Abstract
Francisella tularensis, the causative agent of tularemia, is one of the most infectious bacterial pathogens known and is a category A select agent. We created a sequence-defined, near-saturation transposon mutant library of F. tularensis novicida, a subspecies that causes a tularemia-like disease in rodents. The library consists of 16,508 unique insertions, an average of >9 insertions per gene, which is a coverage nearly twice that of the greatest previously achieved for any bacterial species. Insertions were recovered in 84% (1,490) of the predicted genes. To achieve high coverage, it was necessary to construct transposons carrying an endogenous Francisella promoter to drive expression of antibiotic resistance. An analysis of genes lacking (or with few) insertions identified nearly 400 candidate essential genes, most of which are likely to be required for growth on rich medium and which represent potential therapeutic targets. To facilitate genome-scale screening using the mutant collection, we assembled a sublibrary made up of two purified mutants per gene. The library provides a resource for virtually complete identification of genes involved in virulence and other nonessential processes.
Keywords: essential genes, promoter, tularemia, U112
Francisella tularensis is a Gram-negative, facultative intracellular pathogen that causes the disease tularemia. Infection can occur by inhalation, ingestion, exposure to infected animals, or transmission from arthropod vectors. Clinical presentation can take various forms and depends on the route of infection (1, 2). F. tularensis is one of the most infectious bacterial pathogens known, with as few as 10 organisms causing disease (3, 4). If untreated, mortality in humans can reach 30% (4). Hundreds of aquatic and terrestrial mammals are also susceptible to the disease (2, 3). Because of its high infectivity, severe virulence, ease of aerosol dissemination, and broad geographic and host distribution, F. tularensis is considered a potential biological weapon threat and is classified as a Category A select agent (5).
Four subspecies of F. tularensis are recognized. Subspecies tularensis (the most virulent subspecies) and holarctica are the significant human pathogens (6), whereas subspecies novicida (a.k.a. Francisella novicida) and mediasiatica are relatively nonpathogenic toward people (although human infections with F. novicida have been reported) (7). A subsp. novicida type strain (U112) and an attenuated subsp. holarctica strain (LVS) are commonly used as surrogates for subsp. tularensis in virulence studies using animal models (8, 9). Both strains cause tularemia-like diseases in rodents, and F. novicida is particularly virulent toward mice (8, 10). The genomes of subspecies novicida (U112), holarctica (LVS), and tularensis (Schu S4) have been sequenced (refs. 11 and 12 and L. Rohmer and M.B., unpublished work). The three highly similar genomes are relatively small for bacterial species (1.89–1.91 Mbp), suggesting low genetic redundancy in this versatile pathogen.
The basis of the exceptional virulence of F. tularensis is poorly understood (6, 13). The pathogen appears able to evade host innate immune responses, attributable in part to an unusual LPS structure (10, 14, 15) and an ability to escape the normal endosomal–lysosomal trafficking pathway within phagocytic cells (13, 16). A pair of regulatory genes (mglA and mglB) and a locus known as the Francisella pathogenicity island have been shown to play important roles in the organism's escape from the phagosome and replication within the cytosol of host immune cells (17–19). Other proposed pathogenicity determinants include pili (20) and a siderophore (21).
To facilitate the systematic identification of virulence determinants and other factors associated with Francisella pathogenesis, we have assembled a comprehensive library of sequence-defined transposon insertion mutants of F. novicida. Such defined mutant libraries provide resources for carrying out efficient genome-scale phenotypic screens and serve as sources of individual mutant alleles (22). In addition, the genes refractory to insertional inactivation help define the genes essential for viability of the organism, and essential genes represent potential drug targets (23).
Results
Mutant Library Construction.
We assembled a large collection of individually arrayed transposon insertion mutants of F. novicida U112 [Table 1 and supporting information (SI) Table 6]. Insertion sites were identified by amplification and sequencing of one of the transposon–genome junctions, followed by comparison with the reference genome sequence. Five different transposon Tn5 derivatives were used (Fig. 1).
Table 1.
Composition of the F. novicida transposon mutant library
| Transposon type |
Total | ||
|---|---|---|---|
| Biased* | Unbiased* | ||
| Mutants arrayed and sequence-mapped | 9,312 | 16,794 | 26,106 |
| Insertions mapped to specific locations | 6,760 | 13,670 | 20,430 |
| Duplicate insertions | 1,821 | 2,101 | 3,922 |
| Unique insertions | 4,939 | 11,569 | 16,508 |
| Within ORFs | 4,881 | 10,999 | 15,880 |
| Outside of ORFs | 58 | 570 | 628 |
| Genes hit internally | 743 | 1,427 | 1,490 |
| Average unique hits per gene in genome | 2.8 | 6.5 | 9.3 |
| Average unique hits per gene hit | 6.6 | 8.1 | 11.1 |
*“Biased” transposons (<KAN-2> and T15) produce insertions oriented in a consistent direction relative to the gene of insertion (orientation bias); “unbiased” transposons (T17, T18, and T20) produce insertions oriented in either direction relative to the gene of insertion.
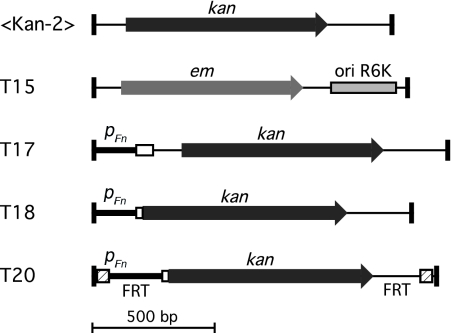
Fig. 1.
Transposons used for mutagenesis. Transposons are Tn5 derivatives with end sequences optimized for in vitro transposition (41). The top two transposons (<KAN-2> and T15) yielded orientation-biased insertions in F. novicida (i.e., the antibiotic-resistance gene was nearly always oriented in the same direction as the chromosomal gene hit), whereas the bottom three (T17, T18, and T20) yielded insertions unbiased in orientation. In T17, T18, and T20, an endogenous F. novicida promoter (pFn) drives expression of the kanamycin-resistance ORF. The open box represents the first few codons of the native F. novicida gene (FTN_1451). T17 carries a transcriptional gene fusion, whereas T18 carries a translational fusion between the initial codons of FTN_1451 and the kanamycin-resistance ORF. T20 was derived from T18 and carries FRT sites at each end (hatched boxes). Cre-mediated recombination excises the sequences between the FRT sites, leaving a 97-bp insertion. Black rectangles at the transposon ends represent the 19-bp Tn5 mosaic end recognition sequences. kan, kanamycin resistance determinant; em, erythromycin resistance determinant; ori R6K, R6K origin of replication.
Initially, mutants were generated by using the transposon <KAN-2> (24). Of 8,928 mutants examined, 6,624 were unambiguously sequence-mapped to 4,831 unique insertion sites. Surprisingly, insertions in only 715 (41%) of the 1,767 genes predicted for the annotated genome sequence were represented, an unexpectedly low fraction (Fig. 2A and ref. 25). In addition, the orientation of these insertions showed a striking bias: for 99.5% of them, the kanamycin-resistance gene of the transposon was oriented in the same direction as the chromosomal gene interrupted. To explain this result, we hypothesized that the transposon kanamycin-resistance marker was poorly expressed from its own promoter in F. novicida and that only insertions with read-through expression from their target genes generated kanamycin-resistant colonies. The hypothesis implies that the genes mutated are ones expressed above a threshold level necessary to confer kanamycin resistance. A transposon (T15) carrying an erythromycin-resistance marker (26) also produced orientation-biased insertions.
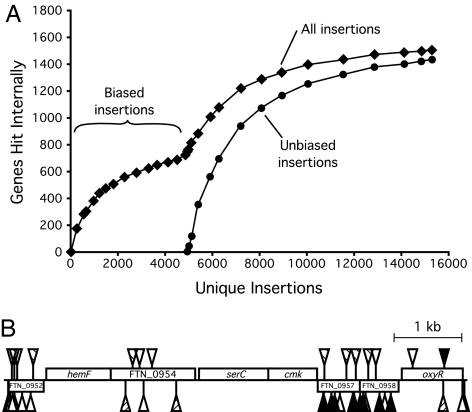
Fig. 2.
Transposon insertions in the F. novicida genome. (A) Number of genes hit with increased mutagenesis. The number of genes hit reached a plateau of ≈750 for the orientation-biased transposons (<KAN-2> and T15), and increased to ≈1,500 after the orientation-unbiased transposons (T17, T18, and T20) were introduced. In total, 1,490 genes (of 1,767 in the genome) received insertions. (B) Insertions in a representative segment of the genome. Rectangles above and below the line represent genes transcribed from the forward and reverse strands, respectively. Triangles above and below the line show positions of transposon insertions in the forward (antibiotic marker transcribed from the forward strand) and reverse orientations, respectively. Solid triangles, transposon <KAN-2>; open triangles, transposon T18; hatched triangles, transposon T20. Genes without insertions (hemF, serC, cmk) are putative essentials. Of the five genes with insertions, only three (FTN_0957, FTN_1958, oxyR) carry <KAN-2> insertions, and all of these insertions are oriented in the same direction as the target gene. T18 and T20 insertions, in contrast, are found in both orientations relative to the target genes.
To overcome the observed orientation bias, we constructed new transposons (T17, T18, and T20) using an endogenous F. novicida promoter to drive kanamycin-resistance expression (Fig. 1 and SI Fig. 4). The promoter was derived from a gene (FTN_1451) in which several <KAN-2> insertions had been recovered and was chosen after screening several candidate promoters for suitability (SI Fig. 4). Mutagenesis with the new transposons produced insertions virtually free of orientation bias (SI Table 6). In addition, the new transposons targeted nearly twice as many genes as <KAN-2> (Table 1 and Fig. 2A).
The entire collection comprised 16,508 unique, sequence-mapped insertions, corresponding to 9.3 insertions per predicted gene (Table 1 and SI Table 6). The insertions were distributed throughout the genome, with the only significant cold spot (around position 260,000) corresponding to a group of essential genes (Fig. 3). A total of 1,490 genes (84% of the genes in the genome) carried insertions. Predictably, larger genes tended to receive more hits (SI Fig. 5). The insertions in a representative segment of the genome illustrate the orientation bias of the <KAN-2> insertions and the greater number of genes hit by the newly constructed transposons (Fig. 2B).
Fig. 3.
Distribution of insertions in the genome. Bars represent number of insertions per 10,000 bp. The regions with relatively few insertions generally correspond to regions rich in essential genes. For example, the region around position 260,000 includes numerous genes encoding ribosomal proteins; the region around position 1,670,000 includes RNA polymerase and ribosomal genes; and the region around position 1,790,000 includes an operon encoding NADH dehydrogenase genes. Position 0 corresponds to the putative replication origin.
Marker Recycling Capability of a Subset of the Mutants.
Approximately 40% (6,226) of the unique mutants in the collection were made by using transposon T20, which carries FRT recombination sites at each end (Fig. 1). Flp-mediated recombination between the FRT sites should therefore result in excision of the antibiotic-resistance gene, facilitating reuse of the resistance marker (27). To test this feature, a shuttle plasmid (pLG72) carrying the flp recombinase gene under control of a Francisella promoter was constructed and transformed into five T20 mutants (Materials and Methods). All of 30 purified transformants were kanamycin-sensitive, and sequence analysis demonstrated the expected precise excision between the FRT sites in five representative colonies examined (data not shown).
Quality Control Tests.
To assess the accuracy of the insertion site assignments, we resequence-mapped 534 T20 insertion mutants using primers corresponding to the end of the transposon opposite to that used for the original mapping. A total of 98.3% (525 of 534) of the insertion assignments were confirmed (SI Table 7). Interestingly, the analysis also revealed deviations from the expected 9-bp target site duplication size (28) for ≈2% of insertions (SI Table 7).
To test for multiple insertions in individual strains, we probed for transposon sequences in Southern blots of genomic DNA cut with a restriction enzyme (RsaI), which cleaves once in each transposon. If two (or more) insertions were present in a single strain, four (or more) bands should have been observed in the corresponding blot. Exactly two strong bands were observed in each of the blots for 41 strains tested (data not shown), suggesting that <2% of the mutants carry multiple insertions.
Genetic instability of transposon insertions in Francisella species has been reported (26). Southern analysis of <KAN-2> insertions in subsp. holarctica strain LVS, however, has shown identically sized bands before and after serial passage, suggesting that Tn5 insertions are stable in LVS (24). To examine transposon stability in F. novicida, eight auxotrophic mutants from the collection (three <KAN-2>, one T18, and four T20 insertions) were serially passaged in rich media without antibiotic selection for ≈50 generations and then plated on minimal media. No colonies were observed for any of the strains, implying that the insertions examined were all stable (reversion frequency <10−12).
Robotic picking of closely spaced colonies and well-to-well contamination from handling library plates are potential sources of contamination in the arrayed cultures. To assess contamination levels, cultures of 45 predicted amino acid or nucleoside auxotrophs were tested for growth on minimal media. Eleven cultures (24%) exhibited growth, with the contamination levels ranging from 10−5 to >90% of total colony-forming units present. Sequence analysis revealed that the auxotrophic colonies (8 of 8 analyzed) corresponded to the expected mutants, whereas the prototrophic colonies carried different transposon insertions. The prototrophs thus apparently represent contamination rather than suppression by secondary mutation.
A Purified “Two-Allele” Mutant Set.
Because the large size of the primary mutant collection could limit genome-scale phenotypic screens, and because a significant fraction of the cultures exhibited detectable contamination, we created a smaller second library made up of single-colony-purified mutants (Table 2 and SI Table 8). The 3,050-member library includes two insertion alleles for the majority of genes. The alleles chosen were primarily insertions positioned between 5% and 70% within the ORF and are thus likely to represent null mutations. In addition, insertions of transposon T20 (which carries FRT sites) were favored. After single-colony purification, the mutants were arrayed in individually sealed tubes in 96-well format, then re-sequence-mapped to confirm their identities. Contamination tests of 49 auxotrophic mutant cultures found prototrophic contaminants in only one (at <10−3 of total titer), implying a high degree of purity in the two-allele set.
Table 2.
The two-allele mutant set
| n | |
|---|---|
| Mutants included | 3,050 |
| Gene representation | |
| Genes represented | 1,448 |
| Genes with 1 internal insertion | 203 |
| Genes with 2 internal insertions | 1,014 |
| Genes with 3 internal insertions | 193 |
| Genes with 4 internal insertions | 29 |
| Genes with 5 internal insertions | 9 |
| Insertions outside of gene ORFs* | 79 |
| Genes not represented | 319 |
| Transposon types | |
| <KAN-2 > insertions | 366 |
| T15 insertions | 3 |
| T17 insertions | 26 |
| T18 insertions | 487 |
| T20 insertions | 2,168 |
| Re-sequence-mapping | |
| Insertion locations confirmed | 2,785 |
| Insertion locations unconfirmed† | 265 |
All mutants were single-colony purified and resequence-mapped to verify their identities.
*Some insertions outside of predicted genes were included for (i) insertions in promoter regions of genes lacking coding sequence insertions and (ii) insertions in long intergenic regions.
†Sequence-mapping quality was insufficient to identify the transposon insertion site. Based on the fraction of sequence confirmed strains that were of expected identity, we estimate that 92% of these strains are correctly identified.
Candidate Essential Genes.
A total of 312 predicted F. novicida genes lacked transposon insertions and therefore appear likely to be essential for growth on nutrient media. Other candidate essential genes are those with insertions only near the termini of the ORFs (the 3′ ends of some essential genes may be functionally dispensable, and some 5′ ends may tolerate insertion if transcription can restart or proceed through the transposon) and large genes with very few insertions (which could be due to insertions in tandemly duplicated genes or at rare permissive sites). Using these criteria, we compiled a list of 396 candidate essential genes (Table 3 and SI Table 9).
Table 3.
F. novicida candidate essential genes
| Criteria* | Genes† | Orthologs in E. coli | % Essential in E. coli |
|---|---|---|---|
| Genes without insertions | 312 | 255 | 62 |
| Genes with insertions only in 3′ 10% | 49 | 46 | 61 |
| Genes with insertions only in 5′ 5% | 15 | 7 | 57 |
| >250 codon genes with only 1 hit | 42 | 31 | 61 |
| >450 codon genes with only 2 hits | 20 | 18 | 56 |
| Total: | 396 | 320 | 61 |
*To ensure a comprehensive list of candidate essentials, the analysis used only insertions with high-quality sequence-mapping data (Materials and Methods); 34 of the 396 genes were included because low-sequence-quality hits were not considered.
†Values indicate all genes fitting each category. Some categories overlap and 32 genes fit multiple categories. The total gene count (396) reports unique genes.
In bacteria, essential genes are more evolutionarily conserved than nonessential genes (29). To help assess the conservation of the F. novicida candidate essential genes, we examined the occurrence of orthologs in two other Francisella strains and in Escherichia coli. The candidate essentials were much more highly conserved than the full complement of F. novicida genes, a result which strongly supports their assignment as a group as candidate essentials (Table 4).
Table 4.
F. novicida gene orthologs in other bacterial species
| Genes with orthologs in specified organisms, % |
|||||
|---|---|---|---|---|---|
| F.t.t. | F.t.h. | Both | E. coli | All 3 | |
| All F.n. genes | 78 | 75 | 70 | 56 | 46 |
| F.n. candidate essentials | 94 | 93 | 92 | 82 | 79 |
F.t.t., F. tularensis tularensis Schu S4; F.t.h., F. tularensis holarctica LVS; F.n., F. novicida.
Based on the functional categories assigned in the genome annotation (L. Rohmer and M.B., unpublished work), the list of candidate essential genes was analyzed for functional representation relative to the complete set of genes (SI Fig. 6). Several functional categories, transcription, translation, and some metabolic functions, are more highly represented among candidate essentials than in the genome as a whole, as has been observed for other bacteria (25, 30). It is noteworthy, however, that 76 (19%) of the F. novicida candidate essential genes are of unknown or hypothetical function.
Comprehensive deletion mutagenesis has defined 303 essential genes in E. coli (30). We compared the F. novicida candidate essential gene set with the E. coli set by analyzing orthologous gene pairs (Table 5 and Materials and Methods). Of the 396 F. novicida candidate essential genes, 320 have orthologs in E. coli and 194 (61%) of these are essential. Note that for each criterion used to define the F. novicida candidate essential genes, a consistent fraction (≈60%) of the E. coli orthologs are essential (Table 3), a result that tends to validate the criteria chosen. Remarkably, 126 of the F. novicida candidate essential genes and 42 of the E. coli essential genes have nonessential orthologs in the other organism (Table 5).
Table 5.
F. novicida and E. coli essential and nonessential genes
| E. coli | F. novicida | |
|---|---|---|
| Total genes | 4,237 | 1,767 |
| Orthologs | 962 | 962 |
| Essential genes | 303 | 396 |
| Essential orthologs | 236 | 320 |
| Shared | 194 | 194 |
| Unique | 42 | 126 |
| Essential nonorthologs | 67 | 76 |
Two complicating factors may have overinflated the number of candidate essential genes identified. First, polar effects on essential genes in operons may have prevented the recovery of insertions in upstream nonessential genes. In fact, there are 27 candidate essential genes that have nonessential orthologs in E. coli yet are upstream in potential operons of candidate essential genes whose E. coli orthologs are essential. For example, the F. novicida candidate essential gene fabH (no insertions recovered) has a nonessential E. coli ortholog; in F. novicia, fabH is upstream in a potential operon of three other candidate essential genes, fabD, fabG, and acpP, all of which have essential E. coli orthologs. Second, some ORFs, particularly small ones, may not have received insertions because of chance. Assuming no essential genes in the genome, a neutral-base pair model of mutagenesis (which assumes every base in the genome has an equal probability of receiving an insertion) (25) predicts that, for the level of saturation obtained, 65 genes would not receive insertions because of chance. Taking into account the effects of polarity and insertional randomness, we estimate the number of true essential genes in F. novicida to be between 300 and 350.
Discussion
We created a large, sequence-defined transposon mutant library of F. tularensis subsp. novicida, a surrogate of the highly infectious pathogen F. tularensis subsp. tularensis. The library includes strains corresponding to an average of 9.3 insertions per gene with coverage of 84% of the predicted genes of the organism. We assume that nearly all of the genes without insertions are required for growth on nutrient medium. In addition to identifying candidate essential genes, the library provides a comprehensive resource for genome-scale identification and analysis of genes involved in virulence and related activities of the bacterium (31–33).
Several different transposons were used in generating the library. The transposon initially used (24) targeted a relatively low fraction of genes and produced insertions showing an extreme orientation bias. These properties apparently reflect the inefficient function of the promoter driving the antibiotic-resistance determinant in the transposon, so that only insertions creating active gene fusions (in nonessential genes) were generated. To provide more complete coverage, new transposons were constructed that use an endogenous F. novicida promoter to drive expression of the transposon antibiotic-resistance gene. One of the new transposons carries FRT recombination sites at its ends, making it possible to eliminate the resistance determinant. This capability allows reutilization of the resistant determinant, e.g., in generating double mutants for epistasis tests.
The F. novicida library is distinguished in its level of coverage and extent of quality control testing. It was possible to assign an average of 9.3 unique insertions per predicted gene, a value nearly twice that of the greatest previously achieved for any bacterial species (22). This high coverage increases the probability that genes lacking insertions are essential rather than having escaped insertions by chance and also increases the likelihood that rare alleles (e.g., those that reduce without eliminating function) are represented. Several levels of quality control testing were carried out. First, to define frequency of insertion location misassignment, we resequenced a sample of the insertions. Southern blot hybridization was also carried out to establish limits on the frequency of double-insertion events. Finally, an analysis of auxotrophic mutants made it possible to specify the level of cross-contamination in the library, a consequence of storage and replication of strains in a multiwell microtiter dish format.
We created a smaller second mutant library made up of two alleles per gene from the complete library (the “two-allele set”). This second library was designed to facilitate genome-scale mutant screens in F. novicida, because it reduces the number of strains that must be analyzed for full coverage by ≈5-fold. The 3,050 strains making up the two-allele set were single-colony-purified and resequenced to confirm their genomic insertion locations. Including two rather than one allele per gene helps ensure that genotype–phenotype associations are not missed because of allele-specific effects or cross-contamination.
Analysis of the mutant library identified 396 candidate essential genes required for growth on rich media. Essential genes are potential antimicrobial drug targets. Although additional experimentation will be required to confirm the essentiality of specific genes, the number of candidate essentials identified is on the order of estimates for other bacteria, e.g., E. coli, Pseudomonas aeruginosa, etc. (25, 30, 34, 35). It is, however, plausible that the number of essential genes is somewhat greater for F. novicida than for E. coli, which has 303 essential genes (30). The 2-fold-larger genome of E. coli may encode more redundant genes for essential functions than does F. novicida. In support of this point, 32% (126 genes) of the F. novicida candidate essentials were orthologous to nonessential E. coli genes, whereas only 14% (42 genes) of E. coli essentials were orthologous to F. novicida nonessential genes.
A significant proportion of the candidate essential genes in each organism –19% (76 genes) of F. novicida candidate essentials and 22% (67 genes) of E. coli essentials – did not have evident orthologs in the other organism. These values are similar to those obtained in an analogous comparison of E. coli and Bacillus subtilis (30) and presumably reflect the divergent lifestyles and evolutionary histories of these organisms.
In summary, we have created a transposon insertion mutant library covering virtually all nonessential genes in F. novicida, a surrogate of F. tularensis, a potential bioweapon because of its exceptionally high virulence and pulmonary route of infection. The collection should help identify potential drug targets, as well as facilitate genome-scale analyses of the mechanisms contributing to the extraordinary infectivity and virulence of this organism.
Materials and Methods
Strains, Plasmids, Primers, and Media.
The strains used were F. novicida U112 (36), E. coli CC118 (37) and TOP10 (Invitrogen, Carlsbad, CA). Oligonucleotide primers are listed in SI Table 10. Plasmid pLG72 was made by replacing the gfp ORF in pKK214GFP (38) with the flp ORF, retaining the groEL promoter to drive flp expression (SI Methods). Nutrient growth media for F. novicida was trypticase soy broth or agar supplemented with 0.1% l-cysteine HCl and 0.2% dextrose (TSBC or TSAC, respectively). TSB freezing media was 36 mM K2HPO4, 13.2 mM KH2PO4, 1.7 mM sodium citrate, 0.4 mM MgSO4, 6.8 mM (NH4)2SO4, and 4.4% glycerol (vol/vol) in TSBC. Minimal media for F. novicida was a modified version of Chamberlain's defined medium (39): 4 g/liter glucose, 10 g/liter NaCl, 0.135 g/liter MgSO4·7H2O, 1 g/liter KH2PO4, 1 g/liter K2HPO4, 4 mg/liter thiamine HCl, 2 mg/liter Ca-pantothenate, 2 mg/liter FeSO4·7H2O, 40 mg/liter spermine, 0.4 g/liter l-arginine, 0.2 g/liter l-histidine, 0.4 g/liter l-methionine, 0.2 g/liter l-cysteine HCl, 0.4 g/liter l-lysine HCl, 0.4 g/liter l-proline, 0.4 g/liter l-leucine, and 0.4 g/liter l-threonine, pH adjusted to 6.2–6.4. Antibiotics used for F. novicida were kanamycin (15 μg/ml), erythromycin (30 μg/ml), and, for plasmid maintenance, tetracycline (10 μg/ml).
Transposons.
Transposon <KAN-2> (EZ-Tn5 <KAN-2>) was purchased as preassembled transposon–transposase complexes (Epicentre Biotechnologies, Madison, WI). Transposon T15 (a.k.a. ISR6K-em) was made by cloning the ermC-bearing EcoRI fragment from plasmid X-em-N (generously supplied by Francis Nano, University of Victoria, Victoria, BC, Canada) into plasmid pMOD-3 <R6Kγori/MCS> (Epicentre Biotechnologies) at the EcoRI site, creating plasmid pLG52a. Transposons T17 (ISFn1), T18 (ISFn2) and T20 (ISFn2/FRT) were derived from a <KAN-2> insertion in the F. novicida gene FTN_1451 (see SI Fig. 4 and Materials and Methods). T17 carries an FTN_1451-kan transcriptional gene fusion, whereas T18 carries an FTN_1451-kan translational gene fusion. T20 was derived from T18 by introducing FRT sites at each end. Transposon–transposase complexes were assembled according to published protocols (Epicentre Biotechnologies, and see SI Methods).
Transposon Mutagenesis.
An overnight culture of F. novicida started from a fresh colony was subcultured 1:100 in 60 ml of TSBC and incubated at 37°C with aeration to OD550 of 0.7–0.9. Cells were pelleted and resuspended twice in 0.5 M sucrose with 1 mM EDTA and twice in 0.5 M sucrose and then pelleted and resuspended in 0.5 M sucrose to a volume of 0.2–0.6 ml. Centrifugations were at ≈7,000 × g for 8 min, and all steps were at room temperature. Concentrated cells (50 μl) were mixed with transposon–transposase complexes (>100 ng/μl DNA), incubated at room temperature for 10 min, and electroporated (Gene Pulser; 0.2 cm cuvette; 2.5 KV; 25 μF; 400 Ω; Bio-Rad, Hercules, CA). After adding 1 ml of TSBC, cells were incubated for 3–5 h at 37°C with aeration and then plated on selective TSAC at a dilution yielding well separated single colonies.
Picking, Arraying, Replicating, Storing, and Mapping Mutants.
Colonies were picked with a Genetix Qpix2 robot into 384-well plates (X7007; Genetix, Boston, MA) containing 90 μl per well TSB freezing media with antibiotic. After picking, plates were covered with Airpore tape sheets (Qiagen, Valencia, CA), placed in a plastic bag containing a wet paper towel, incubated at 37°C overnight with shaking (250 rpm), and then removed from the bag and incubated at room temperature for 24 h without shaking. The Airpore sheets were replaced with aluminum tape sheets (Island Scientific, Bainbridge Island, WA) before storage at −80°C. Working stock replicas of the library were created by inoculating from the original plates by using the robot. Insertion sites were identified (“sequence-mapping”) by using methods similar to those described (25) (and see SI Methods).
Flp Recombination.
Mutants were transformed with pLG72 or pKK214GFP (vector lacking flp). Colonies purified from pLG72 tranformants into five T20 insertion mutants (six per mutant parent) were all kanamycin-sensitive, whereas colonies purified from pKK214GFP transformants were all kanamycin-resistant. Cells spontaneously cured of pLG72 were isolated after growth in TSBC without selection.
Two-Allele Set.
Database scripts and hand curation were used to choose the two insertion alleles per gene that best combined the characteristics described in Results. To assemble the two-allele set, strains from the original library were cherry-picked by using the robot into 96-well plates containing 100 μl of TSBC per well. After 2 h outgrowth at 37°C with shaking, the cultures were serially diluted into three 96-well plates containing 50 μl of TSBC per well by using a grooved-pin replicator (V&P Scientific, San Diego, CA). The dilutions were spotted onto TSAC omnitrays (Cat. no. 242811; Nalge Nunc International, Naperville, IN) and incubated at 37°C overnight. For each strain, an isolated colony from a dilution plate was hand picked into 1.25 ml of TSB freezing media supplemented with antibiotic in 96-well, deep-well culture plates (Cat. no. S1205; Genetix) and incubated for 24 h at 37°C with shaking and 24 h at room temperature without shaking. For strains not yielding isolated colonies, colonies of the same or replacement strains were hand-purified. The deep-well block cultures were aliquotted (250 μl) into 96-well 500-μl Trakmate tube racks (Cat. no. 3735; Matrix Technologies, Hudson, NH), sealed (Cat. no. 4464; Matrix), and stored at −80°C. The sequence-mapping success rate was 91%. The intended mutants were confirmed for 92% of the successfully sequence-mapped strains. The identities of the remaining 8% (unintended but confirmed mutants) were noted. The set was assembled and mapped in portions to allow adjustments for underrepresented genes.
Candidate Essential Genes.
Analyses were carried out by using database and spreadsheet functions and PERL. Candidate essentials were identified based on insertions meeting the following minimum criteria for sequence-mapping data quality (87% of the unique insertions): match lengths of at least 20 bp and 30 bp, respectively, and average phred scores of at least 15 and 18, respectively, for the portions of the sequence reads matching the transposon and the genome, and at least 25 bases of sequence identity for the portion matching the genome. For comparison to E. coli, orthologs were defined as gene pairs that were reciprocal best-hits in BLASTP comparisons between the two genomes (40), which aligned >50% of the F. novicida ORF length and had >15% amino acid identity in the region of alignment. Genes were considered operonic if cooriented and the downstream gene's start codon was within 100 bp of the upstream gene's termination codon.
Supplementary Material
Acknowledgments
We thank Sam Miller, Francis Nano, and Laurence Rohmer for helpful discussions; Tom Kawula for sharing protocols; Ted Freeman for help with computer scripts; and Cheri Turner for technical assistance. This work was supported by Washington, Wyoming, Alaska, Montana, and Idaho (WWAMI) Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research, National Institute of Allergy and Infectious Diseases Grant 5 U54 AI057141-04.
Abbreviations
- TSAC
trypticase soy agar supplemented with 0.1% l-cysteine HCl and 0.2% dextrose
- TSBC
trypticase soy broth supplemented with 0.1% l-cysteine HCl and 0.2% dextrose
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606713104/DC1.
References
- 1.Jensen WA, Kirsch CM. Semin Respir Infect. 2003;18:146–158. [PubMed] [Google Scholar]
- 2.Petersen JM, Schriefer ME. Vet Res. 2005;36:455–467. doi: 10.1051/vetres:2005006. [DOI] [PubMed] [Google Scholar]
- 3.Ellis J, Oyston PC, Green M, Titball RW. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCrumb FR. Bacteriol Rev. 1961;25:262–267. doi: 10.1128/br.25.3.262-267.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darling RG, Catlett CL, Huebner KD, Jarrett DG. Emerg Med Clin North Am. 2002;20:273–309. doi: 10.1016/s0733-8627(02)00005-6. [DOI] [PubMed] [Google Scholar]
- 6.Titball RW, Johansson A, Forsman M. Trends Microbiol. 2003;11:118–123. doi: 10.1016/s0966-842x(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 7.Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CW, Brenner DJ. J Clin Microbiol. 1989;27:1601–1608. doi: 10.1128/jcm.27.7.1601-1608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pammit MA, Raulie EK, Lauriano CM, Klose KE, Arulanandam BP. Infect Immun. 2006;74:2063–2071. doi: 10.1128/IAI.74.4.2063-2071.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santic M, Molmeret M, Abu Kwaik Y. Cell Microbiol. 2005;7:957–967. doi: 10.1111/j.1462-5822.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- 10.Kieffer TL, Cowley S, Nano FE, Elkins KL. Microbes Infect. 2003;5:397–403. doi: 10.1016/s1286-4579(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 11. www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genome&cmd=Retrieve&dopt=Overview&list_uids=19299.
- 12.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, et al. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 13.Santic M, Molmeret M, Klose KE, Abu Kwaik Y. Trends Microbiol. 2006;14:37–44. doi: 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Barker JH, Weiss J, Apicella MA, Nauseef WM. Infect Immun. 2006;74:3277–3284. doi: 10.1128/IAI.02011-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duenas AI, Aceves M, Orduna A, Diaz R, Sanchez Crespo M, Garcia-Rodriguez C. Int Immunol. 2006;18:785–795. doi: 10.1093/intimm/dxl015. [DOI] [PubMed] [Google Scholar]
- 16.Bosio CM, Dow SW. J Immunol. 2005;175:6792–6801. doi: 10.4049/jimmunol.175.10.6792. [DOI] [PubMed] [Google Scholar]
- 17.Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. Proc Natl Acad Sci USA. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. Cell Microbiol. 2005;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- 20.Forslund AL, Kuoppa K, Svensson K, Salomonsson E, Johansson A, Bystrom M, Oyston PC, Michell SL, Titball RW, Noppa L, et al. Mol Microbiol. 2006;59:1818–1830. doi: 10.1111/j.1365-2958.2006.05061.x. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. J Bacteriol. 2006;188:3785–3795. doi: 10.1128/JB.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salama NR, Manoil C. Curr Opin Microbiol. 2006;9:307–311. doi: 10.1016/j.mib.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Chalker AF, Lunsford RD. Pharmacol Ther. 2002;95:1–20. doi: 10.1016/s0163-7258(02)00222-x. [DOI] [PubMed] [Google Scholar]
- 24.Kawula TH, Hall JD, Fuller JR, Craven RR. Appl Environ Microbiol. 2004;70:6901–6904. doi: 10.1128/AEM.70.11.6901-6904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauriano CM, Barker JR, Nano FE, Arulanandam BP, Klose KE. FEMS Microbiol Lett. 2003;229:195–202. doi: 10.1016/S0378-1097(03)00820-6. [DOI] [PubMed] [Google Scholar]
- 27.Schweizer HP. J Mol Microbiol Biotechnol. 2003;5:67–77. doi: 10.1159/000069976. [DOI] [PubMed] [Google Scholar]
- 28.Reznikoff WS. Mol Microbiol. 2003;47:1199–1206. doi: 10.1046/j.1365-2958.2003.03382.x. [DOI] [PubMed] [Google Scholar]
- 29.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, et al. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Mol Syst Biol 2. 2006:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier TM, Pechous R, Casey M, Zahrt TC, Frank DW. Appl Environ Microbiol. 2006;72:1878–1885. doi: 10.1128/AEM.72.3.1878-1885.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin A, Mann BJ. BMC Microbiol. 2006;6:69. doi: 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tempel R, Lai XH, Crosa L, Kozlowicz B, Heffron F. Infect Immun. 2006;74:5095–5105. doi: 10.1128/IAI.00598-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. Mol Microbiol. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson CL, Wicht W, Jellison WL. Public Health Rep. 1955;70:253–258. [PMC free article] [PubMed] [Google Scholar]
- 37.Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abd H, Johansson T, Golovliov I, Sandstrom G, Forsman M. Appl Environ Microbiol. 2003;69:600–606. doi: 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamberlain RE. Appl Microbiol. 1965;13:232–235. doi: 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivera MC, Jain R, Moore JE, Lake JA. Proc Natl Acad Sci USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Bhasin A, Reznikoff WS. J Mol Biol. 1998;276:913–925. doi: 10.1006/jmbi.1997.1579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.