Abstract
Intracellular water dynamics in Haloarcula marismortui, an extremely halophilic organism originally isolated from the Dead Sea, was studied by neutron scattering. The water in centrifuged cell pellets was examined by means of two spectrometers, IN6 and IN16, sensitive to motions with time scales of 10 ps and 1 ns, respectively. From IN6 data, a translational diffusion constant of 1.3 × 10−5 cm2 s−1 was determined at 285 K. This value is close to that found previously for other cells and close to that for bulk water, as well as that of the water in the 3.5 M NaCl solution bathing the cells. A very slow water component was discovered from the IN16 data. At 285 K the water-protons of this component displays a residence time of 411 ps (compared with a few ps in bulk water). At 300 K, the residence time dropped to 243 ps and was associated with a translational diffusion of 9.3 × 10−8 cm2 s−1, or 250 times lower than that of bulk water. This slow water accounts for ≈76% of cell water in H. marismortui. No such water was found in Escherichia coli measured on BSS, a neutron spectrometer with properties similar to those of IN16. It is hypothesized that the slow mobility of a large part of H. marismortui cell water indicates a specific water structure responsible for the large amounts of K+ bound within these extremophile cells.
Keywords: Haloarcula marismortui, water structure and dynamics, extreme haplophile
The study of the specific properties of water in biological systems continues to yield fascinating surprises. Haloarcula marismortui, an archaeal extreme halophile isolated from the Dead Sea, attracted our attention some years ago because of its high selectivity for K+ ([Kin+]:[Nan+]/[Kout+]:[ Naout+] ≈ 20,000), despite a high membrane permeability (1). In distinction to other organisms, K+ was retained within the cell, even in the absence of metabolism, with a half-time of exchange with the outside medium of >24 h (2). At the time, the only systems known with a very high binding selectivity were antibiotics such as nonactin and valinomycin. The structure responsible for their specificity is a “cage” of six to eight carbonyl oxygen atoms arranged in space according to a definite pattern. If the same principle were to be responsible for K+ binding in H. marismortui, 24–32 moles oxygen per liter of cell water would be required, enormous amounts of oxygen atoms that cannot be supplied exclusively by the organic components of the cell. It is therefore suggested that the oxygen atoms required might be furnished by the ordering of water molecules forming a tertiary system of water, KCl, and cell proteins.
Evidence in favor of more than a single phase of water in cell pellets of H. marismortui came initially from H-NMR measurements (3). The visible intensity of water in such pellets accounted for all of the total water present (101 ± 7%). When the pellets were slowly cooled to below −19°C (the temperature at which the culture medium froze), 40% of the water signal remained visible. From analysis of the dynamic properties of the intracellular water, it was concluded that 55% of the cell water had a translational diffusion coefficient of 10−8 cm2 s−1, a figure three orders of magnitude lower than that of pure bulk water, protein hydration water, or water in molar solutions of NaCl or KCl (Table 1). The measurements were consistent with differential scanning calorimetric studies (4) in which the enthalpy of evaporation of water in pelleted cells was measured. The enthalpy of evaporation determines the heat needed to convert liquid water to the gaseous phase, and is, therefore, a measure of the strength of interaction between water molecules in the liquid phase. Two populations of water molecules were found; the first (52% of total water) had an enthalpy similar to that of 3.5 M NaCl, whereas the enthalpy of the other population was 13 KJ/mole higher (4). This is roughly equivalent to the enthalpy of one hydrogen bond and suggests that, in roughly half the cell water of H. marimortui, the water molecules are more tightly bound to each other, and therefore less mobile, than in 3.5 M NaCl.
Table 1.
Translational and rotational diffusion of water in the pure state, in the cell pellet, and in concentrated solutions of NaCl and KCl
| Method | Origin of water (temperature, K) | Translational diffusion coefficient 10−5 cm2 s−1 | τ0*, ps | Rotational diffusion coefficient | τR, ps | 〈R2〉1/2, Å | Ref. |
|---|---|---|---|---|---|---|---|
| NMR | Pure | 2.5 | 1011 s−1 | 3 | |||
| Pure (288) | 3.68† | 27 | |||||
| Pure (room temperature) | 2.4 | ∼4‡ | 50 | ||||
| QENS | Pure (263) | 0.7 | 6.47 | 12 | |||
| Pure (278) | 1.3 | 2.33 | |||||
| Pure (285) | 1.6 | 1.66 | |||||
| Pure (298) | 2.3 | 1.10 | 11 | ||||
| Pure (273) | 1.1 | 3 | 19 | ||||
| Pure (294.4) | 3.3‡ | 51 | |||||
| 4 M NaCl (285) | 1.5 | F.G., unpublished data | |||||
| 3 M KCl (285) | >2 | ||||||
| Pure (room temperature) | 0.40 | 23 | |||||
| Cell pellet (285) | 0.58 | This work | |||||
| Isotopic | Pure (283) | 1.67 | 52 | ||||
| Pure (298) | 2.57 | ||||||
| 3.5 M NaCl (283) | 1.26 | 53 | |||||
| 3 M KCl (283) | 1.76 |
*Translational residence time.
†Orientational time constant.
‡Rotational correlation time.
Dielectric spectroscopy is another method for measuring molecular dynamics. Suspensions of Escherichia coli showed a single β-dispersion centered on 1 MHz with no dispersion at higher frequencies (5). On the other hand, cell pellets of H. marimortui examined by means of time domain reflectometry showed two dispersions; the first had a relaxation time of 1–2 ps, characteristic of cell membranes, whereas the second, at a frequency of 250 MHz, gave a relaxation time, τ, of 636 ps (6). This second dispersion may probably be identified with water because it had a dielectric decrement of 80, characteristic of this substance, even though τ is high compared with that of bulk water (τwater measured by dielectric spectroscopy = 7 ps at 298 K; ref. 6). The volume fraction represented by the second dispersion has recently been calculated to represent 58.5% of total cell water (Y. Hayashi, personal communication).
Thus, observations obtained by three different physical methods led us to conclude that cells of H. marismortui contain water with two different types (7). The first consists of water in the physical state characteristic of 3.5M NaCl, whereas the second involves water with a low rate of mobility. Therefore, we planned more direct measurements of water diffusion in intact H. marismortui cells by measuring neutron scattering in deuterium-labeled cells.
Neutron scattering is a powerful technique for the study of water and protein dynamics (8, 9). Neutron wavelengths (≈1 Å) and energies (0.1–1 meV) are similar to H-bond lengths and thermal excitation energies, respectively. Dynamics information on a sample is derived from an analysis of its incoherent neutron scattering as a function of angle and energy change. Because the scattering cross-sections of natural abundance H and 2H are very different, specific components in a complex system can be examined by labeling them selectively with deuterium. The incoherent neutron scattering cross-section of 1H is 40 times larger than that of 2H (D), so that D-labeling a chemical group significantly reduces its contribution to the scattering. The dynamics of bulk water and confined water have been studied successfully at room temperature (10, 11), in supercooled water (10, 12), water in hydrogels (13), in porous systems (11), aqueous solutions (14), and water in membrane stacks (15, 16). Biological molecules impose restrictions on water structure and movement: the dynamics of water associated with amino acids (17) and proteins (18, 19) has been measured by neutron scattering on deuterium-labeled samples and shown to be slower by about one order of magnitude as compared with bulk water. The mass density of hydration shell water around proteins was measured by small angle x-ray and neutron scattering to be significantly higher than 1 (20).
Here, we describe experiments designed to examine the behavior of H2O in pellets of fully deuterated cells of H. marismortui and E. coli.
Results
The Space-Time Window in Quasielastic Neutron Scattering (QENS).
The length scale of atomic motions examined in a QENS experiment is ≈1/Q, where Q is called the scattering vector magnitude. It depends on scattering angle and radiation wavelength and is defined in Materials and Methods. The time scale of motions examined is ≤1/ΔE, where ΔE is the spectrometer energy resolution, i.e., the ability of the instrument to perceive changes in neutron energy that occur during the scattering process. QENS is measured as a function of both energy transfer and scattering vector. The technique gives access to the correlation function for the atomic motions that are explored for distances of a few angstroms and for times of the order of 10−12 to 10−9 s. On the energy scale, the elastic scattering appears as a peak centered on zero energy transfer whose width corresponds to the spectrometer resolution. The QENS appears as a significantly broader peak, also centered on zero energy transfer. The shape of the QENS peak and its Q variation contain information on the relaxation time and geometry associated with the motion of the scattering particle. In the same way as diffraction by a structure is related to the structure itself by Fourier transformation in crystallography, the energy transfer scattering pattern of a moving particle is related to the time dependence of its motion by Fourier transformation. A diffusing particle is described by an exponential decay function in time; the Fourier transform of such a function is called a Lorentzian, and it corresponds to the peak shape usually observed in QENS. See Materials and Methods for the equations used in data analysis. By applying various models to fit the Lorentzian width as a function of Q, it is possible to calculate rotational relaxation time, translational residence times, and translational and rotational diffusion coefficients for the scattering particle.
“Normal” Cell Water in H. marismortui: IN6 Results.
Incoherent neutron scattering spectra fitted each with an elastic peak and two Lorentzian functions (sharp and broad components) are shown in Fig. 1A.
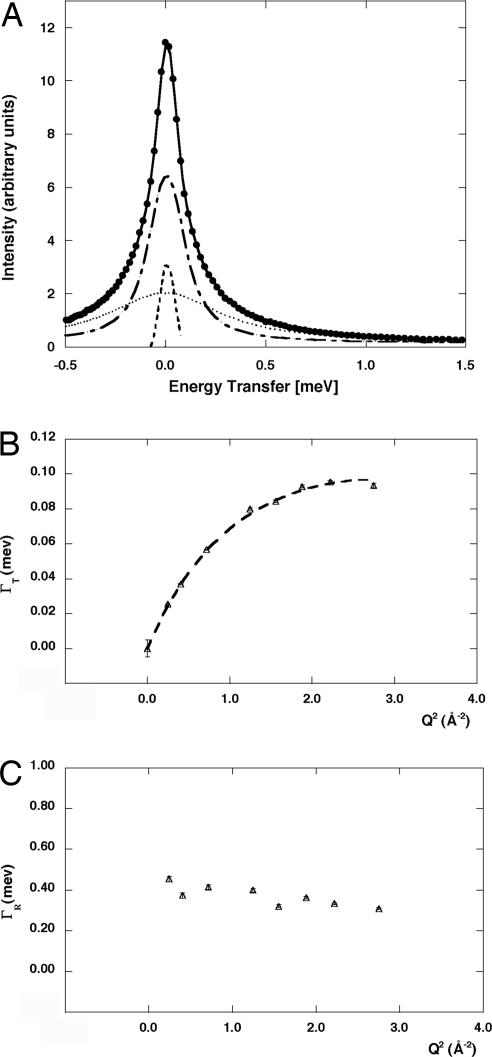
Fig. 1.
IN6 data. (A) Quasielastic spectrum at T = 285 K and Q = 1.65 Å−1. Filled circles indicate the data, and the bold solid line is the fitted curve using Eqs. 5 and 7. The fit of the quasielastic spectra was performed for −0.6 < ħω < 1.5 meV. The different components correspond to the elastic peak (dashed line), the broad Lorentzian (dotted line), and the sharp Lorentzian (dot–dash line). (B) Half-widths at half-maximum of the sharp Lorentzian Γ as a function of Q2 at T = 285 K. The dashed line results from the fit using Eq 1. This component was attributed to the translational motions of water. (C) Half-width at half-maximum of the broad Lorentzian Γ as a function of Q2 at T = 285 K. This component was attributed to the rotational motions of water.
The sharp component.
This component was attributed to translational motions of water. The dependence of the half-width at half-maximum of this component (ΓT) on the scattering vector squared (Q2) (see Materials and Methods) was best fitted by the diffusion model of Singwi and Sjölander (21) in which a molecule, executes oscillatory motions about one position for a mean residence time τ0, then diffuses by continuous motion for a mean time τ1 to another position in a repetitive way. In general, the shape of the quasielastic scattering for this model is not Lorentzian and the quasielastic broadening is represented by a long and complicated equation. However, the equation assumes a simple form in two limiting cases, in which the shape of the quasielastic peak is Lorentzian. In case i, when τ0 ≪ τ1, the equation reduces to the simple diffusion formula; in case ii, when τ0 ≫ τ1 at low momentum transfer, the broadening corresponds to simple diffusion as in case i. An asymptotic value approached at high momentum transfer gives the broadening expected for a jump mechanism (21). With the assumption τ0 ≫ τ1, the ΓT (Q2) dependence reduces to (21)
where DT represents the translational diffusion coefficient and 2W is the Debye–Waller factor describing the oscillations. By plotting ΓT against Q2, a curve is obtained from which the values of the three fit parameters, τ0, DT and 2W, 6.12 ps, 1.29 × 10−5 cm2 s−1 and 0.056 Q2, respectively, can be derived.
The following checks were performed to test the validity of the Singwi and Sjölander model (21, 22).
At the limit of small momentum transfer, Eq. 1 reduces to ΓT = DT Q2, the expression predicted by simple diffusion theory. The same value for DT is obtained from data at low Q2 as from the full fit.
At large Q2 values, the broadening tends toward the asymptotic value ΓT = (1/τ0). Again, the same τ0 was obtained from the data at high Q2 values as from the full fit.
The Debye–Waller factor, 2W, equals 1/6 Q2 〈R2〉, where 〈R2〉 is the mean square radius of the fully developed thermal cloud in the oscillatory motion, with a value of 〈R2〉/6 = 0.056 Å2. If the assumption that τ0 ≫ τ1 is justified, then we expect 2W < Q2DTτ0; this is indeed the case because 2W = 0.056 Q2 < Q2DT τ0 = 0.78 Q2. Brockhouse (23) observed that, in the case of normal water, the angular distribution of the quasielastic scattering is well governed by a factor of the type exp(−2W) and has also estimated the numerical value of 2W. It gives a value for of ≈0.4 Å, which is reasonably consistent with the value observed in this work ().
Moreover, if the assumption that τ0 ≫ τ1 holds, then we can also expect that the diffusion mechanism will not be primarily through jumps corresponding to the mean distance between water molecules; this is verified because the root mean square diffusion distance is given by , which is smaller than the mean distance between water molecules (nearly 3 Å) (21).
Calculated values for DT for water in 3.5 M NaCl at 285 K corresponds to 1.26 × 10−5 cm2/s (Table 1). This value is close to those obtained for H. marismortui, i.e., 1.29 ± 0.12 × 10−5 cm2 s−1. Therefore, we conclude that the sharp Lorentzian corresponds to “normal” water slightly slowed down with respect to bulk because of the presence of ions, chiefly NaCl.
The broad component.
This component was attributed to rotational motions of water. The half-width at half-maximum of the broad Lorentzian line, ΓR, is plotted as a function of Q2 in Fig. 1C. The curve shows a constant value of ΓR0 = 0.36 meV. The rotational relaxation of water has been described, using the Sears model (24), corresponding to hindered rotational diffusion of protons on the surface of a sphere of radius ρ. In the Q range of the measurements (Q ≈ 1 Å−1), the first order dominates the Sears approximation (24), and the dynamics structure factor is approximated as (11, 25)
where j0 is the zero order rotational Bessel function, δ(ω) is the Dirac delta function describing elastic scattering, and L(ΓR, ω) represents a Lorentzian of half-width at half-maximum ΓR = 2DR. DR is the rotational diffusion coefficient, which is approximately independent of Q and increases with temperature (26). The rotational correlation time τR = 1/(2DR) in H. marismortui is 1.83 ps, which is reasonably consistent with values for bulk water measured by neutron scattering and NMR (Table 1) (27). Protein hydration water rotational relaxation times have been measured to be higher by a retardation factor between 5 and 10 (28, 29). This finding suggests that it was the bulk water, and not the macromolecule hydration water, that was measured in the IN6 neutron scattering experiments on H. marismortui.
Slow Cell Water in H. marismortui: IN16 Results.
Fig. 2A shows data and fitted spectra for natural abundance (H cells) and deuterated (D cells) H. marismortui cells in H2O. The incoherent scattering cross-section of water in the sample accounts for 99% of the total for the D cells H2O sample and for 70% of the total in the H cells H2O sample (see Materials and Methods). The Lorentzian components of the two samples are identical, indicating that, in this time scale, the quasielastic signal is due to water and contains negligible contributions from cell components.
Fig. 2.
IN16 data. (A) Quasielastic spectra at T = 300 K and Q = 0.7 Å−1: the crosses indicate the experimental data and the solid lines are the fitted curves obtained by using equations 5 and 7, for D-cells in H2O (black) and H-cells in H2O (red) respectively. The fits of the quasielastic spectra were performed for −0.013 < ħω < +0.013 meV. The different components correspond to the resolution function (dashed line) and one Lorentzian (dotted line). (B) Half widths at half-maximum of the Lorentzian Γ as a function of Q2 at T = 285 K and T = 300 K. At 300 K, the bold solid line results from the fit using Eq. 4 from Q02 = 0.9 Å−2. (Inset) A zoom-in on the data obtained at 300 K.
Each spectrum was fitted reasonably well with an elastic peak and one Lorentzian function. Values of Γ (Lorentzian half-width at half-maximum) were plotted versus Q2 in Fig. 2B. At 285 K, the values of Γ are independent on Q2. This feature has been seen also in the translational line-widths Γ of water in other systems (11, 19). Γ is constant at ≈1.6 μeV, a value that corresponds to a residence time of 1/Γ = 411 ps. At 300 K, Γ shows a constant value Γ0 at low Q2, then increases from about Q2 = Q02 = 0.9 Å−2 to approach asymptotically a constant value Γ∞ at large Q2 (Fig. 2B Inset). The first feature indicates that the observed behavior is not free diffusion, but it is a typical characteristic of diffusion in a confined space (22). It can be approximately accounted for by the model of Volino and Dianoux, in which a particle diffuse inside a sphere of radius a (30)
where Dlocal is the diffusion coefficient inside a confinement sphere of radius a and a is calculated from Q0 = π/a. Estimating the Γ0 and Q0 values from Fig. 2B, we obtain the diffusion coefficient Dlocal = 6.15 × 10−7 cm2 s−1 and a = 3.31 Å.
Beyond Q = 1 Å−1 the line width follows the well-known jump diffusion behavior, given by (22)
where τ0 represents the residence time before the jump and DT is the translational diffusion constant. In the high Q2 region, one observes motion on short length scales. On such scales, local jump motion of the protons becomes dominant. The residence time of a hydrogen atom on a given site between jumps is τ0 = 1/Γ∞ where Γ∞ is obtained from the asymptotic behavior at high Q2; in this region Γ approaches a constant value. At the top of the Q-range examined, the Γ value at high Q2 was still increasing, and had not yet reached a constant value, which could be estimated by curve fitting using Eq. 4 and extrapolating to higher Q2 (Fig. 2). The translational diffusion constant was also extracted using the curve fitted by use of Eq. 4 (Fig. 2). A similar result has been seen in the Γs of hydration water in protein (31); it is also consistent with the restricted jump diffusion model of Hall and Ross (32). Clearly, the Γ of the restricted jump diffusion model exhibits the characters both of diffusion within a restricted volume (here a sphere) model and the jump diffusion model tending to asymptotic values at low and high Q2 (22). At low Q2, we are mainly concerned with a longer length scale, i.e., with the effects of the boundaries, which force the Γ to deviate from the DQ2 law and approach a finite value, Γ0. Conversely, at large Q2 the nature of the local jump motions over shorter length scale predominate and, because the elementary displacements of the particles are not infinitely small, the Γ of the quasielastic component tends to the asymptotic value 1/τ0.
At 300 K, DT is 9.34 × 10−8 cm2 s−1 and τ0 is 243 ps. These values indicate severely reduced water mobility with respect to bulk cell water.
Proportion of “Slow” to “Normal” Cell Water in H. marismortui.
The scattering of vanadium is purely elastic (and therefore independent of the energy resolution of the measuring spectrometer) and is often used to calibrate neutron scattering data. To connect the scattering functions of a sample in the different time windows, we divided the IN6 and IN16 data by the scattering of the same vanadium sample on each instrument. The scattering of the vanadium was then considered as a reference to which the IN6 and IN16 data were renormalized and put on the same scale. This made it possible to make a quantitative comparison of the amounts of water detected by the two instruments. At the same lowest value of Q accessible on both instruments, the proportion of “slow” to total water was then calculated from the ratio of the IN16 to IN6 elastic intensities. The slow water was thus found to represent 55% of the total cell pellet water. Because our former experiments had shown that the bacterial cells contained some normal water, we infer that in the quasielastic scattering of the bacterial pellets examined on IN6, both extracellular 3.5 M NaCl and the faster intracellular water is measured. A correction has therefore to be made for the extracellular water content of the bacterial pellets. This is estimated to be 28% of total pellet water under the conditions of centrifugation used (1). The proportions of slow and normal intracellular water detected by neutron scattering in H. marismortui could then be calculated and were found to be 76% and 24%, respectively, of cell water. The slow water content found by this method is somewhat higher than that detected by former methods.
There Is No Slow Water Component in E. coli.
BSS normalized quasielastic spectra (see Materials and Methods) of deuterated E. coli cells measured in H2O and in D2O are shown in Fig. 3. A flat background was applied, which could be seen as an extremely broad Lorentzian due to normal cell water. Despite the low statistical accuracy of the results, it is clear that neither the Q value nor the isotopic nature of the water had any effect on the width of the signal. These observations demonstrate the absence of a slow water component in E. coli.
Fig. 3.
Normalized quasielastic spectrum of D cells in H2O (gray) and D2O (black) at Q = 0.31 A−1 (A) and Q = 1.86 A−1 (B). The quasielastic experiments were performed at 285 K on the BSS spectrometer (FRJ-2, Jülich) using an incident wavelength of 6.27 Å. The elastic energy resolution (FWHM) is 1.2 μeV.
Discussion and Conclusion
Our results on H. marismortui fall into two distinct groups according to the measuring instrument used. On the ≈10-ps time scale (IN6 data, Fig. 1), at 285 K DT and τ0 are 1.29 × 10−5 cm2 s−1 and 6.12 ps, respectively. The DT value is slightly lower than that of pure water at 285 K, close to the 278 K value. (Table 1). The τ0 value corresponds to that for pure water at 263 K (Table 1). This is consistent with the fact that the water is slightly slowed down with respect to bulk because of the presence of high concentration of ions. The DT value is also close to that for 3.5M NaCl and lower than that of 3M KCl solutions (Table 1). This is consistent with the fact that, in the time window of the spectrometer, the data are sensitive to the principal extracellular component (3.5 M NaCl) and to intracellular water with DT values similar to the ones found in other cells studied. In the few cells in which the diffusion of cell water has been measured by neutron scattering, its value has been found to be reduced by at most one order of magnitude from that of bulk water, with DT in the range from 0.1 to 0.42 of the bulk water value (33–35).
The IN16 and BSS spectrometers (time scale ≈1 ns) detected a large population of protons with very low mobility in H. marismortui (Fig. 2), but not in E. coli (Fig. 3). Control experiments using samples with different deuterium labeling established that the signal was strongly dominated by H2O protons. The IN16 and BSS spectrometers cannot detect bulk water, nor indeed water retarded by dissolved salts or polymers even at high concentrations, because the rates of diffusion of these substances are outside the range of the instrument. Thus, the water detected by IN16 must be intracellular, providing strong evidence for the presence of water with a very low rate of mobility within H. marismortui cells. At 300 K, this water had a DT of 9.34 × 10−8 cm2 s−1 or ≈250 times lower than that of bulk water at the same temperature (Table 1). The residence time, τ0, of ≈240 ps is ≈220 times higher than for bulk water (Table 1). Results showed a strong qualitative effect of temperature (Fig. 2B), with H2O protons becoming unable to diffuse over the “jump distance” when the temperature fell by only 15 K (to 285 K).
A possible reason for a lowering of cell water mobility is that this water functions as water of hydration near protein surfaces. The dynamics of hydration water near proteins has been studied by neutron scattering and by NMR. Long-range diffusion coefficients of water close to protein surfaces were found to be reduced 5–6 times as compared with the bulk solvent (36). Measurements of water mobility at the surface of a protein, C-phycocyanin (19) yielded a DT of 1.2. 10−5 cm2 s−1 at 298 K, in comparison with DT = 2.3 × 10−5 cm2 s−1 in bulk water at the same temperature, i.e., there is a relatively small difference between the two. This observation is consistent with NMR measurements of water mobility at the surface of bovine pancreatic trypsin inibitor, in which the translational retardation factor at 277 K is two (28). Protein solvation has also been studied by hydrodynamics and solution-scattering methods. Of these studies, the most relevant to the present one are those dealing with solvent interactions in purified halophilic proteins. A summary of the 10 halophilic enzymes studied up to 2000 is given in Madern et al. (37). Each enzyme molecule is surrounded by a solvation shell composed of 0.2–0.4 g of water and 0.1–0.2 g of NaCl or KCl per gram of protein (38). This is equivalent to 5 M NaCl in NaCl-containing media. In contrast, the solvation shell of homologous mesophilic proteins contains bound water but no ions, and it was concluded that salt binding appears to be a characteristic specific of molecular adaptation to halophilic conditions (38), although it is relatively weak and unspecific. In halophilic proteins, ion binding appears to exert a positive function in that it contributes to preserve the stability and solubility of folded proteins immersed in highly concentrated salt solutions (39). Significant differences in the dynamics of the NaCl and KCl forms of H. marismortui malate dehydrogenase have also been found by neutron scattering, with the form binding K+ displaying higher flexibility and lower resilience at physiological temperatures (40, 41). Halophilic proteins have also been studied by crystallography (42–47). Water and ions in close association with the enzyme molecule have been found in these studies, although the amounts of water and ions detected were far less than in the solution studies, because most of the water molecules in the solvation shell are in a disordered state and therefore invisible in high-resolution electron density maps.
There are two remarkable similarities between the solvent interactions that have been found in purified halophilic proteins and those known to occur in whole cells of H. marismortui: both purified halophilic proteins and whole cells of H. marismortui bind water and salt, and both require high salt concentrations in the bathing medium for viability (halophilic proteins unfold below 2 M KCl or NaCl). However, there are also important differences. Halophilic enzyme proteins are active in a wide variety of salt solutions, whereas H. marismortui cells have an absolute requirement for NaCl and MgSO4 in the bathing medium while binding K+ within the cells; isolated proteins do not appear to be selective for Na+ or K+, whereas intact cells show very high specificity toward these ions. To conclude, the work done so far on isolated proteins explains neither the high selectivity between Na+ and K+ found in these cells (1), nor the low water mobility found in the present study. This latter finding confirms the indications for the existence of structured water that were deduced from the NMR, dielectric, and calorimetric measurements described in the Introduction.
Recently, in a long series of papers, MacKinnon and coworkers have described a system demonstrating high specificity between K+ and Na+. This occurs in K+ channels, molecular structures responsible for K+-transport within cell membranes. The work is summarized in MacKinnon's Nobel lecture (48). The arrangement of the water–oxygen atoms in the central cavity of the K+ channel may serve as model to explain how water in the vicinity of protein can bind K+ specifically with a very high affinity in H. marismortui.
In conclusion, our data demonstrate that the water of H. marismortui cells is of exceptionally low mobility, as compared with other cells studied up to now. This low mobility cannot be explained by the hydration interactions of halophilic proteins, as far as they are known at present. Structured water around K+ ions has been seen by MacKinnon in the central cavity of K+ channels (48). A similar mechanism might explain our own results.
Materials and Methods
Cell Material.
Natural abundance and deuterated H. marismortui cells were grown at 37°C to an optical density of 0.8–1 (late logarithmic phase) in medium described (40), in which, for the deuterated material, yeast extract was replaced by deuterated algal extract produced at the Max Planck Institute (Martinsreid, Germany). Deuterated E. coli [BLE21(DE3)] were cultivated at 37°C to an optical density value of 1.5, in deuterated Enfors minimum growth medium with deuterated glycerol (d8) as the carbon source.
Preparation of Samples for Neutron Scattering.
Cells were pelleted by centrifugation at 9,000 rpm in a Beckman centrifuge (JA14 rotor) for 30 min at room temperature. The supernatant was discarded, and the cells were washed twice with 200 ml of H2O–saline (200 g/liter NaCl, 46.6 g/liter MgSO4, 0.5 g/liter CaCl2, 2 g/liter KCl, and 10 mM Tris·HCl at pH 7.6 for the halophiles and 8.766 g/liter NaCl, 0.372 g/liter KCl, and 10 mM Tris·HCl at pH 7.6 for E. coli) followed by 15-min centrifugation. Washed pellets of H. marismortui, which were naturally bright red whether the cells were natural abundance or deuterated, were transferred to aluminum sample holders (4 × 3 × 0.03 cm3). Excess liquid was removed with a capillary or by careful wiping with a tissue and the holder immediately sealed. After the experiments, a portion of the pellets was resuspended in medium. They were found to grow to an OD of 1 within a few days, indicating that most cells were still viable after a total beam time of 20 h. Another portion was completely dried and was found to have contained 1.4 g of water per gram of dry cell material for all samples.
There was very good agreement for the ratio of water to dry cell material (i.e., 1.4 g of water per g of dry cell material), whether measured by NMR, calorimetry, or neutron scattering. Samples of natural abundance and deuterated cells suspended in H2O saline were named H cells H2O and D cells H2O, respectively.
Neutron Scattering Measurements.
Experiments on H. marismortui were performed on the IN6 time of flight and the IN16 backscattering spectrometer at the Institut Laue Langevin (ILL), Grenoble, France. Experiments on E. coli were performed on the BSS spectrometer, also a backscattering spectrometer, at the Jülich neutron scattering centre, with a time scale of 1 ns (www.neutronscattering.de). Incident wavelengths for the three instruments are 5.12, 6.27, and 6.27 Å, respectively. The elastic energy resolutions (FWHM) are 100 μeV for IN6 and 0.9 μeV for IN16 and BSS, corresponding to time scales of 10 ps and 1 ns, respectively. The scattering was measured over a wave-vector range of 0.3 < Q < 2 Å−1 for IN6 and 0.02 < Q < 1.9 Å−1 for IN16 and BSS (Q = 4πsinθ/λ, where 2θ is the scattering angle and λ is the neutron wavelength). A vanadium sample (a purely elastic scatterer) was measured to define the instrument resolution and to correct for detector efficiency. The measured spectra were corrected, normalized, grouped, and transformed into energy transfer spectra by using the standard ILL data reduction programs INX and SQW. Correction for multiple scattering was estimated to be negligible because the transmission of all samples was >90%.
Incoherent Neutron Scattering.
An exhaustive description of QENS can be found in Bée (22). Application of this method to protein dynamics has been reviewed by Smith (49) and Gabel et al. (9). Hydrogen nuclei dominate incoherent neutron scattering because their cross section is more than an order of magnitude larger than that of other nuclei in biological samples. The approximate incoherent scattering cross sections of H2O, H cell, and D cell material were calculated to be 5.3 cm2/g, 3.2 cm2/g, and 0.08 cm2/g, respectively. If all of the sample material scatters in the same time window, we expect 99% of the signal to be from water in D cell H2O samples and 70% of the signal to be from water in the H cell H2O samples. In incoherent neutron scattering experiments, dynamic information is contained in the scattering function S(Q, ω).
Data Analysis.
In the quasielastic incoherent approximation, the theoretical scattering function has been described by Bée (22)
 |
A0(Q) is the elastic incoherent structure factor (EISF).
The quasielastic component Σi=1n Ai(Q)L(Γi, ω) is a sum of Lorentzian functions
where Γi is the half-width at half-maximum of a Lorentzian peak. Incoherent quasielastic neutron scattering, in the experimental conditions used here, mainly measures motions on the picosecond time scale.
The function in Eq. 5 was fitted to the data with the standard ILL fitting program Profit by using the following relation
 |
In Eq. 7 a convolution with the spectrometer resolution function Sres(Q, ω) and a detailed balance factor e−ħω/2kBT are applied. B0 is the inelastic background due to the vibrational modes of lowest energy (the “lattice phonons”; ref. 22). The fits were performed over the energy transfer range −0.6 to +1.5 meV using two Lorentzian functions for the IN6 spectra. For the IN16 spectra, the fits were performed with one Lorentzian function over the energy transfer range −0.013 to +0.013 meV.
The Γi values derived from the analysis were subsequently interpreted in the context of models described in Results.
Acknowledgments
H. marismortui culture was obtained from Prof. Aharon Oren (Institute of Life Sciences, The Hebrew University of Jerusalem, Givat Ram, Jerusalem, Israel). We thank Martine Moulin and Michael Haertlein from the DLAB and the Dynamics Club at Institut Laue Langevin, Grenoble, France. M.T. was supported by a doctorate grant from the Région Rhône-Alpes, France, and by the Istituto Nazionale Fisica della Materia, Italy. The work was supported by the Centre National de la Recherche Scientifique GEOMEX program. We acknowledge support from the European Union under the DLAB contracts HPRI-CT-2001-50035 and RII3-CT-2003-505925, and The Integrated Infrastructure Initiative for Neuron Scattering and Muon Spectroscopy (NMI3).
Abbreviation
- QENS
quasielastic neutron scattering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Ginzburg M, Sachs L, Ginzburg BZ. J Gen Physiol. 1970;55:187–207. doi: 10.1085/jgp.55.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginzburg BZ, Ginzburg M. In: Biophysics of Water. Franks F, Mathias SF, editors. London: Interscience; 1982. pp. 340–343. [Google Scholar]
- 3.Edzes HT. Groningen, The Netherlands: Univ of Groningen; 1976. PhD thesis. [Google Scholar]
- 4.Ginzburg BZ. Thermochim Acta. 1981;46:249–258. [Google Scholar]
- 5.Morgan H, Ginzburg M, Ginzburg BZ. Biochim Biophys Acta. 1987;924:54–66. [Google Scholar]
- 6.Bone S, Ginzburg BZ, Morgan H, Wilson G, Zaba B. Phys Med Biol. 1996;41:45–54. doi: 10.1088/0031-9155/41/1/004. [DOI] [PubMed] [Google Scholar]
- 7.Ginzburg M, Ginzburg BZ. In: Biomembranes. Eisenberg H, Katchalsi-Katzir E, Manson LA, editors. Vol 7. New York: Plenum; 1975. pp. 219–251. [Google Scholar]
- 8.Zaccai G. Science. 2000;288:1604–1607. doi: 10.1126/science.288.5471.1604. [DOI] [PubMed] [Google Scholar]
- 9.Gabel F, Bicout D, Lehnert U, Tehei M, Weik M, Zaccai G. Q Rev Biophys. 2002;35:327–367. doi: 10.1017/s0033583502003840. [DOI] [PubMed] [Google Scholar]
- 10.Zanotti JM, Bellissent-Funel MC, Chen SH. Phys Rev E. 1999;59:3084–3093. [Google Scholar]
- 11.Bellissent-Funel M, Chen SH, Zanotti J. Phys Rev E. 1995;51:4558–4569. doi: 10.1103/physreve.51.4558. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira J, Bellissent-Funel M, Chen SH, Dianoux AJ. Phys Rev A. 1985;31:1913–1917. doi: 10.1103/physreva.31.1913. [DOI] [PubMed] [Google Scholar]
- 13.Paradossi G, Cavalieri F, Chiessi E, Telling MTF. J Phys Chem B. 2003;107:8363. [Google Scholar]
- 14.Bellissent-Funel M-C, Kahn R, Dianoux AJ, Fontana MP, Maisano G, Migliardo P, Wanderlingh F. Mol Phys. 1984;52:1479–1486. [Google Scholar]
- 15.Zaccai G. Philos Trans R Soc London B. 2004;359:1269–1275. doi: 10.1098/rstb.2004.1503. and discussion 1275, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weik M, Lehnert U, Zaccai G. Biophys J. 2005;89:3639–3646. doi: 10.1529/biophysj.104.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo D, Hura G, Head-Gordon T. Biophys J. 2004;86:1852–1862. doi: 10.1016/S0006-3495(04)74252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellissent-Funel MC, Teixeira J, Bradley K, Chen S. J Phys I. 1992;2:995–1001. [Google Scholar]
- 19.Bellissent-Funel MC, Zanotti JM, Chen SH. Faraday Dis. 1996;103:281–294. [Google Scholar]
- 20.Svergun DI, Richard S, Koch MH, Sayers Z, Kuprin S, Zaccai G. Proc Natl Acad Sci USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singwi KS, Sjolander A. Phys Rev. 1960;119:863–871. [Google Scholar]
- 22.Bée M. Quasielastic Neutron Scattering: Principles and Applications in Solid State Chemistry, Biology and Materials Science. Philadelphia: Adam Hilger; 1988. [Google Scholar]
- 23.Brockhouse BN. Suppl Nuovo Cimento. 1958;9:45. [Google Scholar]
- 24.Sears VF. Can J Phys. 1966;44:1299–1311. [Google Scholar]
- 25.Zanotti JM. Paris: Université de Paris XI; 1997. PhD thesis. [Google Scholar]
- 26.Volino F. In: Microscopic Structure and Dynamics of Liquids. Dupuy J, Dianoux AJ, editors. New York: Plenum; 1978. pp. 221–300. [Google Scholar]
- 27.Sposito G. J Chem Phys. 1981;74:6943–6949. [Google Scholar]
- 28.Modig K, Liepinsh E, Otting G, Halle B. J Am Chem Soc. 2004;126:102–114. doi: 10.1021/ja038325d. [DOI] [PubMed] [Google Scholar]
- 29.Halle B. Philos Trans R Soc London B. 2004;359:1207–1223. doi: 10.1098/rstb.2004.1499. and discussion 1223–1224, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volino F, Dianoux AJ. Mol Phys. 1980;41:271–279. [Google Scholar]
- 31.Bellissent-Funel MC, Texeira J, Bradley KF, Chen SH, Crespi HL. Physica B. 1992;180–181:740–744. [Google Scholar]
- 32.Hall PL, Ross DK. Mol Phys. 1981;42:673–682. [Google Scholar]
- 33.Trantham EC, Rorschach HE, Clegg JS, Hazlewood CF, Nicklow RM, Wakabayashi N. Biophys J. 1984;45:927–938. doi: 10.1016/S0006-3495(84)84239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rorschach HE, Bearden DW, Hazlewood CF, Heidorn DB, Nicklow RM. Scanning Microsc. 1987;1:2043–2049. [Google Scholar]
- 35.Neville MC, Paterson CA, Rae JL, Woessner DE. Science. 1974;184:1072–1074. doi: 10.1126/science.184.4141.1072. [DOI] [PubMed] [Google Scholar]
- 36.Bon C, Dianoux AJ, Ferrand M, Lehmann MS. Biophys J. 2002;83:1578–1588. doi: 10.1016/S0006-3495(02)73927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madern D, Ebel C, Zaccai G. Extremophiles. 2000;4:91–98. doi: 10.1007/s007920050142. [DOI] [PubMed] [Google Scholar]
- 38.Ebel C, Costenaro L, Pascu M, Faou P, Kernel B, Proust-De Martin F, Zaccai G. Biochemistry. 2002;41:13234–13244. doi: 10.1021/bi0258290. [DOI] [PubMed] [Google Scholar]
- 39.Ebel C, Zaccai G. J Mol Recognit. 2004;17:382–389. doi: 10.1002/jmr.697. [DOI] [PubMed] [Google Scholar]
- 40.Tehei M, Madern D, Pfister C, Zaccai G. Proc Natl Acad Sci USA. 2001;98:14356–61. doi: 10.1073/pnas.251537298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tehei M, Zaccai G. Biochim Biophys Acta. 2005;1724:404–410. doi: 10.1016/j.bbagen.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Besir H, Zeth K, Bracher A, Heider U, Ishibashi M, Tokunaga M, Oesterhelt D. FEBS Lett. 2005;579:6595–6600. doi: 10.1016/j.febslet.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 43.Zeth K, Offermann S, Essen LO, Oesterhelt D. Proc Natl Acad Sci USA. 2004;101:13780–5. doi: 10.1073/pnas.0401821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frolow F, Harel M, Sussman JL, Mevarech M, Shoham M. Nat Struct Biol. 1996;3:452–458. doi: 10.1038/nsb0596-452. [DOI] [PubMed] [Google Scholar]
- 45.Richard SB, Madern D, Garcin E, Zaccai G. Biochemistry. 2000;39:992–1000. doi: 10.1021/bi991001a. [DOI] [PubMed] [Google Scholar]
- 46.Irimia A, Ebel C, Madern D, Richard SB, Cosenza LW, Zaccai G, Vellieux FM. J Mol Biol. 2003;326:859–873. doi: 10.1016/s0022-2836(02)01450-x. [DOI] [PubMed] [Google Scholar]
- 47.Bieger B, Essen LO, Oesterhelt D. Structure (London) 2003;11:375–385. doi: 10.1016/s0969-2126(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 48.MacKinnon R. Nobel Lecture. Stockholm: Stockholm University; 2003. [Google Scholar]
- 49.Smith JC. Q Rev Biophys. 1991;24:227–291. doi: 10.1017/s0033583500003723. [DOI] [PubMed] [Google Scholar]
- 50.Teixeira J, Luzar A. In: Hydration Processes in Biology: Theoretical and Experimental Approaches. Bellissent-Funel MC, editor. Vol 305. Amsterdam: IOS; 1999. pp. 35–65. [Google Scholar]
- 51.Chen SH, Texeira J, Nicklow R. Phys Rev A. 1982;26:3477–3482. [Google Scholar]
- 52.Wang JH. J Phys Chem. 1965;69:442. [Google Scholar]
- 53.Wang JH. J Phys Chem. 1954;58:686–692. [Google Scholar]