Abstract
The 466-aa tail of the heavy chain of Acanthamoeba myosin IC (AMIC) comprises an N-terminal 220-residue basic region (BR) followed by a 56-residue Gly/Pro/Ala-rich region (GPA1), a 55-residue Src homology 3 (SH3) domain, and a C-terminal 135-residue Gly/Pro/Ala-rich region (GPA2). Cryo-electron microscopy of AMIC had shown previously that the AMIC tail is folded back on itself, suggesting the possibility of interactions between its N- and C-terminal regions. We now show specific differences between the NMR spectrum of bacterially expressed full-length tail and the sum of the spectra of individually expressed BR and GPA1-SH3-GPA2 (GSG) regions. These results are indicative of interactions between the two subdomains in the full-length tail. From the NMR data, we could assign many of the residues in BR and GSG that are involved in these interactions. By combining homology modeling with the NMR data, we identify a putative pleckstrin homology (PH) domain within BR, and show that the PH domain interacts with the SH3 domain.
Keywords: NMR, homology modeling
The myosin superfamily consists of 24 different classes distinguished by the sequences of their catalytic (head) domains, but also differing in the structure of their tails (1, 2). A total of 35 members of the myosin-I family have been identified in 19 species at the protein and/or DNA levels, and many organisms express multiple myosin-Is with structurally and functionally similar catalytic domains. The tails are thought to target myosin-I isoforms to different intracellular locations and to adapt them to different functions.
Acanthamoeba myosin IC (AMIC), one of three Acanthamoeba myosin-Is (3), consists of a single heavy chain (3) and one light chain (4). The heavy chain has a head domain (which contains the ATPase site and an ATP-sensitive actin-binding site), a neck domain (to which the light chain binds), and a 466-residue tail domain (5). The tail domain (Fig. 1) has four subdomains (5): a 220-residue basic region (BR), which binds acidic phospholipids (6) and probably membranes (7, 8); two Gly/Pro/Ala-rich regions (56-residue GPA1 and 135-residue GPA2), which bind F-actin in an ATP-independent manner (9, 10); and a 55-residue Src homology 3 (SH3) domain between GPA1 and GPA2. The SH3 domain binds CARMIL/Acan125 (11, 12), a protein that also binds capping protein and the Arp2/3 complex (13).
Fig. 1.
AMIC heavy chain tail. The tail domains are shown schematically with their sequences below. GPA1 and GPA2, glycine/proline/alanine-rich regions; SH3, Src homology 3 domain.
Recently, the spatial arrangement of the subdomains in the tail was determined by cryo-electron microscopy of actin filaments decorated with wild-type AMIC or tail-truncated mutants of various lengths (14). Interpretations of the reconstructed images suggest that the tail is folded back on itself with possible interactions between the BR and GPA1–SH3–GPA2 (GSG) subdomains. A better understanding of the nature of the interactions between these subdomains might provide useful insight into the roles of the various tail subdomains in linking AMIC to potential interaction partners such as membranes, organelles, and actin filaments (10, 15, 16).
X-ray crystallography and NMR spectroscopy are the principal mechanisms for determining the structures of biological macromolecules. Attempts to crystallize the AMIC heavy chain tail failed, not surprisingly given the number of glycines (27%) and prolines (38%) in GPA1 and GPA2, and thus, despite the size of the protein, NMR was the remaining option. We studied three bacterially expressed proteins: the full-length tail, and its N- and C-terminal halves, BR and GSG, respectively. Because the size and segmental flexibility of the AMIC tail limited our ability to complete an NMR structure determination, we combined the NMR results with structural homology modeling.
Results
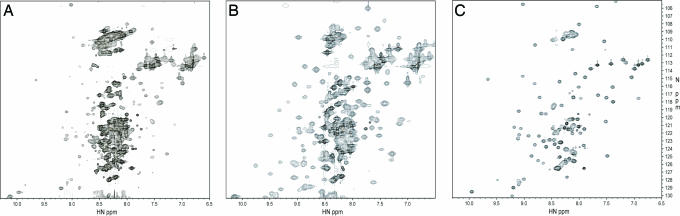
An 15N HSQC spectrum of the full-length tail of AMIC, residues 721-1186 (Fig. 1) with C-terminal FLAG, showed a moderate number of well dispersed amide resonances, indicative of a folded protein, as well as a large number of poorly dispersed resonances in the 7.9–8.6 1H chemical shift range, typical of flexible random coil (Fig. 2A). A TROSY experiment resulted in narrower line widths for the well dispersed resonances but did not improve the resolution in the flexible random coil regions of the spectrum (Fig. 3A, black). Many of the well dispersed resonances were quite weak, and attempts to increase the protein concentration beyond 200 μM failed due to precipitation. To simplify resonance assignments and detect any interdomain interactions in the full-length tail, the tail was divided into two smaller constructs, BR and GSG (Fig. 1).
Fig. 2.
800 MHz 2D 15N HSQC spectra of the full-length tail of AMIC (A), the BR (B), and the GSG region (C). The BR spectrum is of the sample expressed in 65% D2O. The full-length tail and GSG constructs were expressed in 100% H2O.
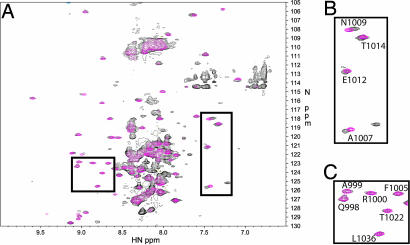
Fig. 3.
Superposition of the 800 MHz 2D TROSY spectra of the amplitude-adjusted GSG region and full-length tail of the heavy chain of AMIC. (A) The GSG region is in red and the full-length tail in black. (B) Expansion of the region of A showing residues where 1H shift changes occurred between the full-length tail and the GSG region. (C) Expansion of a region where there were no differences between the full-length tail and the GSG region.
BR.
NMR.
In the CBCA(CO)NH and HNCACB experiments, no Cα and Cβ signals could be detected for 40% and 50% of the BR construct residues, respectively. Residues lacking Cα and Cβ signals corresponded to weaker, well dispersed amide peaks in the 15N HSQC spectrum. The BR construct with a C-terminal FLAG-tag precipitated at concentrations above 200 μM. Moving the FLAG-tag from the C terminus to the N terminus increased solubility to at least 680 μM. However, the 15N HSQC spectra at the higher concentration showed evidence of aggregation, the signal-to-noise ratio was not significantly better, and many peaks were broader. Neither construct could tolerate temperatures above 25°C. Therefore, a deuterated (65%) version of the N-FLAG-BR construct was expressed to reduce 1H dipolar interactions (Fig. 2B). With deuteration, Cα and Cβ signals were seen for an additional 40 residues (16% of the total). Ultimately, 142 of 238 residues were assigned, leaving 57 observed backbone amide peaks unassigned and 39 undetected.
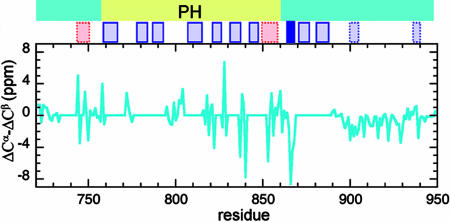
Fig. 4 shows the ΔCα–ΔCβ chemical shift index values (17) for the assigned BR residues. The smaller ΔCα–ΔCβ values from Q891 to G949 are indicative of flexible random coil. However, this flexible C-terminal region might have some structure because five residues have split amide signals, indicating slow exchange between two conformations. Although two of these residues (A905, G907) flank a proline, which can undergo slow cis-trans isomerization, the other three split amides (K898, T900, and T929) are not close to prolines in the sequence.
Fig. 4.
ΔCα–ΔCβ chemical shift index for the basic region construct. Large negative values (less than −4 ppm) are typical of β-strands, extended stretches of large positive values (>4 ppm) are indicative of α-helices, and flexible random coil regions have values closer to zero. Unassigned regions are shown with zero values. Above the graph are β-strands (blue) and helices (pink) from a consensus secondary structure prediction. β-strands and helices contradicted by NMR are shown with dashed rectangles. The strand with the dark blue rectangle is confirmed by NMR. The remaining β-strands lie in unassigned regions. A PH domain predicted by the Phyre threading program is shown at the top.
Most of the BR construct, from F733 to P890, appears to be folded, with larger ΔCα–ΔCβ values on average and well dispersed amide resonances (Fig. 4). The assigned residues in this region appear to be mostly in loops, with sequential amide–amide NOESY cross-peaks that are neither consistently weak nor strong. A consensus secondary structure prediction (see Materials and Methods) for the BR construct is shown in Fig. 4. Twelve β-strands are predicted. One predicted β-strand (the solid blue rectangle in Fig. 4) is supported by the ΔCα–ΔCβ values (R865–F868); two predicted β-strands in the flexible C-terminal region are contradicted by the NMR data. The other nine predicted β-strands fall in segments where no, or only partial, assignments could be made; seven of these are in a putative PH domain (see below). Neither of the two predicted α-helices is supported by either ΔCα–ΔCβ values or NOESY.
Modeling.
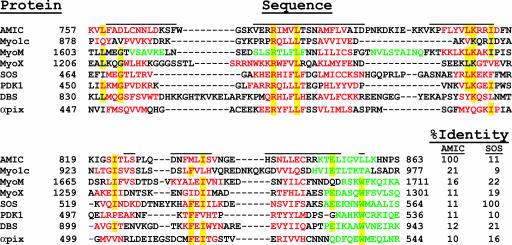
A BLAST comparison of the BR sequence with sequences of known structures produced no significant hits, but the Phyre threading program (18) identified residues V758–S863 as a putative PH domain (Fig. 4). The best matches to PH domains of known structure were SOS (Son of Sevenless) [Protein Data Bank (PDB) ID code 1DBH; ref. 19], PDK1 (phosphoinositide-dependent protein kinase 1) (PDB ID code 1RJ2; ref. 20), Dbs (Dbl's Big Sister) (PDB ID code 1W1D; ref. 21), and murine α pix GEF (PDB ID code 1V61). Fig. 5 shows the alignment of the putative PH domain of BR with the PH domains of these proteins, and to three myosins in which putative PH domains have been identified by sequence homology.
Fig. 5.
Sequence alignments of the putative PH domain of AMIC with PH domains of three other myosins and four nonmyosin proteins. β-strands are in red, helices in green, unassigned regions are indicated by lines above the sequence, and conserved residues are highlighted in yellow. The myosin PH domain secondary structures are predicted, and the nonmyosin secondary structures are experimentally determined.
The canonical PH domain includes seven β-strands followed by an α-helix (22, 23). Eight NMR-assigned residues of BR lie within predicted PH domain β-strands (Fig. 5). Four of these residues (822–824 and 837 in the fifth and sixth strands) are in β-strand conformation by NMR, which agrees with the conformations of the corresponding residues in the PH domains of known structure in Fig. 5 (SOS, PDK1, DBS, and αpix). Although the predicted β conformations of the other four residues (757–759 and 816 at the beginning and end of the first and fourth β-strands, respectively) are contradicted by the NMR data, these residues are also predominantly in loop, not β-strand, conformation in the PH domains of known structure.
Fig. 6A shows a homology model structure of the BR PH domain superposed with the PH domain of SOS. Like the SOS PH domain, the aligned BR sequence has its longest loop between the third and fourth β-strands. Also, like the SOS PH domain (24), this region appears to be the most mobile of the loops in the putative PH domain of BR, with narrower line widths and chemical shifts closer to flexible random coil values. The model structure shows the electrical polarization typical of PH domains (22, 23), with a large positive potential opposite the C-terminal side (Fig. 6B).
Fig. 6.
Homology modeling of the putative PH domain of the BR of AMIC. (A) The putative PH domain of AMIC (blue) is superposed on the NMR structure of the PH domain of SOS (pink). The seven β-strands are numbered. The PH domain of AMIC BR lacks the C-terminal helix and contains a six residue insert between strands one and two (bottom) not seen in the SOS PH domain. Both PH domains have a mobile loop between strands three and four (left). (B) Electrostatic potential energy surface for the putative PH domain of AMIC BR, shown in the same orientation as in A. The polarization between the mildly negative C-terminal area of the domain (right) and the very positive opposite side and adjoining mobile loop region (left) is typical of PH domains.
There is no clear NMR evidence for helix in the C-terminal region of the putative PH domain in BR, although the first four residues of the “missing” helix (K850–L853) are unassigned. Known PH domains have a very highly conserved tryptophan residue in this helix, but there is no tryptophan residue at or near this position (L857) in AMIC, or in the BRs of any myosin-I (pfam: PF06017). In addition, there is a proline (P862) in this region, and a proline cannot occur within an α-helix without significant distortion; the NMR results confirm that the three residues preceding the proline are not α-helical. On the other hand, the Cα chemical shift of G855 in the “missing” helix is shifted downfield by almost 2 ppm, as expected for an α-helix but uncommon for glycines, which are seldom found in helices. This observation suggests that this region of the “missing” helix in the putative BR PH domain might have some helical character, or be in equilibrium between helical and nonhelical conformations, possibly destabilized by the presence of glycine or absence of the conserved tryptophan.
GSG Region.
NMR.
The GSG construct, with C-terminal FLAG, showed no evidence of aggregation or temperature instability. Cα and Cβ resonances were observable for all of the backbone amides, so deuteration was unnecessary. All of the well dispersed peaks in the 15N HSQC spectrum (Fig. 2C) could be assigned to the SH3 domain, E997–I1051, predicted by a BLAST search (Fig. 7), with only two SH3 residues, K1028 and N1037, unaccounted for. The five predicted β-strands of the SH3 domain (Fig. 7) are confirmed by five corresponding negative stretches of ΔCα–ΔCβ values (Fig. 7). One additional potential strand, E1012–N1016, implied by the ΔCα–ΔCβ values does not appear to be a β-strand in the 15N-edited NOESY spectrum, and corresponds to an extended loop region in the SH3 structure.
Fig. 7.
ΔCα–ΔCβ chemical shift index for the GSG construct. For significance of values, see legend to Fig. 3. Predicted β-strands confirmed by NMR (dark blue rectangles) are shown above the graph. The position of the SH3 domain predicted by BLAST and confirmed by NMR is shown at the top.
All of the remaining amides of the GSG construct have chemical shifts in the 1H 7.9–8.6 ppm range typical of flexible random coil. The GPA1 (G941–P996) and GPA2 (P1052–M1186) regions consist of multiple sequences of primarily glycine, proline, and alanine residues. Due to their lack of folded structure and repetitive sequences, the majority of GPA1 and GPA2 resonances are degenerate, appearing as broad single peaks or highly overlapped clusters of peaks in the 15N HSQC spectrum. The amino acid type corresponding to each degenerate peak could be determined from the characteristic random coil Cα and Cβ values in the HNCACB experiment, and, similarly, the preceding amino acid type could also be determined from the CBCA(CO)NH experiment. Residues preceding prolines were identified by characteristic upfield Cα and Cβ shift changes (25). Including degenerate amide assignments, 93% of GPA1 and 88% of GPA2 was assigned. Unlike the flexible C-terminal region of the BR construct, no structure other than flexible random coil is evident for GPA1 and GPA2.
Modeling.
Comparison of the 3D 15N-edited NOESY spectrum of the GSG construct with distances in the homology model showed excellent agreement, with no indication of any additional structure beyond the SH3 domain in the GSG construct.
Evidence for Interactions Between BR and GSG.
Differences between the full-length tail and GSG are illustrated in the superposed TROSY spectra (Fig. 3A), which contrast perturbed SH3 amides (Fig. 3B) with relatively unperturbed SH3 amides (Fig. 3C). Differences in amide chemical shifts for the GSG and BR constructs compared with the full-length AMIC tail are shown in Fig. 8A and B, respectively. For GSG, the greatest changes are confined to a small stretch of residues in the first loop of the SH3 domain, Y1003–N1009 (Fig. 8A). The GPA1 (G941–P996) and GPA2 (P1052–M1186) regions of GSG are relatively unperturbed (see below).
Fig. 8.
Comparison of the backbone amide chemical shift differences between the 15N HSQC spectra of full-length AMIC tail and the BR and GSG regions. Unassigned regions and proline residues are shown with zero values. (A) GSG construct: The accuracy of the chemical shift differences is <0.01 ppm. The dominant differences occur in the SH3 domain. (B) BR construct: The accuracy of the chemical shift differences is ≈0.01 ppm; the largest differences occur in the putative PH domain.
Comparing the chemical shifts in BR and full-length tail (Fig. 8B), the largest changes (>0.03 ppm) are seen in the putative PH domain, with smaller, but still significant, perturbations (>0.02 ppm) outside the PH domain region, including the flexible C-terminal region. The greater number of significantly perturbed BR amides, compared with the GSG construct, suggests possible allosteric effects in the BR construct beyond the implied PH/SH3 interface.
The GPA regions of the GSG construct appear not to interact with BR (Fig. 8A); however, the data cannot completely rule this out. For example, perturbations of a small subset of degenerate GPA residues might not be detected, especially if their signals were also weakened by the perturbing interaction. Another potential complicating factor is that the BR construct has an N-terminal FLAG-tag, whereas the FLAG-tag is at the C-terminus of the full-length tail construct; hence, some of the BR perturbations could be due to the FLAG-tag.
The NMR signals of the SH3 domain are considerably stronger than the signals of the BR structured region, even in the full-length tail spectrum. This finding implies that the SH3 domain undergoes faster molecular tumbling, unaffected for the most part by the slower tumbling or exchange processes that lead to the weaker BR signals. The relatively small perturbations and the stronger SH3 signals are most consistent with a weak, transient interaction between SH3 and BR. However, the covalently linked BR and SH3 domains would be expected to interact more frequently than nonlinked domains, and, in the right context, even weak interactions can have biological importance (26).
Discussion
SH3 and PH Domains in Myosin Tails.
Myosin-I tails fall into two groups, long (classic) and short (truncated) (27). Short-tailed myosin-Is contain only the basic region, whereas in long-tailed myosin-Is the basic region is followed by a GPA region either terminating in or, as in AMIC, divided by an SH3 domain. In addition to long-tailed myosin-Is, myosins-IV, -VII, and -XV contain SH3 domains in their tails (28). Although the SH3 domain in Dictyostelium homologues of AMIC is known to interact with CARMIL (13), little is currently known of the functions of SH3 domains in the other myosin classes. The SH3 domain of yeast myo3, a long-tailed class-I myosin like AMIC, interacts with verprolin and has been structurally characterized (29).
PH domains are found in myosin-X, which possesses three PH domains (30), and Dictyostelium myoM, which has a PH domain at the end of its tail (31) immediately after a Dbl homology domain (32). In myosin-X, the first PH domain is split by the second PH domain, which binds to phosphatidylinositol-3,4,5-trisphosphate (30, 32). The sequence of the myoM PH domain suggests that it may bind to proteins rather than to acidic phospholipids (31). The PH domain of vertebrate-specific myosin-X is required for localization of myosin-X to the phagocytic cup of macrophages (33). In addition, a putative PH domain in the basic region of mouse myo1c (34), a short-tailed myosin-I, is required for binding myo1c to phosphatidylinositol-4,5, bisphosphate in vitro and to membranes, and proper localization in vivo (34, 35). Interestingly, like AMIC, the putative PH domain of mouse myo1c seems not to have a C-terminal α-helical cap (34).
Interacting PH and SH3 Domains.
PH and SH3 domains are the 10th and 12th most common domain superfamilies in humans, and 29th and 23rd most common in yeast (36), with numerous examples of both domains in the same protein. For example, both Dbs and αPix GEF, whose PH domains are predicted by the Phyre threading program to be similar to the putative AMIC PH domain, also contain SH3 domains. However, there are no previous examples in the literature of interactions between PH and SH3 domains, and such interactions have been explicitly ruled out for some proteins (37, 38).
The canonical ligand of the SH3 domain is the PXXP motif (39). In the basic region of AMIC, this motif occurs only in residues 905–910, APGLPP. The NMR spectrum of this region is interesting because A905 and G907 are two of the amides with split signals in the flexible random coil N-terminal region of the BR construct; however, these amide signals are only moderately perturbed in the full-length tail construct.
Other parts of the basic region are more strongly perturbed, specifically the putative PH domain (Fig. 8B). Thus, the NMR chemical shift perturbation results imply an interaction between the SH3 domain and the putative PH domain of the basic region. The basic regions of long-tailed myosin-Is invariably have a long, primarily basic, insertion between putative PH domain strands 3 and 4, whereas the short-tailed myosin-Is, which do not have SH3 domains, typically lack this insertion (pfam: PF06017). This observation suggests a role for this interstrand region in the interaction of the putative PH domain with the SH3 domain in AMIC. The NMR results indicate that this insertion is in a loop conformation in the AMIC BR construct.
The putative PH domain does not contain a PXXP motif, the canonical ligand for SH3 domains, but there are proteins in which SH3 domains interact with domains that lack PXXP motifs (39, 40). One case might be particularly relevant to the interaction between the AMIC basic region and SH3 domain. A 15-residue basic amino acid sequence of Bin1/M-amphiphysin-II, located in a linker region between a Bar domain and an SH3 domain (40), interacts with the SH3 domain and blocks interaction of the SH3 domain with a canonical PXXP ligand. Although the basic sequence is not part of a PH domain, it interacts with phosphoinositides, typical ligands of PH domains, and binding of phosphatidylinositol-4,5-bisphosphate to the basic region of Bin1 releases the SH3 domain. Interestingly, one of the most strongly perturbed regions of the putative PH domain of AMIC is the long loop between β-strands 3 and 4, and this loop is mostly basic (NKDKIEKKVK), although less so than the longer basic sequence of Bin1.
The region of the model AMIC SH3 domain surface perturbed by BR interaction is shown in Fig. 9, along with the hypothetical PXXP ligand binding surface. The two regions overlap, similar to the overlap seen for the interactions of Bin1 SH3 domain with a typical PXXP ligand and with its basic sequence (40). By analogy to Bin1, it is plausible that interaction between the putative PH and the SH3 domains of AMIC could interfere with their binding to other ligands (for example, SH3 to CARMIL, GPA1 and GPA2 to F-actin, and BR to membranes), and that binding of a ligand to one of the domains would release the other domain to bind to its ligand.
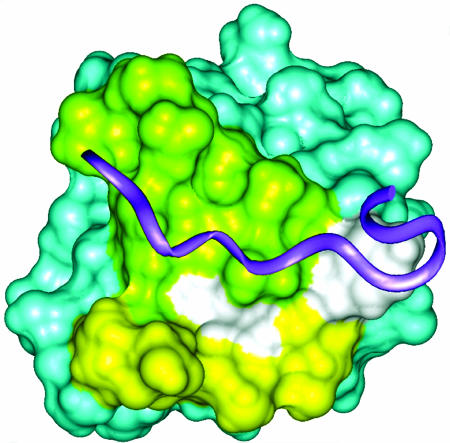
Fig. 9.
Surface view of the SH3 domain of AMIC. The region perturbed by interactions with residues in the basic region is shown in yellow, the region that is expected to be the binding site for proline-rich ligands is in green, and the overlap between these two regions is in white. The position of a hypothetical ligand (purple ribbon) is modeled by superposition of the Fyn proto-oncogen tyrosine kinase SH3 domain/ligand complex (Protein Data Bank ID Code 1A0N).
Concluding Comment.
The NMR data in this paper provide strong support for interactions between the N-terminal BR and the C-terminal GSG region of AMIC, as suggested by cryo-electron microscopy data. In addition, NMR data and homology modeling provide evidence for a PH domain within the BR of this myosin, and the NMR data indicate that the putative PH domain is the principal site within the basic region that interacts with the SH3 domain.
Materials and Methods
Expression and Purification of Proteins.
The cDNAs for the full-length tail, BR, and GSG were prepared by standard PCR methods using the full-length myosin as template and appropriate primers. To facilitate purification of the expressed proteins, C-terminal FLAG-tags were added to the full-length tail and GSG cDNAs, and either a C- or N-terminal FLAG-tag was added to the BR cDNA. All PCR products were gel-purified. The full-length tail and GSG DNAs were cloned into pTYB12 of the IMPACT (Intein Mediated Purification with an Affinity Chitin-binding Tag) vector, and the BR DNA was cloned into pTYB1. All clones were verified by restriction enzyme digestion and DNA sequencing.
Plasmids were expressed in Escherichia coli (BL21-codonplus) cultured in Luria–Bertani medium supplemented with 100 μg/ml of ampicillin and either 2.5 g/liter of [13C]glucose, 1.0 g/liter of [15N]NH4Cl, or both; in some experiments cells were grown in 65% D2O. Isopropyl-β-d-thiogalactopyranoside (0.5 mM) was added when the cultures reached an A600 of 0.5–0.6, and the cultures were placed in an incubator at 15°C with continued shaking for 16 h. Cells were harvested by centrifugation, resuspended in lysis buffer, 20 mM Hepes (pH 8.0), 1 mM diethylenetriaminepentaacetate, 500 mM NaCl, 0.1 mM PMSF, and 1 tablet of Roche protease inhibitor. Cells were broken by two passes through a French Press at 1,000 psi, and the lysates were clarified by centrifugation. Streptomycin (1%) was added to the supernatants and, after shaking for 30 min at 4°C, DNA was removed by centrifugation. The supernatants containing full-length tail and GSG were loaded onto a 5-ml chitin column (New England BioLabs, Ipswich, MA) that had been equilibrated with lysis buffer. After washing with 10 volumes of lysis buffer, the column was stored for 3–4 days at 4°C in lysis buffer containing 50 mM DTT for intein-mediated cleavage (41). Proteins were recovered by elution with 4 volumes of the same buffer without DTT. All three expressed proteins were purified by FLAG-affinity chromatography; concentrated by Centricon filtration; and dialyzed against 40 mM KCl, 20 mM phosphate buffer (pH 7.0), and 0.5 mM Tris(2-carboxyethyl)phosphine. The solutions were clarified by centrifugation. Final protein concentrations were 15–18 mg/ml; each protein was a single band by SDS/PAGE stained with Coomassie blue.
NMR Experiments and Computational Modeling.
NMR spectra acquired at 25°C for full-length AMIC tail, BR (protonated and 65% deuterated forms), and GSG constructs in buffer containing 40 mM KCl, 20 mM phosphate (pH 6.5), 0.5 mM Tris(2-carboxyethyl)phosphine, and 10% D2O include: 2D 15N-1H HSQC and 15N-1H TROSY (42); 3D HNCACB, HN(CO)CACB; and 15N-edited NOESY-HSQC (tmix of 100 ms) with water flipback/WATERGATE (43). All experiments were performed on Avance DRX600 and 800 MHz spectrometers (Bruker BioSpin, Billerica, MA) with triple-resonance, triple-axis-gradient 5-mm TXI probes; all spectra were processed and analyzed using NMRPipe (spin.niddk.nih.gov/NMRPipe). Chemical shift changes were calculated from HSQC spectra using the equation
 |
where Δδ1H and Δδ15N are the amide proton and nitrogen chemical shift changes, respectively, in ppm.
Consensus secondary structure predictions were generated by Psipred (44), JNet (45), and SSPro (46). Threading analyses were performed with Phyre (www.sbg.bio.ic.uk/phyre; ref. 18). Homology model and electrostatic potential surface generation were carried out with the Homology and DELPHI modules of InsightII (Accelrys, San Diego, CA).
Abbreviations
- AMIC
Acanthamoeba myosin IC
- BR
basic region
- SH3
Src homology 3
- GPA
Gly/Pro/Ala-rich region
- PH
pleckstrin homology domain
- GSG
GPA1–SH3–GPA2.
Footnotes
The authors declare no conflict of interest.
References
- 1.Foth BJ, Goedecke MC, Soldati D. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korn ED. Proc Natl Acad Sci USA. 2000;97:12559–12564. doi: 10.1073/pnas.230441597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, Brzeska H, Miyata H, Korn ED. J Biol Chem. 1989;264:19333–19339. [PubMed] [Google Scholar]
- 4.Wang ZY, Sakai J, Matsudaira PT, Baines IC, Sellers JR, Hammer JA, III, Korn ED. J Musc Res Cell Motil. 1997;18:395–398. doi: 10.1023/a:1018686428955. [DOI] [PubMed] [Google Scholar]
- 5.Jung G, Korn ED, Hammer JA., III Proc Natl Acad Sci USA. 1987;84:6720–6724. doi: 10.1073/pnas.84.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doberstein SK, Pollard TD. J Cell Biol. 1992;117:1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams RJ, Pollard TD. Nature. 1989;340:565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- 8.Miyata H, Bowers B, Korn ED. J Cell Biol. 1989;109:1519–1528. doi: 10.1083/jcb.109.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch TJ, Albanesi JP, Korn ED, Robinson EA, Bowers B, Fujisaki H. J Biol Chem. 1986;261:7156–7162. [PubMed] [Google Scholar]
- 10.Liu X, Brzeska H, Korn ED. J Biol Chem. 2000;275:24886–24892. doi: 10.1074/jbc.M004287200. [DOI] [PubMed] [Google Scholar]
- 11.Xu P, Mitchelhill KI, Kobe B, Kemp BE, Zot HG. Proc Natl Acad Sci USA. 1997;94:3685–3690. doi: 10.1073/pnas.94.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zot HG, Bhaskara V, Liu L. Arch Biochem Biophys. 2000;375:161–164. doi: 10.1006/abbi.1999.1648. [DOI] [PubMed] [Google Scholar]
- 13.Jung G, Remmert K, Wu XF, Volosky JM, Hammer JA., III J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa T, Cheng NQ, Liu X, Korn ED, Steven AC. Proc Natl Acad Sci USA. 2004;101:12189–12194. doi: 10.1073/pnas.0404835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baines IC, Korn ED. J Cell Biol. 1990;111:1895–1904. doi: 10.1083/jcb.111.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baines IC, Brzeska H, Korn ED. J Cell Biol. 1992;119:1193–1203. doi: 10.1083/jcb.119.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS, Sykes BD. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 18.Kelley LA, MacCallum RM, Sternberg MJE. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 19.Soisson SM, Nimnual AS, Uy M, Bar-Sagi D, Kuriyan J. Cell. 1998;95:259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- 20.Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Downes CP, Safrany ST, Alessi DR, van Aalten DMF. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worthylake DK, Rossman KL, Sondek J. Structure (London) 2004;12:1079–1086. doi: 10.1016/j.str.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 22.Lemmon MA, Ferguson KM. Biochem J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 23.Rebecchi MJ, Scarlata S. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- 24.Koshiba S, Kigawa T, Kim JH, Shirouzu M, Bowtell D, Yokoyama S. J Mol Biol. 1997;269:579–591. doi: 10.1006/jmbi.1997.1041. [DOI] [PubMed] [Google Scholar]
- 25.Lippens G, Wieruszeski JM, Leroy A, Smet C, Sillen A, Buee L, Landrieu I. Chembiochem. 2004;5:73–78. doi: 10.1002/cbic.200300763. [DOI] [PubMed] [Google Scholar]
- 26.Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, Gelmann EP, Gruschus JM. J Mol Biol. 2006;360:989–999. doi: 10.1016/j.jmb.2006.05.064. [DOI] [PubMed] [Google Scholar]
- 27.Coluccio LM. Am J Physiol. 1997;273:C347–C359. doi: 10.1152/ajpcell.1997.273.2.C347. [DOI] [PubMed] [Google Scholar]
- 28.Sellers JR. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 29.Musi V, Birdsall B, Fernandez-Ballester G, Guerrini R, Salvatori S, Serrano L, Pastore A. Protein Sci. 2006;15:795–807. doi: 10.1110/ps.051785506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geissler H, Ullmann R, Soldata T. Traffic. 2000;1:399–410. doi: 10.1034/j.1600-0854.2000.010505.x. [DOI] [PubMed] [Google Scholar]
- 32.Rossman KL, Sondek J. Trends Biochem Sci. 2005;30:163–165. doi: 10.1016/j.tibs.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, Greenberg S. Nat Cell Biol. 2002;4:469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- 34.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hokanson DE, Ostap EM. Proc Natl Acad Sci USA. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller A, MacCallum RM, Sternberg MJ. Genome Res. 2002;12:1625–1641. doi: 10.1101/gr.221202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamanian JL, Kelly RB. Mol Biol Cell. 2003;14:1624–1637. doi: 10.1091/mbc.E02-08-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu YZ, Liang L, Frank SJ, Wu CY. Biochem J. 2001;354:315–322. doi: 10.1042/0264-6021:3540315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer BJ. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 40.Kojima C, Hashimoto A, Yabuta I, Hirose M, Hashimoto S, Kanaho Y, Sumimoto H, Ikegami T, Sabe H. EMBO J. 2004;23:4413–4422. doi: 10.1038/sj.emboj.7600442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong SR, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, et al. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 42.Zhu G, Kong XM, Sze KH. J Biomol NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
- 43.Gruschus JM, Ferretti JA. J Magn Reson. 1999;140:451–459. doi: 10.1006/jmre.1999.1852. [DOI] [PubMed] [Google Scholar]
- 44.Jones DT. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 45.Cuff JA, Barton GJ. Proteins Struct Funct Genet. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 46.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. Nucleic Acids Res. 2005;33:72–76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]