren Kreamalmayer for expertise in animal care and Richard Cadagan for technical assistance. This work was supported by National Institutes of Health Grants K08 AI059390 (to D.J.L.), U54 AI057160 Project 6 (to H.W.V.), R01 AI46954 (to A.G.-S.), P01 AI058113 (to A.G.-S.), and R01 EY09083 (to D.A.L.); a Lew Wasserman Scholarship for Research to Prevent Blindness (to D.A.L.); Core Grants P30 EY02687 from the National Institutes of Health (to the Department of Ophthalmology, Washington University School of Medicine) and from Research to Prevent Blindness (to D.A.L.); and by Deutsche Forschungsgemeinschaft/German Research Council Grant Wo554/3-2 (to T.W.).
Abbreviations
- γHV68
gammaherpesvirus 68
- HIV-1
human immunodeficiency virus type 1
- HSV
herpes simplex virus 1
- i.c.
intracranial/intracranially
- IFNαβR
IFN-αβ receptor
- i.n.
intranasal/intranasally
- ISG
interferon-stimulated gene
- LCMV
lymphocytic choriomeningitis virus
- MDCK
Madin–Darby canine kidney
- MEF
murine embryonic fibroblast
- VSV
vesicular stomatitis virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607038104/DC1.
References
- 1.Samuel CE. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foy E, Li K, Sumpter R, Loo YM, Johnson CL, Wang CF, Fish PM, Yoneyama M, Fujita T, Lemon SM, et al. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou A, Paranjape JM, Der SD, Williams BR, Silverman RH. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]
- 4.Narasimhan J, Wang M, Fu Z, Klein JM, Haas AL, Kim JJ. J Biol Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 5.D'Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight E, Jr, Borden EC. J Immunol. 1996;157:4100–4108. [PubMed] [Google Scholar]
- 6.Recht M, Borden EC, Knight E., Jr J Immunol. 1991;147:2617–2623. [PubMed] [Google Scholar]
- 7.D'Cunha J, Knight E, Jr, Haas AL, Truitt RL, Borden EC. Proc Natl Acad Sci USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owhashi M, Taoka Y, Ishii K, Nakazawa S, Uemura H, Hambara H. Biochem Biophys Res Commun. 2003;309:533–539. doi: 10.1016/j.bbrc.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 9.Padovan E, Terracciano L, Certa U, Jacobs B, Reschner A, Bolli M, Spagnoli GC, Borden EC, Heberer M. Cancer Res. 2002;62:3453–3458. [PubMed] [Google Scholar]
- 10.Yuan W, Krug RM. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. Proc Natl Acad Sci USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou W, Zhang DE. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 14.Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 15.Giannakopoulos NV, Luo JK, Papov V, Zou WG, Lenschow DJ, Jacobs BS, Borden EC, Li J, Virgin HW, Zhang DE. Biochem Biophys Res Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Proc Natl Acad Sci USA. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie KJ, Zhang DE. Semin Cell Dev Biol. 2004;15:237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Osiak A, Utermohlen O, Niendorf S, Horak I, Knobeloch KP. Mol Cell Biol. 2005;25:6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knobeloch KP, Utermohlen O, Kisser A, Prinz M, Horak I. Mol Cell Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Proc Natl Acad Sci USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HW. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Sastre A, Durbin RK, Zheng H, Palese P, Gertner R, Levy DE, Durbin JE. J Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholl MJ, Robinson LH, Preston CM. J Gen Virol. 2000;9:2215–2218. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- 24.Dunn GP, Bruce AT, Sheehan KCF, Shankaran V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM, et al. Nat Immunol. 2005;6:722–729. doi: 10.1038/ni1213. [DOI] [PubMed] [Google Scholar]
- 25.Muller U, Steinhoff S, Reis LFL, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 26.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 27.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, et al. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. J Virol. 2002;76:1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dauber B, Heins G, Wolff T. J Virol. 2004;78:1865–1872. doi: 10.1128/JVI.78.4.1865-1872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. Proc Natl Acad Sci USA. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton ES, Lutzke ML, Rochford R, Virgin HW. J Virol. 2005;79:14149–14160. doi: 10.1128/JVI.79.22.14149-14160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCown MF, Pekosz A. J Virol. 2005;79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.