Abstract
To assess the role of the macrophage scavenger receptor type A (SRA) in immune activation by CpG DNA, cytokine induction and DNA uptake were tested in vitro and in vivo using SRA knockout (SRA−/−) and wild type (WT) mice. As a source of CpG DNA, Escherichia coli DNA (EC DNA) and a 20-mer phosphorothioate oligodeoxynucleotide with two CpG motifs (CpG ODN) were used. In vitro, both EC DNA and the CpG ODN induced dose-dependent increases of interleukin (IL)-12 production by spleen cells and bone-marrow-derived macrophages (BMMΦ) from both SRA−/− and WT mice. The levels of cytokines produced by SRA−/− spleen cells and BMMΦ were similar to those of WT spleen cells and BMMΦ. When injected intravenously with CpG ODN and EC DNA, both SRA−/− and WT mice showed elevated serum levels of IL-12. To investigate further the role of the SRA, flow cytometry and confocal microscopy were performed to examine the uptake of fluorescently labelled oligonucleotides. SRA−/− and WT BMMΦ showed similarity in the extent of uptake and distribution of oligonucleotides as assessed by these two techniques. Together, these findings indicate that, while the SRA may bind DNA, this receptor is not essential for the uptake of CpG DNA or its immunostimulatory activity.
Introduction
DNA is a large macromolecule whose immunological properties depend on sequence microheterogeneity.1–5 While mammalian DNA is inactive, DNA from bacteria displays potent immunostimulatory activities that resemble those of lipopolysaccharide (LPS).6,7 These activities depend on short sequence motifs called CpG motifs or immunostimulatory sequences (ISS).8–11 These motifs, which consist of a cytosine–guanosine dinucleotide flanked by two 5′ purines and two 3′ pyrimidines, are much more common in bacterial DNA than in mammalian DNA.12 In vitro and in vivo, DNA containing CpG motifs (CpG DNA) can activate B lymphocytes, macrophages, dendritic cells and natural killer cells as well as induce cytokines such as interleukin (IL)-6, IL-12, interferon-γ (IFN-γ) and tumour necrosis factor-α (TNF-α).6–9,13–17 As such, CpG DNA may serve as a ‘danger’ signal to stimulate the innate immunity, and has the potential to be used as a stimulant and an adjuvant.18,19
Because of the potential use of CpG DNA as an immunomodulator, there has been intense investigation to elucidate signal transduction pathways critical to these responses. In the murine system, these studies have shown that immune cell activation requires internalization of DNA, with DNA entrance into cells occurring by an endocytic pathway.9,20 Subsequent events in stimulation include activation of stress kinases as well as translocation of nuclear factor (NF)-κB.21,22
Various cell-surface proteins have been shown to play a role in the binding and uptake of natural and synthetic DNA.23,24 Among these molecules, the scavenger receptor type A (SRA) on macrophages has attracted interest as a putative ‘DNA receptor’.25,26 The SRA was initially discovered by Brown, Goldstein and colleagues for its role in the uptake of chemically modified lipoproteins, including oxidized and acetylated low-density lipoproteins (LDL).27,28 The role of the SRA in macrophage responses to DNA derives from studies showing that DNA uptake and immune stimulation can be inhibited by SRA ligands, including poly G, poly I, dextran sulphate, and polyvinyl sulphate.24,25,29–32
While studies based on inhibitors suggest that the SRA on macrophages binds DNA, the analysis of SRA function is complicated by the binding of SRA ligands to other surface molecules and the existence of other ‘scavenger receptors.’ Recent studies using SRA knockout (SRA−/−) mice have suggested that the SRA accounts for only a limited portion of DNA binding to macrophages.33,34 These studies, however, do not address whether the uptake of DNA by the SRA, even if it accounts for only part of the total uptake of DNA, is critical for activation of macrophages by CpG DNA. To clarify the role of the SRA in responses to CpG DNA, we therefore investigated the production of cytokines by macrophages and spleen cells from SRA−/− and wild type (WT) mice. In data presented herein, we show that cytokine production and DNA uptake by cells lacking the SRA are similar to those of cells that express this receptor. These findings indicate that the SRA, while able to bind DNA, is not essential for immune stimulation by CpG DNA and that other receptors can mediate this process.
Materials and methods
Synthetic oligonucleotides and bacterial and mammalian DNA
Oligonucleotides were purchased from Midland Certified Reagent Company (Midland, TX). The compounds were synthesized using cyanoethyl phosphoramidite chemistry and purified by gel filtration. The sequences for a CpG-containing phosphorothioate oligonucleotide (TCCATGACGTTCCTGACGTT, CpG ODN) and a non-CpG-containing phosphorothioate oligonucleotide (TCCATGAGCTTCCTGAGTCT, control ODN) were taken from the literature.35 E. coli DNA (EC DNA) and calf thymus DNA (CT DNA) were purchased from Sigma Chemical Co. (St. Louis, MO). The dried DNA compounds were dissolved to a concentration of 1 mg/ml in distilled water and were sterilized by filtering through a 0·22-µm Millex-GV filter (Millipore, Bedford, MA). Optical density readings at 260 nm before and after filtration showed no loss of material.
To prepare fluorescently labelled oligonucleotides, oligonucleotides containing an amino terminus were mixed with fluorescein isothiocyanate (FITC; Sigma) in a ratio of 0·1 mg FITC/10 mg oligonucleotides in carbonate–bicarbonate buffer (pH 9·5). After a 2-hr reaction at room temperature, unreacted FITC was removed by gel filtration through a Sephadex G-25 column (NAP10 column, Phamarcia, Piscataway, NJ) followed by ethanol precipitation. Boron dipyrromethene difluoride (BODIPY-Fl)-labelled oligonucleotides were prepared with FluoReporter®BODIPY®FL Oligonucleotide Amine Labeling Kits (Molecular Probes, Inc., Eugene, OR) following the manufacturer's instructions.
Mice
BALB/c and 129 mice were purchased from Jackson Laboratory (Bar Harbor, ME) and ICR mice were purchased from Harlan Laboratories (Indianapolis, IN). SRA−/− mice, generated as described elsewhere,36 were the generous gift of Dr Tatsuhito Kodama (University of Tokyo, Japan) and were obtained from Dr Mason Freeman (Massachusetts General Hospital, Boston, MA). Mice were housed under conventional conditions in the animal facility of the Durham VA Hospital.
SRA−/− mice were genetically produced by transfecting murine A3-1 ES cells of 129 origin with vectors containing disrupted SRA I/II allele and developed in ICR mice.37,38 These SRA−/− mice possess a mixed genetic background of 129 mice and ICR mice. Preliminary experiments were conducted to compare DNA uptake and DNA-induced cytokine production by spleen cells and bone-marrow-derived macrophages (BMMΦ) from SRA−/−, BALB/c, ICR and 129 mice. No obvious differences were observed among the four type mice in their uptake of, or responses to DNA. Therefore, in this report, control WT mice include BALB/c, ICR and 129 mice. Strains of the mice used in individual experiments are specified in each figure.
Cell culture
To prepare spleen cells, SRA−/− and control WT mice were killed by cervical dislocation and their spleens removed aseptically. Non-stromal cells were expressed with flame-sterilized microscope slides into RPMI-1640 medium (Life Technologies, Grand Island, NY). Suspended cells were transferred to a 15-ml conical centrifuge tube and large debris was allowed to settle. The overlying cell suspension was carefully removed and centrifuged at 400 g for 5 min. The cell pellet was resuspended in lysis medium (1 volume of 0·17 m Tris, pH 7·6: 9 volume of 0·16 m NH4Cl) to eliminate red blood cells. Cells were then washed twice with RPMI-1640 medium. The final pellets were suspended in complete medium consisting of RPMI-1640 supplemented with 5% heat inactivated fetal bovine serum (HyClone, Logan, UT) and 50 µg/ml gentamicin (Life Technologies). Cells were counted using a haemocytometer and cell concentrations adjusted with complete medium. Cells were plated at 1 × 106 cells/well in 96-well cell culture plates (Costar Incorporated, Corning, NY).
BMMΦ were cultured from SRA−/− and WT mice. After killing the mice, intact femurs and tibias were removed. The bone marrow cells were harvested by repeated flushing of the bone shaft with RPMI-1640 medium. Bone marrow cell culture was established at a concentration of 1 × 106 cells/ml in medium consisting of RPMI-1640, 10% fetal calf serum (FCS), 50 µg/ml gentamicin and 20% L929 conditioned medium as a source of macrophage colony-stimulating factor (M-CSF).39 Absence of the SRA on BMMΦ from SRA−/− mice was confirmed by staining with monoclonal anti-SRA I/II antibodies conjugated with FITC (clone 2F8, Serotec Ltd, Raleigh, NC) and examined by flow cytometry. Abundant surface expression of the SRA was observed on BMMΦ from WT mice (data not shown).
Cytokine enzyme-linked immunosorbent assays (ELISA)
IL-6, Il-12 p40/p70, and TNF-α secretion was measured by ELISA using capture immunoassays. Culture supernatants were removed at 6 hr (TNF-α) or 24 hr (IL-6 and IL-12 p40/p70), and frozen at −20° until analysis. ELISA plates (Immulon II HB, Dynex Technologies, Chantilly, VA) were prepared by coating each well with 100 µl of capture antibody diluted to 1–5 µg/ml in phosphate-buffered saline (PBS) pH 8·5. After overnight incubation at 4°, plates were washed with PBS pH 7·4 using an automated plate washer (Skatron Instruments Inc., Sterling, VA). 100 µl of culture supernatants or serum samples from in vivo experiments diluted in PBS pH 7·4 containing 0·5% bovine serum albumin (BSA; Sigma) and 0·5% Tween-20 (Sigma) were added to each well. Recombinant cytokines were diluted in duplicate and run on each plate as standards.
Samples and standards were allowed to incubate 2 hr at room temperature. After washing of the plates, 100 µl of diluted biotinylated detection antibody (0·1–1 µg/ml) was added to each well. Following another 2 hr incubation, plates were washed and 100 µl of diluted avidin peroxidase (Zymed, San Francisco, CA) was added to each well. After a 30-min incubation, plates were washed and 100 µl of a solution containing 0·015% 3,3′,5,5′-tetramethylbenzidine hydrochloride (Sigma) and 0·01% hydrogen peroxide in 0·1 m sodium citrate buffer pH 4·0 was added. Plates were read at 380 nm with an automated microplate reader (Molecular Devices, Menlo Park, CA) and cytokine concentrations derived from standard curves. All the capture and detection antibodies for cytokine ELISA described here were purchased from PharMingen (San Diego, CA).
Flow cytometry
For oligonucleotide uptake assays, 100 µl of cells at the concentration of 1 × 107 cells/ml were incubated at 37° with 1–16 µg/ml of FITC-labelled oligonucleotides in RPMI-1640 with 5% FCS. After 30 minute incubation, unbound oligonucleotides were stripped by incubating cells in 0·2 m acetic acid (pH 2·5) on the ice for 10 min, and washed three times in cold PBS−2% BSA−0·1% NaN3 by centrifugation at 400 g for 5 min. Cells were fixed in 1% paraformaldehyde–PBS−0·1% NaN3, and examined by FACScan flow cytometer (Becton Dickinson, Mansfield, MA). The acquisition was performed with 10 000 events per sample. The list mode data were analysed by using LYSYSTM II software (Becton Dickinson Immunocytometry Systems, San Jose, CA).
Confocal microscopy
BMMΦ were cultured on glass slide coverslips in a 6-well plate overnight. The coverslips with the adherent BMMΦ were washed three times with Dulbecco's PBS (Life Technologies), followed by incubation with culture medium with or without 10 µg/ml of BODIBY-FL-labelled CpG ODN or control ODN, at 37° for 30 min. To assess the endocytosis pathway of oligonucleotides, BMMΦ were simultaneously incubated with 10 µg/ml BODIBY-FL labelled oligonucleotides and 1 mg/ml of Texas red-labelled dextran 70 000 MW (Molecular Probes, Inc., Eugene, OR), a marker for pinocytosis (fluid phase endocytosis).40 After three washes with cold PBS−2% BSA−0·1% NaN3 and fixed in 1% paraformaldehyde–PBS−0·1% NaN3, the slides were viewed for intracellular distribution of labelled oligonucleotides and dextran under a confocal microscope (Zeiss LSM 510, Carl Zeiss, Oberkochen, Germany).
In vivo induction of cytokines
SRA−/− and WT mice were injected via their tail veins with 100 µl/mouse of Dulbecco's PBS (Life Technologies) with or without 20 µg of CpG ODN or control ODN, or 200 µg of EC DNA or CT DNA. Blood samples were collected by retro-orbital bleeding 4 hr after DNA injection. Serum from the blood samples was harvested for cytokine assays. This experiment was performed three times, with two or three mice in each treatment group.
Statistical analysis
All results are presented as mean±SEM. Data are analysed by one way anova, and difference between experimental treatment groups was assessed by an unpaired Student's t-test. P-values < 0·05 were considered significant.
Results
Cytokine induction with CpG ODN and bacterial DNA
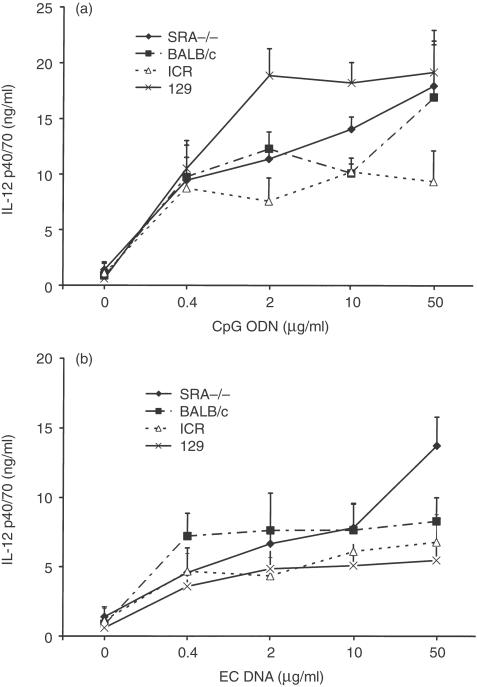
To evaluate the role of the SRA in the response to CpG DNA, cytokine production by spleen cells of SRA−/− and WT mice treated with various doses of CpG ODN and EC DNA were tested. Spleen cells from SRA−/− and WT mice demonstrated a dose dependent response to CpG ODN and EC DNA, and produced similar levels of IL-12 p40/p70 (Fig. 1). Spleen cells from both types of mice failed to respond to either CT DNA or control ODN under the same conditions (data not shown).
Figure 1.
Effects of DNA on IL-12 production from spleen cells. SRA–/– and WT murine spleen cells were stimulated with various doses of a CpG-containing oligonucleotide (CpG ODN) and E. coli DNA (EC DNA), together with a non-CpG oligonucleotide (control ODN) and calf thymus DNA (CT DNA) as controls. Supernatants were harvested after 24 hr and assayed for IL-12 p40/70 by ELISA. CpG ODN (a) and EC DNA (b) induced increased production of IL-12, while control ODN and CT DNA did not (data not shown). The different strains of mice used as WT mice are indicated. Results are presented as means±SEM.
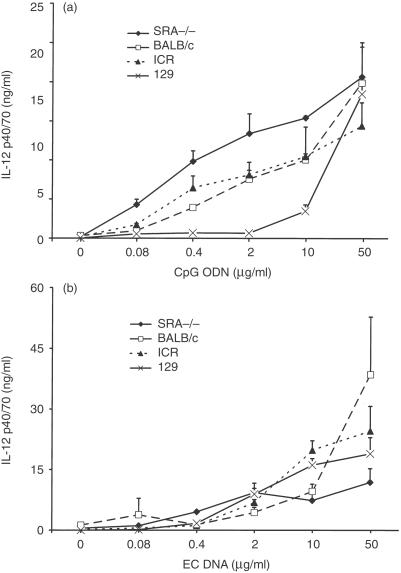
To assess further the role of the SRA in mediating DNA-induced responses, cytokine production by BMMΦ treated with CpG ODN and EC DNA were measured. BMMΦ from SRA−/− mice showed a dose-dependent production of IL-12 p40/p70 in response to both the CpG ODN and EC DNA stimulation, as did by BMMΦ from WT mice (Fig. 2). The levels of cytokine production by the BMMΦ from SRA−/− and WT mice were similar, indicating that the SRA is not major factor in mediating DNA induced activation of the murine macrophages. BMMΦ from both types of mice failed to respond to either CT DNA or control ODN under the same conditions (data not shown).
Figure 2.
Effects of DNA on IL-12 production from BMMΦ. SRA−/− and WT BMMΦ were stimulated with various doses of CpG ODN and EC DNA, together with the control ODN and CT DNA as controls. Supernatants were harvested after 24 hr and assayed for IL-12 p40/70 by ELISA. CpG ODN (a) and EC DNA (b) induced increased production of IL-12, while control ODN and CT DND did not (data not shown). The different strains of mice used as WT mice are indicated. Results are presented as means±SEM.
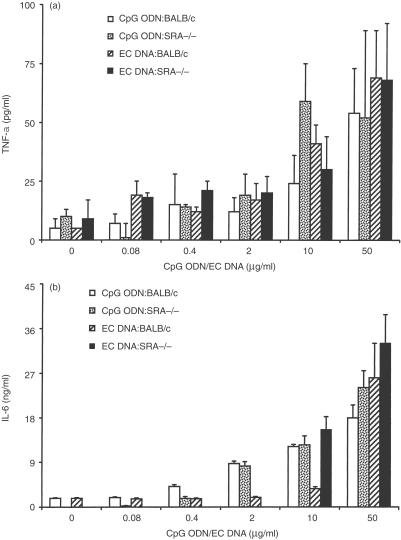
Like IL-12 p40/p70, TNF-α plays an important role in innate immunity, and is produced by cells stimulated by CpG oligonucleotides and bacterial DNA.16 When stimulated with EC DNA or CpG ODN in the range between 0·08 µg/ml to 50 µ/ml, BMMΦ from SRA−/− mice produced similar levels of TNF-α as their WT counterparts (Fig. 3a). IL-6 was also induced in SRA−/− BMMΦ at similar levels to their WT counterparts in response to either CpG ODN or EC DNA (Fig. 3b). The amounts of IL-6 and TNF-α production by SRA−/− spleen cells were comparable to those by WT spleen cells in their response to the stimulation by either CpG ODN or EC DNA (data not shown).
Figure 3.
Effects of DNA on IL-6 and TNF-α production from BMMΦ. SRA−/− and BALB/c murine BMMΦ were stimulated with various doses of CpG ODN (CpG ODN-SRA−/− and CpG ODN-BALB/c) or EC DNA (EC DNA-SRA−/− and EC DNA-BALB/c), together with control ODN and CT DNA as controls. Supernatants were harvested after 6 hr for TNF-α assay and after 24 hr for IL-6 assay by ELISA. CpG ODN and EC DNA induced increased production of TNF-α (a) and IL-6 (b), while control ODN and CT DND did not (data not shown). Results are presented as means±SEM.
Uptake of oligonucleotides by BMMΦ
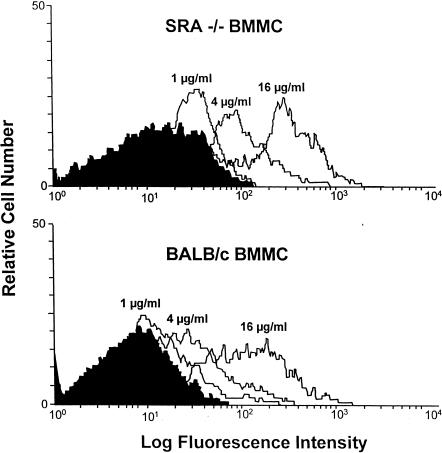
To determine whether the SRA influences the extent of uptake of oligonucleotides, we incubated SRA−/− and WT BMMΦ with 1–16 µg/ml of a dG oligonucleotide (dG30), which displays preferential binding to the SRA.25 The oligonucleotide uptake was examined by flow cytometry 30 min after incubation at 37°. SRA−/− and WT BMMΦ exhibited a similar degree of dose-dependent uptake of dG30 (Fig. 4). A similar time course of dG30 uptake was also found in these two types of macrophages (data not shown).
Figure 4.
Uptake of oligonucleotide dG30 by BMMΦ. SRA−/− and BALB/c BMMΦ were incubated with 1–16 µg/ml of FITC-labelled dG30 at 37° for 30 min, dG30 uptake was assessed by flow cytometry. Both SRA−/− (upper panel) and BALB/c BMMΦ (lower panel) displayed similar degrees of dG30 uptake.
Intracellular distribution of oligonucleotides
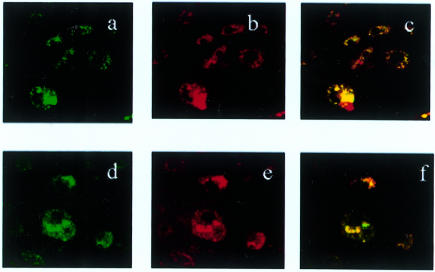
To evaluate further the possible involvement of the SRA in the uptake of DNA, we examined the intracellular distribution of fluorescently labelled oligonucleotides in SRA−/− and WT BMMΦ by confocal microscopy. To test the possibility that oligonucleotides are internalized through a pinocytosis pathway, we incubated BMMΦ with BODIBY-FL-labelled CpG ODN or control ODN together with Texas red-labelled dextran, a hydrophilic polysaccharide that enters cells via pinocytosis.40 Oligonucleotides and dextrans were peripherally distributed in BMMΦ in similar patterns, and the two types of molecules were mostly colocalized in same areas (Fig. 5). There was no prominent difference between SRA−/− and WT BMMΦ in the extent of uptake and the pattern of intracellular distribution of oligonucleotides. These findings indicate that the SRA is not essential for the uptake of oligonucleotides, and that a major portion of oligonucleotide uptake into murine BMMΦ occurs by pinocytosis.
Figure 5.
The intracellular distribution of CpG ODN in BMMΦ. SRA−/− (a, b, c) and BALB/c BMMΦ (d, e, f) were simultaneously incubated with 10 µg/ml of BODIPY-labelled CpG ODN and Texas red labelled dextran at 37° for 30 min. The distribution of CpG ODN (green) and dextran (red) was assessed by laser scanning confocal microscopy after incubation. Photographs (c and f) show the colocalization of CpG ODN and dextran (yellow) in the cells. Magnification, ×630.
In vivo induction of IL-12 by CpG-ODN and bacterial DNA
Although in vitro experiments indicate at most a limited role of the SRA in DNA responses, we decided to assess its role in vivo. SRA−/− and WT mice were injected intravenously with 20 µg/mouse of CpG ODN or control ODN, or 200 µg/mouse of EC DNA or CT DNA. Serum IL-12 levels in both SRA−/− and WT mice receiving CpG ODN were higher than those of mice injected with control ODN, with SRA−/− mice having significantly higher response to CpG ODN than WT mice (P < 0·05) (Table 1). A similar pattern was observed in the response to EC DNA. Control ODN and CT DNA did not induce IL-12 from either strain of the mice. These data thus suggest that SRA−/− mice may be more responsive to in vivo DNA stimulation than WT counterparts.
Table 1.
Serum IL-12 p40/70 levels in mice injected with synthetic and natural DNA
| SRA−/− mice | BALB/c mice | |
|---|---|---|
| PBS | 25 ± 10 | 10 ± 6 |
| CpG ODN | 255 ± 45*† | 133 ± 46* |
| Control ODN | 7 ± 4 | 4 ± 3 |
| EC DNA | 291 ± 197* | 146 ± 109* |
| CT DNA | 61 ± 48 | 14 ± 9 |
Groups of SRA−/− and BALB/c mice (n = 3) were injected via their tail veins with 100 µl/mouse of PBS containing 20 µg of CpG-oligodeoxynucleotide (CpG ODN) or 200 µg of E. coli DNA (EC DNA). Blood was collected 4 hr after injection and serum IL-12 p40/70 levels were assessed by ELISA. Control mice were injected with 100 µl/mouse of PBS or 100 µl/mouse of PBS containing 20 µg of non-CpG-oligodeoxynucleotide (control ODN) or 200 µg of calf thymus DNA (CT DNA). The data represent one of three independent experiments. Results are represented as the mean±SEM.
P < 0·05 when compared with PBS group.
P < 0·05 when compared between SRA−/− and BALB/c mice.
Discussion
Results presented in this study clarify the role of the SRA in immune stimulation and indicate that this receptor is not essential for either uptake of DNA into cells or cytokine induction by CpG DNA. Previous studies on the role of the SRA in mediating cellular effects of DNA were based primarily on the ability of SRA ligands to inhibit the stimulation and uptake of DNA.25,26,29–31 These studies suggested that the SRA or a receptor with similar specificity is involved in these processes. By characterizing the in vitro and in vivo responses of SRA−/− mice, we have provided evidence that, while the SRA may bind DNA, other receptors can mediate DNA-induced immune stimulation. In concert with other studies on SRA−/− mice, these findings suggest that DNA as well as other polyanions can bind to a variety of cell surface molecules with ‘scavenger’ properties.41–44
The SRA was originally identified on the basis of the binding and uptake of oxidized or acetylated LDL.27,28 In addition to binding modified lipoproteins, the SRA interacts with a variety of polyanionic ligands, including LPS, polynucleotides, fucoidan and dextran sulphate.42 Although the binding between the SRA and its ligands most likely reflects ionic interactions, the preference of the SRA for certain nucleic acids (e.g. poly G) as well as its failure to bind polyanions such as chondroitin sulphate suggests that ligand structure as well as charge contributes to binding specificity. In the case of poly G, the ability to associate into high molecular weight aggregates as well as form the quadruplex structure may influence binding properties.45–47 As reflected in the expression of scavenger receptors among species, the SRA appears part of an ancient protein family that may play a role in host defence as a pattern recognition structure binding foreign or degraded self-molecules.42
A number of studies have indicated that the SRA mediates in vivo and in vitro binding of DNA.25,26,29–31 These studies were based mainly on competitive inhibition experiments using SRA ligands, including polyanionic compounds (fucoidan and dextran sulphate) and polynucleotides (poly G and poly I). As we showed previously, SRA ligands including poly dG as well as mammalian DNA can block the in vitro production of IL-12 and IFN-γ by murine spleen cells.32 The inhibitory activity of mammalian DNA suggested that the SRA is not specific for dG-rich DNA among nucleic acids and may be a major cell surface receptor for DNA, with or without CpG motifs or dG sequences.
Despite evidence pointing to the SRA as a DNA receptor, recent studies by Takakura et al. and Butler et al. using SRA−/− mice indicate that this receptor is not essential for DNA uptake.33,34 Takakura et al. observed similar levels of uptake of plasmid DNA into macrophages and multiple organs, including the kidney, spleen and lungs, from SRA−/− and WT mice.33 Similarly, Butler et al. reported that the in vivo organ distribution of oligonucleotides in SRA−/−mice was similar to that of WT mice.34 In the current study, we examined the cellular uptake of oligonucleotides by BMMΦ cultured from SRA−/− and WT mice. Using flow cytometry, we showed that SRA−/− and WT BMMΦ exhibited similar levels of the uptake of dG30, an oligonucleotide considered to be a preferred SRA ligand. Furthermore, using confocal microscopy, we showed similar intracellular distribution of oligonucleotides in SRA−/− and WT BMMΦ. These findings suggest that uptake pathways for oligonucleotides are similar in SRA−/− and WT mice.
As demonstrated using confocal microscopy, oligonucleotides and dextran, a marker for fluid phase endocytosis, are for the most part colocalized inside cells, These observations are consistent with the conclusion that the uptake of oligonucleotides into cells occurs predominantly through absorptive endocytosis and pinocytosis rather than passive diffusion. Although a large number of cell surface molecules, including Mac-1 (CD11b/CD18) and the SRA, can bind oligonucleotides,20,23–25,42,48 the identity of the receptor critical for the uptake process remains uncertain. This uncertainty reflects the charged nature of oligonucleotides and the ability, especially of phosphorothioates, to interact with proteins.49 Furthermore, the pathways involved in DNA uptake in a given cell type may be redundant.50
The results from our studies and those by Takakura et al.33 and Butler et al.34 indicate the limitations in assessing the role of the SRA on the basis of uptake inhibition by putative SRA ligands. Takakura et al. and Butler et al., in addition to demonstrating similar DNA uptake by SRA−/− and WT mice, showed that SRA ligands, such as dextran sulphate and poly I, are equally efficient in inhibiting DNA uptake by macrophages from SRA−/− and WT mice.33,34 Studies on the uptake of modified LDL by SRA−/− and WT mice have led to a similar conclusion.51 Together, these results suggest caution in using certain ligands or molecules to identify receptor interaction as a result of the cross-specificity.
While studies by Takakura et al. and Butler et al. investigated DNA uptake in SRA−/− mice, they did not address the issue of immune stimulation in these mice.33,34 The studies presented herein, in addition to confirming the similarity of DNA uptake in SRA−/− and WT BMMΦ, also demonstrate that SRA−/− and WT BMMΦ or spleen cells display similar responses to CpG-containing oligonucleotides and bacterial DNA. As shown for several cytokines, including IL-6, IL-12, and TNF-α, the levels of the cytokines produced by SRA−/− BMMΦ or spleen cells were similar to those of their WT counterparts; the dose response for stimulation was also similar. These findings suggest that the SRA is not essential for immune stimulation and that other pathways of cellular uptake are involved in macrophage stimulation by CpG DNA.
While the in vitro responses of SRA−/− and WT cells to DNA were similar, in our in vivo experiments, we observed that CpG oligonucleotides and bacterial DNA induced higher levels of IL-12 in SRA−/− mice than those in WT mice. These observations are similar to those from study of Haworth et al.37 These investigators showed that SRA−/− mice infected with bacille Calmette–Guérin (BCG) were more sensitive to endotoxin shock and produced higher levels of cytokines in response to LPS stimulation than WT mice.37 While these findings suggest a role of the SRA in the scavenging of LPS and DNA in the circulation in a non-activating process,29–31,52–55 other studies indicate that the clearance of LPS and organ distribution of oligonucleotides of SRA−/− and WT are similar.33,34,56 These results, however, do not exclude a more subtle alteration in the manner in which LPS and DNA distribute in vivo in the SRA−/− mice and contact macrophages and other cytokine-producing cells. In this regard, processes such as infection may alter the number or activation state of macrophages and thereby influence the role of the SRA.
The ability of the SRA to bind LPS without causing cell activation has suggested that this receptor could serve a protective role in host defence by ‘scavenging’ immunostimulatory molecules with potentially deleterious effects. We are interested therefore in the possibility that the SRA may function as a scavenger for immunostimulatory nucleic acids as well as for LPS, with a failure to remove CpG DNA from the circulation leading to a greater response from the host. Current studies are underway to address any protective function of the SRA and to characterize further the pathways for DNA uptake.
Acknowledgments
We are indebted to Drs Tatsuhito Kodama and Mason Freeman for providing SRA−/−mice. We thank Mr Robert Nelson for technical assistance in confocal microscopy. This study was supported by VA Merit Review and Arthritis Foundation grants.
Abbreviations
- BMMΦ
bone marrow-derived macrophage
- BODIPY-FL
boron dipyrromethene difluoride
- CpG
cytosine guanosine dinucleotide
- CT DNA
calf thymus DNA
- EC DNA
Escherichia coli DNA
- FITC
fluorescein isothiocyanate
- LDL
low-density lipoprotein
- ODN
oligodeoxynucleotide
- SRA
scavenger receptor type A
- WT
wild type
References
- 1.Pisetsky DS, Reich C, Crowley SD, Halpern MD. Immunological properties of bacterial DNA. Ann N Y Acad Sci. 1995;772:152–63. doi: 10.1111/j.1749-6632.1995.tb44740.x. [DOI] [PubMed] [Google Scholar]
- 2.Krieg AM. An innate immune defense mechanism based on the recognition of CpG motifs in microbial DNA. J Lab Clin Med. 1996;128:128–33. doi: 10.1016/s0022-2143(96)90004-9. [DOI] [PubMed] [Google Scholar]
- 3.Lipford GB, Heeg K, Wagner H. Bacterial DNA as immune cell activator. Trends Microbiol. 1998;6:496–500. doi: 10.1016/s0966-842x(98)01408-5. 10.1016/s0966-842x(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 4.Klinman DM, Verthelyi D, Takeshita F, Ishii KJ. Immune recognition of foreign DNA. a cure for bioterrorism? Immunity. 1999;11:123–9. doi: 10.1016/s1074-7613(00)80087-4. [DOI] [PubMed] [Google Scholar]
- 5.Raz E, Spiegelberg HL. Deviation of the allergic IgE to an IgG response by gene immunotherapy. Int Rev Immunol. 1999;18:271–89. doi: 10.3109/08830189909043030. [DOI] [PubMed] [Google Scholar]
- 6.Tokunaga T, Yamamoto H, Shimada S, et al. Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J Natl Cancer Inst. 1984;72:955–62. [PubMed] [Google Scholar]
- 7.Messina JP, Gilkeson GS, Pisetsky DS. Stimulation of in vitro murine lymphocyte proliferation by bacterial DNA. J Immunol. 1991;147:1759–64. [PubMed] [Google Scholar]
- 8.Tokunaga T, Yano O, Kuramoto E, Kimura Y, Yamamoto T, Kataoka T, Yamamoto S. Synthetic oligonucleotides with particular base sequences from the cDNA encoding proteins of Mycobacterium bovis BCG induce interferons and activate natural killer cells. Microbiol Immunol. 1992;36:55–66. doi: 10.1111/j.1348-0421.1992.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 9.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–9. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 10.Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc Natl Acad Sci USA. 1996;93:2879–83. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato Y, Roman M, Tighe H, et al. Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science. 1996;273:352–4. doi: 10.1126/science.273.5273.352. [DOI] [PubMed] [Google Scholar]
- 12.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 13.Roman M, Martin-Orozco E, Goodman JS, et al. Immunostimulatory DNA sequences function as T helper-1-promoting adjuvants [see comments] Nat Med. 1997;3:849–54. doi: 10.1038/nm0897-849. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto S, Kuramoto E, Shimada S, Tokunaga T. In vitro augmentation of natural killer cell activity and production of interferon-alpha/beta and -gamma with deoxyribonucleic acid fraction from Mycobacterium bovis BCG. Jpn J Cancer Res. 1988;79:866–73. doi: 10.1111/j.1349-7006.1988.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S, Beard C, Jaenisch R, Jones P, Sprent J. Mitogenicity of DNA from different organisms for murine B cells. J Immunol. 1997;159:3119–25. [PubMed] [Google Scholar]
- 16.Sparwasser T, Miethke T, Lipford G, Borschert K, Hacker H, Heeg K, Wagner H. Bacterial DNA causes septic shock [letter] Nature. 1997;386:336–7. doi: 10.1038/386336a0. [DOI] [PubMed] [Google Scholar]
- 17.Sparwasser T, Koch ES, Vabulas RM, Heeg K, Lipford GB, Ellwart JW, Wagner H. Bacterial DNA and immunostimulatory CpG oligonucleotides trigger maturation and activation of murine dendritic cells. Eur J Immunol. 1998;28:2045–54. doi: 10.1002/(SICI)1521-4141(199806)28:06<2045::AID-IMMU2045>3.0.CO;2-8. 10.1002/(sici)1521-4141(199806)28:06<2045::aid-immu2045>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 18.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 19.Pisetsky DS. The immunologic properties of DNA. J Immunol. 1996;156:421–3. [PubMed] [Google Scholar]
- 20.Yakubov LA, Deeva EA, Zarytova VF, Ivanova EM, Ryte AS, Yurchenko LV, Vlassov VV. Mechanism of oligonucleotide uptake by cells: involvement of specific receptors? Proc Natl Acad Sci USA. 1989;86:6454–8. doi: 10.1073/pnas.86.17.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacker H, Mischak H, Miethke T, et al. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 1998;17:6230–40. doi: 10.1093/emboj/17.21.6230. 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi AK, Krieg AM. CpG DNA rescue from anti-IgM-induced WEHI-231 B lymphoma apoptosis via modulation of I kappa B alpha and I kappa B beta and sustained activation of nuclear factor-kappa B/c-Rel. J Immunol. 1998;160:1240–5. [PubMed] [Google Scholar]
- 23.Bennett RM, Gabor GT, Merritt MM. DNA binding to human leukocytes. Evidence for a receptor–mediated association, internalization, and degradation of DNA. J Clin Invest. 1985;76:2182–90. doi: 10.1172/JCI112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loke SL, Stein CA, Zhang XH, Mori K, Nakanishi M, Subasinghe C, Cohen JS, Neckers LM. Characterization of oligonucleotide transport into living cells. Proc Natl Acad Sci USA. 1989;86:3474–8. doi: 10.1073/pnas.86.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura Y, Sonehara K, Kuramoto E, Makino T, Yamamoto S, Yamamoto T, Kataoka T, Tokunaga T. Binding of oligoguanylate to scavenger receptors is required for oligonucleotides to augment NK cell activity and induce IFN. J Biochem (Tokyo) 1994;116:991–4. doi: 10.1093/oxfordjournals.jbchem.a124658. [DOI] [PubMed] [Google Scholar]
- 26.Takagi T, Hashiguchi M, Mahato RI, Tokuda H, Takakura Y, Hashida M. Involvement of specific mechanism in plasmid DNA uptake by mouse peritoneal macrophages. Biochem Biophys Res Commun. 1998;245:729–33. doi: 10.1006/bbrc.1998.8521. 10.1006/bbrc.1998.8521. [DOI] [PubMed] [Google Scholar]
- 27.Brown MS, Goldstein JL, Krieger M, Ho YK, Anderson RG. Reversible accumulation of cholesteryl esters in macrophages incubated with acetylated lipoproteins. J Cell Biol. 1979;82:597–613. doi: 10.1083/jcb.82.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein JL, Ho YK, Basu SK, Brown MS. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc Natl Acad Sci USA. 1979;76:333–7. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bijsterbosch MK, Manoharan M, Rump ET, et al. In vivo fate of phosphorothioate antisense oligodeoxynucleotides: predominant uptake by scavenger receptors on endothelial liver cells. Nucl Acids Res. 1997;25:3290–6. doi: 10.1093/nar/25.16.3290. 10.1093/nar/25.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawabata K, Takakura Y, Hashida M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm Res. 1995;12:825–30. doi: 10.1023/a:1016248701505. [DOI] [PubMed] [Google Scholar]
- 31.Sawai K, Mahato RI, Oka Y, Takakura Y, Hashida M. Disposition of oligonucleotides in isolated perfused rat kidney: involvement of scavenger receptors in their renal uptake. J Pharmacol Exp Ther. 1996;279:284–90. [PubMed] [Google Scholar]
- 32.Wloch MK, Pasquini S, Ertl HC, Pisetsky DS. The influence of DNA sequence on the immunostimulatory properties of plasmid DNA vectors. Hum Gene Ther. 1998;9:1439–47. doi: 10.1089/hum.1998.9.10-1439. [DOI] [PubMed] [Google Scholar]
- 33.Takakura Y, Takagi T, Hashiguchi M, et al. Characterization of plasmid DNA binding and uptake by peritoneal macrophages from class A scavenger receptor knockout mice. Pharm Res. 1999;16:503–8. doi: 10.1023/a:1018842210588. [DOI] [PubMed] [Google Scholar]
- 34.Butler M, Crooke RM, Graham MJ, et al. Phosphorothioate oligodeoxynucleotides distribute similarly in class A scavenger receptor knockout and wild-type mice. J Pharmacol Exp Ther. 2000;292:489–96. [PubMed] [Google Scholar]
- 35.Chace JH, Hooker NA, Mildenstein KL, Krieg AM, Cowdery JS. Bacterial DNA-induced NK cell IFN-gamma production is dependent on macrophage secretion of IL-12. Clin Immunol Immunopathol. 1997;84:185–93. doi: 10.1006/clin.1997.4380. 10.1006/clin.1997.4380. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki H, Kurihara Y, Takeya M, et al. The multiple roles of macrophage scavenger receptors (MSR) in vivo: resistance to atherosclerosis and susceptibility to infection in MSR knockout mice. J Atheroscler Thromb. 1997;4:1–11. doi: 10.5551/jat1994.4.1. [DOI] [PubMed] [Google Scholar]
- 37.Haworth R, Platt N, Keshav S, et al. The macrophage scavenger receptor type A is expressed by activated macrophages and protects the host against lethal endotoxic shock. J Exp Med. 1997;186:1431–9. doi: 10.1084/jem.186.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saburi S, Azuma S, Sato E, Toyoda Y, Tachi C. Developmental fate of single embryonic stem cells microinjected into 8-cell-stage mouse embryos. Differentiation. 1997;62:1–11. doi: 10.1046/j.1432-0436.1997.6210001.x. [DOI] [PubMed] [Google Scholar]
- 39.Fischer HG, Opel B, Reske K, Reske-Kunz AB. Granulocyte–macrophage colony-stimulating factor-cultured bone marrow-derived macrophages reveal accessory cell function and synthesis of MHC class II determinants in the absence of external stimuli. Eur J Immunol. 1988;18:1151–8. doi: 10.1002/eji.1830180802. [DOI] [PubMed] [Google Scholar]
- 40.Klein G, Satre M. Kinetics of fluid-phase pinocytosis in Dictyostelium discoideum amoebae. Biochem Biophys Res Commun. 1986;138:1146–52. doi: 10.1016/s0006-291x(86)80402-8. [DOI] [PubMed] [Google Scholar]
- 41.Benimetskaya L, Loike JD, Khaled Z, et al. Mac-1 (CD11b/CD18) is an oligodeoxynucleotide-binding protein [see comments] Nat Med. 1997;3:414–20. doi: 10.1038/nm0497-414. [DOI] [PubMed] [Google Scholar]
- 42.Pearson AM. Scavenger receptors in innate immunity. Curr Opin Immunol. 1996;8:20–8. doi: 10.1016/s0952-7915(96)80100-2. [DOI] [PubMed] [Google Scholar]
- 43.Platt N, Gordon S. Scavenger receptors: diverse activities and promiscuous binding of polyanionic ligands. Chem Biol. 1998;5:R193–R203. doi: 10.1016/s1074-5521(98)90156-9. [DOI] [PubMed] [Google Scholar]
- 44.Freeman M, Ekkel Y, Rohrer L, Penman M, Freedman NJ, Chisolm GM, Krieger M. Expression of type I and type II bovine scavenger receptors in Chinese hamster ovary cells: lipid droplet accumulation and nonreciprocal cross competition by acetylated and oxidized low density lipoprotein. Proc Natl Acad Sci USA. 1991;88:4931–5. doi: 10.1073/pnas.88.11.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson AM, Rich A, Krieger M. Polynucleotide binding to macrophage scavenger receptors depends on the formation of base-quartet-stabilized four-stranded helices. J Biol Chem. 1993;268:3546–54. [PubMed] [Google Scholar]
- 46.Suzuki K, Doi T, Imanishi T, Kodama T, Tanaka T. Oligonucleotide aggregates bind to the macrophage scavenger receptor. Eur J Biochem. 1999;260:855–60. doi: 10.1046/j.1432-1327.1999.00233.x. 10.1046/j.1432-1327.1999.00233.x. [DOI] [PubMed] [Google Scholar]
- 47.Peyman A, Ryte A, Helsberg M, Kretzschmar G, Mag M, Uhlmann E. Enhanced cellular uptake of G-rich oligonucleotides. Nucleosides Nucleotides. 1995;14:1077–88. [Google Scholar]
- 48.Yao GQ, Corrias S, Cheng YC. Identification of two oligodeoxyribonucleotide binding proteins on plasma membranes of human cell lines. Biochem Pharmacol. 1996;51:431–6. doi: 10.1016/0006-2952(95)02198-1. 10.1016/0006-2952(95)02198-1. [DOI] [PubMed] [Google Scholar]
- 49.Stein CA. Controversies in the cellular pharmacology of oligodeoxynucleotides. Ciba Found Symp. 1997;209:79–89. [PubMed] [Google Scholar]
- 50.Hanss B, Stein CA, Klotman PE. Cellular uptake and biodistribution of oligodeoxynucleotides. In: Stein CA, Krieg AM, editors. Applied Antisense Oligonucleotide Technology. New York: Wiley-Liss, Inc; 1998. pp. 111–27. [Google Scholar]
- 51.van Berkel TJ, Van Velzen A, Kruijt JK, Suzuki H, Kodama T. Uptake and catabolism of modified LDL in scavenger-receptor class A type I/II knock-out mice. Biochem J. 1998;331(1):29–35. doi: 10.1042/bj3310029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hampton RY, Golenbock DT, Penman M, Krieger M, Raetz CR. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature. 1991;352:342–4. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]
- 53.Shnyra A, Lindberg AA. Scavenger receptor pathway for lipopolysaccharide binding to Kupffer and endothelial liver cells in vitro. Infect Immun. 1995;63:865–73. doi: 10.1128/iai.63.3.865-873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunne DW, Resnick D, Greenberg J, Krieger M, Joiner KA. The type I macrophage scavenger receptor binds to gram-positive bacteria and recognizes lipoteichoic acid. Proc Natl Acad Sci USA. 1994;91:1863–7. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Oosten MBE, van Berkel TJ, Kuiper J. New scavenger receptor-like receptors for the binding of lipopolysaccharide to liver endothelial and Kupffer cells. Infect Immun. 1998;66:5107–12. doi: 10.1128/iai.66.11.5107-5112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amersfoort ESV, van Berkel TJ, Kuiper J. Clearance of lipopolysaccharide in scavenger receptor knockout mice. J Leukocyte Biol. 1996;210(Suppl):48. [Google Scholar]