Abstract
The coiled-coil protein NuMA is an important contributor to mitotic spindle formation and stabilization. A potential role for NuMA in nuclear organization or gene regulation is suggested by the observations that its pattern of nuclear distribution depends upon cell phenotype and that it interacts and/or colocalizes with transcription factors. To date, the precise contribution of NuMA to nuclear function remains unclear. Previously, we observed that antibody-induced alteration of NuMA distribution in growth-arrested and differentiated mammary epithelial structures (acini) in three-dimensional culture triggers the loss of acinar differentiation. Here, we show that in mammary epithelial cells, NuMA is present in both the nuclear matrix and chromatin compartments. Expression of a portion of the C terminus of NuMA that shares sequence similarity with the chromatin regulator HPC2 is sufficient to inhibit acinar differentiation and results in the redistribution of NuMA, chromatin markers acetyl-H4 and H4K20m, and regions of deoxyribonuclease I-sensitive chromatin compared with control cells. Short-term alteration of NuMA distribution with anti-NuMA C-terminus antibodies in live acinar cells indicates that changes in NuMA and chromatin organization precede loss of acinar differentiation. These findings suggest that NuMA has a role in mammary epithelial differentiation by influencing the organization of chromatin.
INTRODUCTION
The nuclear mitotic apparatus protein (NuMA) was first described in 1980 (Lydersen and Pettijohn, 1980) and observed to concentrate at the spindle poles during mitosis. Subsequently NuMA, variously named SPN antigen, p240 antigen, centrophilin, 1F1/1H1 antigen, SP-H antigen, and W1 antigen (Compton et al., 1991, 1992; Kallajoki et al., 1991, 1993; Maekawa et al., 1991; Tousson et al., 1991; Yang et al., 1992; Maekawa and Kuriyama, 1993; Tang et al., 1993), was shown to be a critical player in the formation and stabilization of the mitotic spindle, notably by associating with the minus end of spindle microtubules and interacting with dynein, dynactin, and LGN (Compton and Cleveland, 1993; Gaglio et al., 1995; Merdes et al., 2000; Du et al., 2001; Gehmlich et al., 2004). In addition to its location at the poles of the mitotic spindle, NuMA has been reported in the nucleus of both cycling and growth-arrested cells, as shown by immunostaining of cells in culture and tissue biopsy sections (Lelièvre et al., 1998; Merdes and Cleveland 1998; Gribbon et al., 2002; Taimen et al., 2004). However, a role for NuMA in interphase remains to be determined. The hypothesis that NuMA might organize nuclear structure (Compton and Cleveland, 1994) is supported by the existence of a long central coiled-coil region in the protein and the prevalence of NuMA in the nucleus upon detergent extraction. Furthermore, NuMA binds DNA matrix attachment regions (MARs) in vitro (Ludérus et al., 1994), and the cleavage of NuMA precedes DNA degradation during apoptosis (Weaver et al., 1996). The likelihood of a link between NuMA and chromatin structure is increased by the observation that expression of NuMA constructs lacking C-terminal residues beyond 2005 or 2030 in HeLa cells is accompanied by a redistribution of nuclear components such as DNA and histone H1 to the nuclear rim (Gueth-Hallonet et al., 1998). NuMA also has been shown to colocalize with the multifunctional, DNA-binding, high-mobility group proteins I/Y (HMG I/Y) (Tabellini et al., 2001) and bind the putative transcription factor GAS41 (Harborth et al., 2000). Despite these previous observations, the precise role of NuMA in nuclear organization remains unclear.
Strikingly, cell and tissue phenotypes have been linked to specific patterns of NuMA distribution. In lymphocytes, the formation of NuMA aggregates in the cell nucleus is associated with increased sensitivity to heat-induced apoptosis (Sodja et al., 1997). NuMA forms distinct foci in mammary epithelial cells differentiated into phenotypically normal glandular structures (acini) upon three-dimensional (3D) culture in the presence of laminin-rich Matrigel (Lelièvre et al., 1998). During differentiation of lens epithelial cells, NuMA colocalizes with the Cajal bodies and with nuclear speckles that contain components of the spliceosome (Gribbon et al., 2002). There is also evidence for a functional relationship between NuMA distribution and establishment of a differentiated phenotype. Expression of a fusion protein of NuMA truncated at its C terminus (CT) and the retinoic acid receptor has been associated with alterations in the distribution of full-length NuMA and differentiation of neutrophils (Sukhai et al., 2004). When antibodies directed against the C terminus of NuMA are introduced into mammary acini in 3D culture, the distinct NuMA foci that characterize differentiated mammary epithelial cells are replaced rapidly by the diffuse pattern characteristic of nondifferentiated cells. This redistribution is accompanied by loss of acinar differentiation, as shown by degradation of the basement membrane and alteration of acinar polarity (Lelièvre et al., 1998). Thus, a large body of data about NuMA in the interphase nucleus points to a role for this protein in differentiation and suggests a relationship between NuMA and chromatin.
In this study, we investigated the hypothesis that NuMA participates in mammary epithelial differentiation by influencing chromatin organization. We used microscopy to analyze NuMA expression in sections of differentiated human adult epithelia, including resting and lactating mammary tissues. We used the 3D cell culture model of breast acinar differentiation described above and different nuclear fractionation procedures to explore the link between NuMA and the chromatin compartment during interphase. We performed computational studies to examine whether the C-terminal region of NuMA that is highly conserved in vertebrates (Abad et al., 2004) has sequence similarity to regions present in chromatin-associated proteins. Finally, based on results from the computational studies, we expressed NuMA-CT peptides of interest in human mammary epithelial cells (HMECs) to induce a dominant-negative effect on endogenous NuMA and analyzed the consequences on mammary epithelial differentiation and chromatin organization.
MATERIALS AND METHODS
Cell Culture and Induction of Acinar Differentiation
Nonneoplastic S1 HMT-3522 HMECs (Briand et al., 1987), between passages 52 and 60, were plated at 2.3 × 104 cells/cm2 for propagation as monolayers (two-dimentsional [2D] culture) on plastic (Falcon; BD Biosciences Discovery Labware, Bedford, MA) in chemically defined H14 medium (Blaschke et al., 1994; Plachot and Lelièvre, 2004). To induce acinar differentiation in 3D culture, S1 cells were plated in the presence of 5% drip laminin-rich extracellular matrix (Matrigel; BD Biosciences Discovery Labware), as described previously, and kept for 10 d in H14 medium (Plachot and Lelièvre, 2004; the 3D cell culture procedure is reviewed in Lelièvre and Bissell, 2005).
Preparation of NuMA cDNA Constructs and Transfection
NuMA cDNAs corresponding to 6151–7183 base pairs (coding for a C-terminal peptide containing the nuclear localization signal [NLS], referred to as NuMA1965–2101) or 6261–7183 base pairs (coding for a C-terminal peptide without the NLS, referred to as NuMA2002–2101) with a 5′ FLAG tag sequence were produced by polymerase chain reaction (PCR) by using primers 5′-CGGGATCCATGGCAGACTATAAGGACGACGA CGACAAGCACATGACTGGCATCACCACCCGGCAG or 5′-CGGGATCCATGGCAGACT ATAAGGACGACGACGACAAGCACATGCCCCGAGACCGACATGAAGGG, respectively, and 3′-CCCAAGCTTGAAAAGATCCATCCCCGGCCC, and N terminus truncated NuMA as a template (kindly provided by Dr. Duane Compton, Dartmouth College, Hanover NH; Compton et al., 1992). Sequencing of cDNAs was performed at facilities of the University of California at Berkeley and Iowa State University; cDNA inserts were introduced into the multiple cloning site of the vector plasmid pcDNA3.1 (Invitrogen, Carlsbad, CA) at BamHI and HindIII restriction sites. S1 cells were transfected with the NuMA-CT constructs or the insertless vector pcDNA3.1 by using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) and selected for neomycin resistance with 100 μg/ml G418 sulfate (Mediatech, Herndon, VA). Clones were isolated using Pyrex cloning cylinders (Corning Life Sciences, Acton, MA) following the manufacturer's instructions and expanded as monolayers. Expression of the transgene was assessed by Western blot analysis using a mouse monoclonal antibody (mAb) against FLAG (clone M2; Sigma-Aldrich, St. Louis, MO) and by immunostaining using a rabbit polyclonal antibody against FLAG (Cayman Chemical, Ann Arbor, MI). Stable transfectants were used for only two passages, because expression of transgenes is labile in S1 cells.
Small Interfering RNA (siRNA) Transfection
S1 cells at 20% confluence (day 3 of 2D culture) were transfected with 10, 50, or 100 nM siRNA NuMA (ON-TARGETplus SMARTpool; Dharmacon RNA Technologies, Lafayette, CO), 50 and 100 nM nontargeting siRNA (ON-TARGETplus siCONTROL nontargeting pool; Dharmacon RNA Technologies), and 50 nM siRNA glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ON-TARGETplus siCONTROL GAPD pool; Dharmacon RNA Technologies) by using Lipofectamine transfection reagent (Invitrogen). Twenty hours posttransfection, cells were detached and plated in 3D drip culture as described above and cultured for 8 d with H14 medium change every 2–3 d.
Deoxyribonuclease (DNase) I Treatment
Cells were cultured in 3D in four-well chamber slides (Nalge Nunc International, Rochester, NY) and permeabilized for 10 min at room temperature with 0.5% Triton X-100 (Sigma-Aldrich) in cytoskeleton buffer (CSK) (100 mM NaCl, 300 mM sucrose [Sigma-Aldrich], 10 mM PIPES [Sigma-Aldrich], pH 6.8, and 5 mM MgCl2), including protease and phosphatase inhibitors (PI) (10 μg/ml aprotinin [Sigma-Aldrich], 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, hydrochloride [Roche Diagnostics], and 250 μM NaF). After two washes for 5 min with CSK-PI, cells were incubated with 130 μg/ml DNase I (Worthington Biochemical, Lakewood, NJ) in CSK-PI for 30 min at 37°C. After two washes with CSK-PI, cells were fixed for 20 min at room temperature with 4% formalin (Sigma-Aldrich) and further processed for immunofluorescence.
Immunofluorescence and Horseradish Peroxidase (HRP) Labeling
Cells cultured in 3D in four-well chamber slides or in four-well plates were either directly immunostained or embedded in Tissue-Tek OCT (Sakura Finetek, Torrance, CA), frozen, and sectioned for immunostaining. Fresh cultures or 20-μm sections of frozen cultures were washed briefly with phosphate-buffered saline (PBS) (130 mM NaCl, 13.2 mM Na2HPO4, and 3.5 mM NaH2PO4) and permeabilized for 10 min at room temperature with 0.5% Triton X-100 in CSK-PI (see DNase I Treatment). After washing twice for 5 min with CSK-PI, cells were fixed for 20 min at room temperature with 4% formalin. After rinsing thrice for 15 min at room temperature with PBS containing 50 mM glycine (Bio-Rad, Hercules, CA), cells were incubated for 1 h at room temperature with 10% goat serum (Invitrogen) in immunofluorescence labeling buffer (130 mM NaCl, 13.2 mM Na2HPO4 [Sigma-Aldrich], 3.5 mM NaH2PO4, 0.1% (wt/vol) radioimmunoassay [RIA] grade bovine serum albumin [Sigma-Aldrich], 0.05% [wt/vol] NaN3, 0.2% [vol/vol] Triton X-100, and 0.05% [vol/vol] Tween 20 [Sigma-Aldrich]). Cells were incubated overnight at 4°C in immunofluorescence labeling buffer containing 10% goat serum, and mouse monoclonal antibodies against collagen IV (clone CIV 22; Dako North America, Carpinteria, CA), PML (clone PG-M3; Santa Cruz Biotechnology, Santa Cruz, CA), NuMA (clone B1C11; kindly provided by Dr. Nickerson, University of Massachusetts, Worcester, MA), NuMA (clone 107-7; EMD Biosciences, San Diego, CA), lamin B (clone 101-B7; EMD Biosciences), SC35 (Sigma-Aldrich), and β-catenin (clone 14; BD Biosciences); rat polyclonal antibody against α 6-integrin (clone NKI-GoH3; Chemicon International, Temecula, CA); and rabbit polyclonal antibodies against acetyl-H4 (Upstate Biotechnology, Lake Placid, NY), histone 4 methylated on lysine 20 (H4K20m; Abcam, Cambridge, MA), nucleophosmin/B23 (H-106; Santa Cruz Biotechnology), and FLAG (Cayman Chemical) with concentrations of 1.88 μg/ml, 4 μg/ml, a 1:1 dilution of the culture supernatant from the mouse hybridoma used to produce the antibody, 5 μg/ml, 2 μg/ml, 9 μg/ml, 5 μg/ml, 20 μg/ml, 5 μg/ml, 1.2 μg/ml, 4 μg/ml and 30 μg/ml, respectively. After incubation with the primary antibody, cells were washed thrice for 15 min at room temperature with immunofluorescence labeling buffer, and incubated for 50 min at room temperature with fluorochrome-tagged antibodies: fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG1 (Southern Biotechnology Associates, Birmingham, AL), Alexa Fluor 488-conjugated or Alexa Fluor 594-conjugated goat anti-mouse IgG (Invitrogen), FITC-conjugated donkey anti-rat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), FITC-conjugated or Texas Red-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories). After three 15-min washes with immunofluorescence labeling buffer, nuclei were counterstained for DNA with 0.5 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride:hydrate (DAPI) (Sigma-Aldrich) in PBS for 10 min. After removal of excess DAPI, samples were mounted with the ProLong antifade kit (Invitrogen).
For immunofluorescence staining of NuMA in normal adult human tissues, Histo-Array tissue array slides (Imgenex, San Diego, CA) and archival normal breast tissue sections were used. Deparaffination and rehydration of tissue samples was achieved by washing three times for 5 min at room temperature with xylene (Mallinckrodt, Hazelwood, MO), three times for 2 min with 100% ethanol (Mallinckrodt), once for 2 min with 95% ethanol, once for 2 min with 70% ethanol, and twice for 5 min with Tris-buffered saline (TBS) (10 mM Tris base and 150 mM NaCl, pH 8.0). Then, cells were incubated for 10 min at 100°C followed by 20 min at room temperature with a 1:9 dilution of Target Retrieval Solution (Dako North America) in water. Samples were washed twice for 5 min with TBS. Samples were then incubated for 15 min with avidin blocking solution (Vector Laboratories, Burlingame, CA), washed once for 2 min with TBS, incubated for 15 min with biotin blocking solution (Vector Laboratories), and washed twice for 2 min with TBS and once for 5 min with TBS containing 0.05% (vol/vol) Tween 20 (Sigma-Aldrich) (TBST). Samples were incubated for 30 min at room temperature with blocking reagent (TSA biotin system; PerkinElmer Life and Analytical Sciences, Boston, MA), followed by incubation overnight at 4°C with a 1:1 dilution of NuMA antibody (clone B1C11) (see above) in blocking reagent. Samples were incubated twice for 10 min in TBS and once for 10 min in TBST, followed by incubation for 1 h with 15 μg/ml biotin-conjugated horse anti-mouse IgG antibodies (Vector Laboratories). After incubation twice for 5 min with TBS and once for 10 min with TBST, samples were then incubated for 1 h with 8 μg/ml fluorescein (DTAF)-conjugated streptavidin (Jackson ImmunoResearch Laboratories) in blocking reagent. Samples were then washed once for 5 min with TBS and once for 5 min with TBST. DNA was labeled for 10 min with 0.5 μg/ml DAPI (Sigma-Aldrich) in PBS. After removal of excess DAPI, samples were mounted with the ProLong antifade kit (Invitrogen). For HRP immunostaining of NuMA in breast tissue, 4-μm paraffin sections of archival, normal resting or lactating breast tissues were processed using an avidin-biotin-peroxidase method as described (Lögdberg et al., 2000). Tissue samples were used according to Institutional Review Board approval 03-135E.
Preparation of Chromatin Fractions
For 2D cultures, cells were rinsed with PBS and scraped from flasks in 5 ml of PBS-PI. For 3D cultures, multicellular structures were released from Matrigel by incubation with 0.75 ml of dispase (5000 caseinolytic units per 100 ml; BD Biosciences) per 1 ml of Matrigel used in the culture for 30 min at 37°C, and washed thrice in DMEM/F-12 medium (Invitrogen) at 37°C and once in PBS-PI at 4°C (each wash was followed by a 5-min centrifugation at 450 × g). After centrifugation, cell pellets obtained from 2D and 3D cultures were resuspended in 0.75 ml of solution containing 10 mM HEPES, pH 7.4, 1 mM EGTA (Sigma-Aldrich), 2 mM MgCl2, 250 mM sucrose (Sigma-Aldrich), and PI; then. an equal volume of 1 mM HEPES, pH 7.4, containing PI, was added. Cell suspensions from 2D and 3D cultures were incubated on ice for 30 and 45 min, respectively, mixing the suspension occasionally. Cells were lysed by performing 150 strokes with a type B pestle dounce homogenizer (Kimble/Kontes, Vineland, NJ) to obtain >80% lysis efficiency. Separation of nuclei from cytoplasm was monitored by phase contrast microscopy. Suspensions were centrifuged at 3200 × g for 15 min at 4°C. The procedure for further isolation of chromatin fractions was performed as described previously (Wysocka et al., 2001) with only minor modifications. Briefly, nuclear pellets were resuspended in buffer X [10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% (vol/vol) glycerol, 1 mM dithiothreitol, and PI]. Triton X-100 was added at 0.1% (vol/vol) final concentration and nuclear suspensions were incubated on ice for 8 min. Nuclei were collected by centrifugation at 1300 × g for 5 min at 4°C, washed in buffer X, and centrifuged at 1300 × g for 5 min at 4°C. Nuclei were lysed by 30-min incubation in buffer Y (3 mM EDTA disodium salt: dehydrate [EDTA], 0.2 mM EGTA, 1 mM dithiothreitol, and PI). After centrifugation at 1650 × g for 5 min at 4°C, pellets were resuspended in buffer Y and centrifuged again at 1300 × g for 5 min at 4°C before preparation of the soluble chromatin fraction. Pellets were resuspended in solution containing 10 mM Tris, pH 8.8, 10 mM KCl, and 1 mM CaCl2. One unit (5 μl/U) of micrococcal nuclease (Sigma-Aldrich) was added and the suspension was incubated for 5 min at 37°C. The reaction was stopped with EGTA (1 mM final concentration). The nuclease-sensitive (chromatin) fraction was separated from the nuclease-insensitive (undigested) nuclear fraction by centrifuging at 1650 × g for 5 min at 4°C. The pellet (undigested nuclear fraction) was resuspended in Laemmli buffer and incubated for 10 min at 95°C. The chromatin fraction and the undigested nuclear fraction were used for SDS-polyacrylamide gel electrophoresis followed by Western blot analysis with mouse monoclonal antibodies against lamin B (clone 101-B7; EMD Biosciences) and NuMA (clone 204-41 [EMD Biosciences] and clone B1C11), and rabbit polyclonal antibody against MCM3 (kindly provided by Dr. Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
Preparation of Nuclear Matrices
Cells obtained from 2D cultures were rinsed with PBS and scraped from flasks in 5 ml of PBS and PI. On centrifugation at 450 × g for 5 min at 4°C, cell pellets were resuspended in CSK-PI and treated as described previously to obtain nuclear matrix fractions (Nickerson et al., 1997). Cells were permeabilized with 0.5% Triton X-100 for 5 min at room temperature. After centrifugation at 3200 × g, pellets were resuspended in CSK-PI (see DNase I Treatment). DNase I (Worthington Biochemical) was added to a final concentration of 130 μg/ml. Samples were incubated for 30 min at 37°C. To aid the removal of cut DNA, (NH4)2SO4 was added to a final concentration of 0.25 M and the samples were incubated for 5 min at room temperature. Samples were centrifuged at 1300 × g for 5 min at room temperature. On centrifugation, the supernatants corresponding to the DNase I-sensitive fractions were kept. The pellets were resuspended in CSK-PI containing 2 M NaCl and incubated for 5 min at 4°C. On centrifugation at 5200 × g for 5 min at 4°C, the pellet corresponding to the nuclear matrix fraction was resuspended in Laemmli buffer and incubated for 10 min at 95°C. DNase I-sensitive fractions and nuclear matrix fractions were used for Western blot analysis.
In Situ Nick Translation
This technique was modified from Krystosek and Puck (1990). Slides were washed briefly with PBS and permeabilized for 10 min at room temperature with 0.5% Triton X-100 in CSK-PI (see DNase I Treatment). After washing twice for 5 min with CSK-PI, cells were fixed for 20 min at room temperature with 4% formalin. After rinsing three times for 15 min at room temperature with PBS containing 50 mM glycine (Bio-Rad), cells were incubated for 30 min at room temperature with 10 μg/ml unconjugated streptavidin (Jackson ImmunoResearch Laboratories) in PBS containing 10% goat serum (Invitrogen). After washing briefly with PBS, cells were incubated for 30 min at room temperature with the in situ nick translation reaction (50 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 10 mM β-mercaptoethanol [Sigma-Aldrich], 50 μg/ml RIA grade bovine serum albumin [Sigma-Aldrich], 100 U/ml Escherichia coli DNA polymerase I [MBI Fermentas, Hanover, MD], 100 μM of each dATP, dCTP, and dGTP [MBI Fermentas], and 10 μM biotin-16-dUTP [Roche Diagnostics]) without DNase I or containing 33 ng/ml DNase I (Worthington Biochemical). The reaction was stopped with 20 mM EDTA, pH 8.0. After a 5-min wash with PBS, cells were incubated for 1 h at room temperature with 10% goat serum (Invitrogen) in PBS. Then, cells were incubated for 1 h at room temperature with PBS containing 10% goat serum and 5 μg/ml fluorescein (DTAF)-conjugated streptavidin (Jackson ImmunoResearch Laboratories). Cells were washed thrice for 5 min with PBS. DNA was labeled for 10 min with 0.5 μg/ml DAPI (Sigma-Aldrich) in PBS. After removal of excess DAPI, samples were mounted with the ProLong antifade kit (Invitrogen).
Imaging and Data Processing
Images of immunofluorescence labeling were recorded using a laser scanning MRC-1024 UV (Bio-Rad) linked to a Diaphot 300 inverted microscope (Nikon, Tokyo, Japan) and oil immersion 60×, numerical aperture (NA) 1.4 apochromatic and 40×, NA 1.4 fluor lenses. Images were converted into tiff files using Confocal Assistant 4.02 (Bio-Rad) and assembled using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA) for presentation.
To analyze chromatin organization, the distribution of chromatin markers H4K20m and acetyl-H4 was visually assessed using the fluorescence microscope. Based on the observations, nuclei were classified in two groups: nuclei that showed accumulation of fluorescence at the nuclear periphery (i.e., accumulation of large staining foci) and those that did not present this pattern. Then the distribution of H4K20m and acetyl-H4 was quantified using the criteria described for visual classification. The quantitative assessment was carried out using two different methods applied independently by two investigators who are experts in imaging analysis. In the first method, the automated radial local bright feature (radial-LBF) analysis was used as described previously (Knowles et al., 2006). Briefly, bright fluorescent features of H4K20m and acetyl-H4 staining were extracted from recorded images using an adaptive algorithm that compares staining foci to their nearest neighbors. Then, the density of bright features was measured relative to specific areas in the cell nucleus. Because the changes observed upon impairing NuMA consist of an accumulation of H4K20m and acetyl-H4 domains at the nuclear periphery, the density of bright features was calculated within a peripheral area corresponding to 50% of the nuclear cross-sectional area. A density of at least 25% of the bright features in the peripheral area was considered an accumulation of H4K20m and acetyl-H4 domains. In the second approach, for each recorded image a binary mask of the cell nucleus was created using the “segment image” feature of MetaMorph (Molecular Devices, Sunnyvale, CA). The mask was smoothed using a median filter and holes were filled in. An erosion filter with a diameter of 15 pixels was applied to the smoothed mask to create a mask of the interior of the nucleus. This smaller mask was then subtracted from the original mask to create a mask for the peripheral portion of the nucleus. This ring shaped mask was applied to the original image to create an image of only the nuclear periphery (see Figure 7C). The areas of the masks and the integrated fluorescence intensities for the images of the whole nucleus and the peripheral nuclear area were calculated. To obtain a measure of the portion of staining in the periphery of the nucleus, the integrated fluorescence intensity of the ring, relative to that of the whole nucleus, was divided by the portion of the nuclear area occupied by the ring.
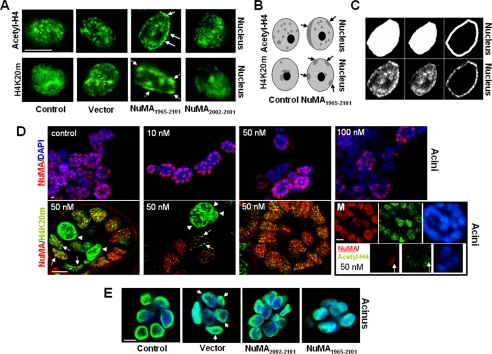
Figure 7.
Altered NuMA induces changes in higher order chromatin organization. (A) NuMA1965–2101-S1, NuMA2002–2101-S1, insertless vector-S1, and (nontransfected) control-S1 cells were cultured for 10 d in 3D to induce acinar morphogenesis. Shown is immunostaining for chromatin markers acetyl-H4 and H4K20m. Arrows point to alterations (i.e., peripheral accumulation) in the staining of chromatin markers. (B) Drawings representing the alterations in the higher order organization of acetyl-H4 and H4K20m commonly observed in NuMA1965–2101- S1 cells. Light gray indicates the diffuse staining for these proteins and dark gray indicates the concentration of acetyl-H4 and H4K20m in specific areas of the cell nucleus. Arrows indicate acetyl-H4 and H4K20m domains that concentrate at the nuclear periphery upon expression of the C terminus of NuMA. (C) Images depicting the procedure followed using MetaMorph (method 2) to quantify alterations in chromatin organization upon alteration of NuMA. Immunostaining for acetyl-H4 (bottom) and masks created using MetaMorph (top), corresponding from left to right to the whole nucleus, interior of the nucleus, and peripheral portion of the nucleus are shown for one nucleus (for a full explanation of the procedure, see Materials and Methods). (D) Twenty hours posttransfection of S1 monolayer cultures with siRNA. S1 cells were placed in 3D culture for 8 d to induce acinar morphogenesis. Top, immunostaining for NuMA (red) and DNA counterstaining (blue) in S1 acini untreated (control), and S1 acini incubated with increasing concentrations of NuMA siRNA (10, 50, and 100 nM). The number of nuclei only stained in blue (i.e., no NuMA staining) increases as cells are treated with higher concentrations of NuMA siRNA. Bottom, dual staining for NuMA (red) and H4K20m (green) in cells transfected with 50 nM NuMA siRNA. Three distinctive types of multicellular structures are shown from left to right: One acinus in which NuMA is expressed in some cells and absent in two cells (first image; arrowheads point to cells in which NuMA is absent); one cellular structure that did not develop into an acinus, from which NuMA is absent (top of the second image; arrowheads); and an acinus in which NuMA was not silenced (third image). Arrowheads indicate the strong peripheral concentration of H4K20m in the nucleus of cells that lack NuMA staining (one arrowhead per cell). Arrows indicate the presence of H4K20m domains at the nuclear periphery in cells that have a weak staining for NuMA. The montage (M) shows two multicellular structures taken from the same recorded image of a cell population transfected with 50 nM NuMA siRNA and stained for NuMA (red), acetyl-H4 (green), and DNA (DAPI, blue). The large multicellular structure (top three images) has strong staining for both NuMA and acetyl-H4, whereas the small multicellular structure (bottom three images) in which NuMA is predominantly absent shows very weak acetyl-H4 staining. Arrows point to a nucleus in which a remnant of NuMA expression coincides with a stronger staining for acetyl-H4. (E) NuMA1965–2101-S1, NuMA2002–2101-S1, insertless vector-S1 cells, and (nontransfected) control-S1 cells were cultured for 10 d in 3D to induce acinar morphogenesis. Shown is the distribution of dUTP (green) upon in situ nick translation. Nuclei are counterstained with DAPI (blue). Bar, 5 μm.
Results from Western blot analyses were recorded using the Epi Chemi II Darkroom system and Labworks 3.0.02.00 analysis software (Ultra-Violet Products, Upland, CA) and assembled using Adobe Photoshop 6.0 for presentation.
RESULTS
NuMA Adopts a Specific Distribution in Differentiated Human Epithelial Tissues
Previously, NuMA was found to be absent from the cell nuclei of certain differentiated nonepithelial human cells, namely, spermatozoa and granulocytes (Merdes and Cleveland, 1998). By contrast, NuMA was observed in many epithelial tissues, including resting mammary luminal cells (Taimen et al., 2000). To assess whether NuMA was present in highly differentiated and active human breast epithelium, we compared the HRP-staining patterns of resting and lactating mammary gland and found that NuMA was abundant in both physiological states (Figure 1, A and B). Immunofluorescence has been used to show that NuMA is distributed into distinct nuclear foci on frozen sections of normal human breast epithelium but has a diffuse distribution in nondifferentiated cells cultured on plastic (Lelièvre et al., 1998). Using immunofluorescence, we confirmed the abundant foci-like distribution of NuMA in paraffin-embedded mammary tissue (Figure 1, C and D). To assess whether distinct NuMA foci were also present in other differentiated epithelia, paraffin-embedded tissues from various adult organs were examined using immunofluorescence staining. NuMA was widely expressed in the epithelia of human colon, stomach, and lung where it occurred as few distinct foci (Figure 1, E and F; data not shown), and it was less abundant in kidney and prostate epithelia but still occurred as distinct foci (data not shown). NuMA was not detected in liver, placenta, and spleen. Thus, NuMA is distributed into distinct foci in a number of functionally and structurally differentiated epithelia and is particularly abundant in the mammary epithelium.
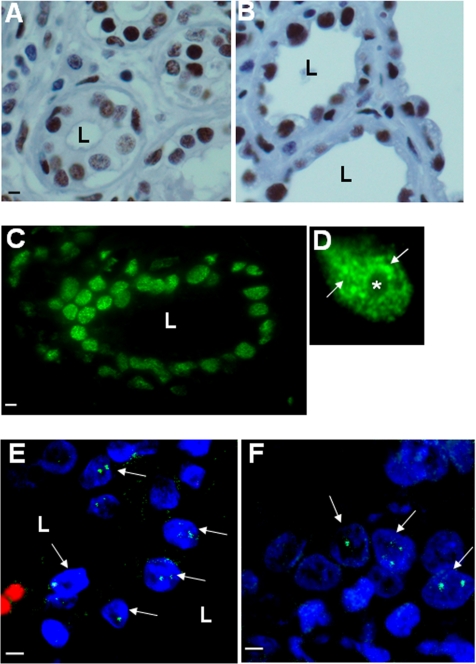
Figure 1.
NuMA is present in various human epithelia. (A–F) Immunostaining for NuMA on paraffin sections of adult human organs. (A and B) HRP-stained NuMA (brown) in resting (A) and lactating (B) mammary gland; the tissue is counterstained with hematoxylin. (C and D) Fluorescently stained NuMA in the mammary gland (green) in a section of one acinus (C) and in one nucleus (D) showing abundant NuMA foci in the midnucleus area (arrows) and around the nucleolus (*). (E and F) Fluorescently stained NuMA (green) in lung (E) and stomach (F). NuMA staining occurs as distinct nuclear foci (arrows indicate nuclei showing NuMA foci in the optical section plane). DNA is stained with DAPI (blue). Erythrocytes fluoresce red (E) due to nonspecific reaction during the staining procedure. L, lumen. Bar, 5 μm.
A Fraction of NuMA Is Present in the Chromatin Compartment during Interphase
The cell nucleus can be roughly subdivided into three compartments: the soluble compartment from which components are easily extractable by detergents, the chromatin compartment from which components are released upon DNA digestion, and the insoluble compartment or nuclear matrix from which components are resistant to detergent extraction as well as DNA digestion. NuMA has generally been considered a nuclear matrix protein because of its presence in fractions resistant to detergent extraction and DNA digestion (Lydersen and Pettijohn, 1980; Zeng et al., 1994a, b). However, there is evidence that NuMA colocalizes with the chromatin-associated architectural protein HMG I/Y (Tabellini et al., 2001)—thought to play a role in chromatin structure and transcriptional regulation—and also interacts with the putative transcription factor GAS41 (Harborth et al., 2000). Because these data raise the possibility that NuMA may be associated with the chromatin compartment, we assessed the nuclear compartmentalization of NuMA in the HMT-3522 breast model by subcellular fractionation and in situ degradation of DNA.
Nonneoplastic HMT-3522 S1 HMECs (Briand et al., 1987) were cultured as monolayers (2D culture) to produce a high number of nondifferentiated cells, and nuclear matrix fractions were prepared according to classical protocols (Nickerson et al., 1997). In this method, the supernatant obtained after DNase I digestion and the pellet obtained after incubation with 2 M NaCl correspond to the DNase I-sensitive fraction and the nuclear matrix fraction, respectively. Western blot analysis revealed that, as expected, NuMA was present in the nuclear matrix fraction. However, a large portion of NuMA (∼75% of the total amount proceeding from the DNase I-sensitive and nuclear matrix fractions, as measured by densitometry) was present in the DNase I-sensitive fraction (Figure 2A). Conversely, on the same nitrocellulose membrane, lamin B, a nuclear matrix protein considered the cornerstone of the insoluble nuclear shell (Vlcek et al., 2001), was predominantly found (∼98% of the total amount proceeding from the DNase I-sensitive and nuclear matrix fractions, as measured by densitometry) in the nuclear matrix fraction and barely detected in the DNase I-sensitive fraction (Figure 2A). To confirm that NuMA was present in the chromatin fraction, nuclei were isolated from cells cultured under 2D conditions and treated to separate the chromatin from the nondigestible nuclear remnant according to classical chromatin fractionation methods (Wysocka et al., 2001). Western blot analysis indicated that NuMA was present in the nondigestible nuclear compartment obtained upon micrococcal nuclease treatment, in agreement with previous observations (Lydersen and Pettijohn, 1980; Zeng et al., 1994b). However, NuMA was also abundantly present (∼73% of the total amount proceeding from the chromatin and nondigestible nuclear fractions, as measured by densitometry) in the chromatin compartment (Figure 2B). To verify that no cross-contamination between fractions had occurred during preparation and that NuMA was indeed behaving similarly to chromatin-associated proteins and differently from nuclear matrix proteins, the same nitrocellulose membranes were blotted for chromatin-associated protein MCM3 (Takei et al., 2001) and nuclear matrix protein lamin B. MCM3 was enriched in the chromatin compartment (∼81% of the total amount proceeding from the chromatin and nondigestible nuclear fractions, as measured by densitometry) and lamin B was enriched in the nondigestible nuclear fraction (∼93% of the total amount proceeding from the chromatin and nondigestible nuclear fractions, as measured by densitometry), indicating that the fractionation had been successful (Figure 2B).
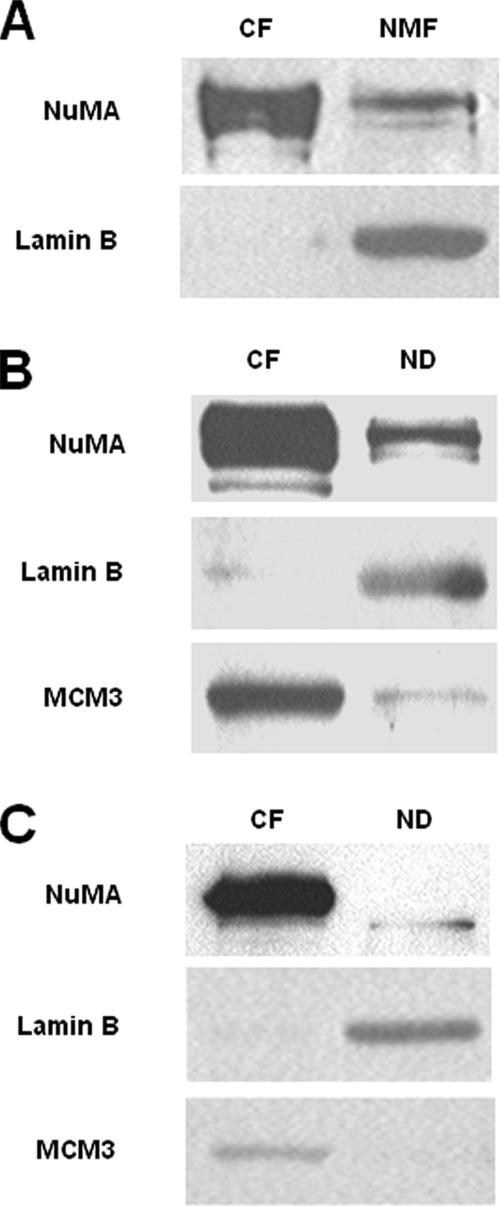
Figure 2.
NuMA is associated with the chromatin compartment. (A–C) Western blot for NuMA, Lamin B, and MCM3. (A) S1 cells were cultured as a monolayer (2D) for 10 d. Cells were fractionated using a classical protocol to obtain nuclear matrices, including treatment with 130 μg/ml DNase I for 30 min. The entire content of each fraction [DNase I-sensitive (chromatin fraction) and nuclear matrix fractions] was loaded on the gel. (B and C) S1 cells were cultured as a monolayer (2D) (B) or in 3D (C) for 10 d, and fractionated using a classical protocol for chromatin isolation, including 5-min incubation with 1 U of micrococcal nuclease. Twenty micrograms of each fraction were loaded on the gel. CF, chromatin fraction; ND, nondigestible nuclear fraction; NMF, nuclear matrix fraction.
A change in the distribution of NuMA upon breast acinar differentiation is shown by the formation of distinct and large foci (Figure 1D; Lelièvre et al., 1998). This process is recapitulated upon culture of nonneoplastic S1 HMECs under 3D conditions in the presence of Matrigel. To assess whether NuMA was also present in the chromatin compartment in differentiated cells, chromatin fractions were prepared from acini. Similar to the observations in 2D culture, NuMA was abundantly present in the chromatin fraction of acinar cells (Figure 2C).
In Mammary Acinar Cells NuMA Distribution Changes upon DNA Degradation and Displays Perinucleolar Accumulation
Because NuMA was found in the chromatin fraction obtained upon nuclease treatment of differentiated HMECs (Figure 2C), the distribution of NuMA in these cells should be dependent on DNA integrity. We incubated live cells organized into acini with DNase I, according to previously used methods (Szekely et al., 1999; Kaminker et al., 2005). Immunostaining for NuMA in control (Triton-permeabilized only) and DNase I-treated 3D cultures, revealed that a large portion of NuMA staining was lost upon DNA digestion. In DNase I-treated cells, the remaining NuMA staining was located mainly to the nuclear perimeter and as a small ring-like structure within the cell nucleus (Figure 3, compare A with B). DNase I degrades easily accessible DNA first; hence, there usually remain areas of unaffected DNA upon incomplete enzymatic digestion. Dual staining for DNA and NuMA in DNase I-treated acinar cells showed that, in cases of incomplete degradation of DNA, NuMA staining was immediately next to DNA staining (Figure 3B). Staining for NuMA (although the pattern was different from that seen in cells nontreated with DNase I) was still observed upon complete degradation of DNA. This observation is consistent with the fact that NuMA has been shown to be part of the nuclear matrix (Figure 3H). In contrast, PML, another coiled-coil protein found in nuclear matrix fractions, showed no dramatic alteration in its distribution upon DNase I treatment and no specific localization relative to the remaining DNA (Figure 3, C and D). On DNase I treatment staining for nuclear matrix protein lamin B remained at the periphery of the nuclei and was mostly located immediately outside of peripheral, nondigested DNA (Figure 3, E and F).
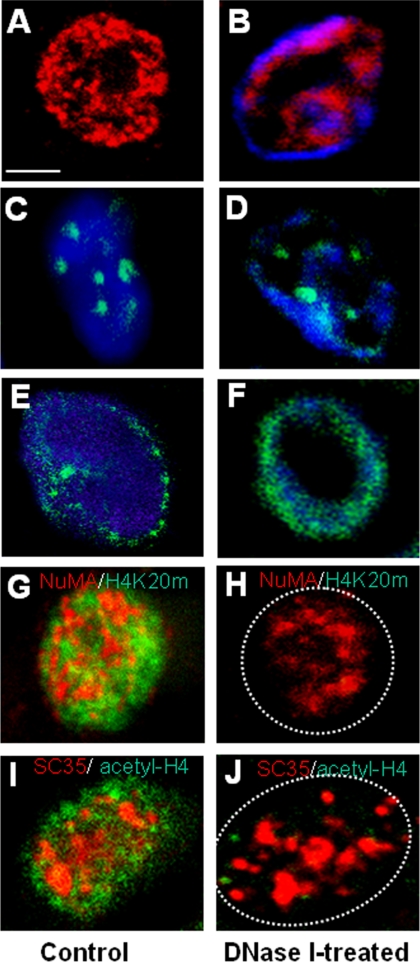
Figure 3.
NuMA distribution is altered by DNase I treatment. (A–J) S1 cells were cultured in 3D for 10 d to induce acinar differentiation. (A–F) Acinar cells were permeabilized with Triton X-100 without DNase I treatment (A, C, and E) or treated with DNase I for 30 min (B, D, and F) before fixation and immunostaining for NuMA (red; A and B), PML (green; C and D), and lamin B (green; E and F). (G–J) Acinar cells were permeabilized with Triton X-100 without DNase I treatment (G and I) or treated with DNase I for 30 min (H and J) before fixation and dual immunostaining for NuMA (red) and H4K20m (green) (G and H), or SC35 (red) and acetyl-H4 (green) (I and J). DAPI was used for DNA staining and is shown in images B–F. One nucleus is shown per image; in H and J, a dotted white circle indicates approximate nuclear boundary. Bar, 2.5 μm.
To verify that the DNase I treatment mentioned in the previous paragraph indeed affected chromatin components, we analyzed the staining pattern of chromatin markers acetyl-H4 and H4K20m (Strahl and Allis, 2000; Fischle et al., 2003) after DNase I treatment. Staining for these proteins was almost totally eliminated in cells treated with DNase I, whereas the staining pattern for SC35, indicative of nonchromatin splicing speckles, seemed unaltered (Figure 3, G–J).
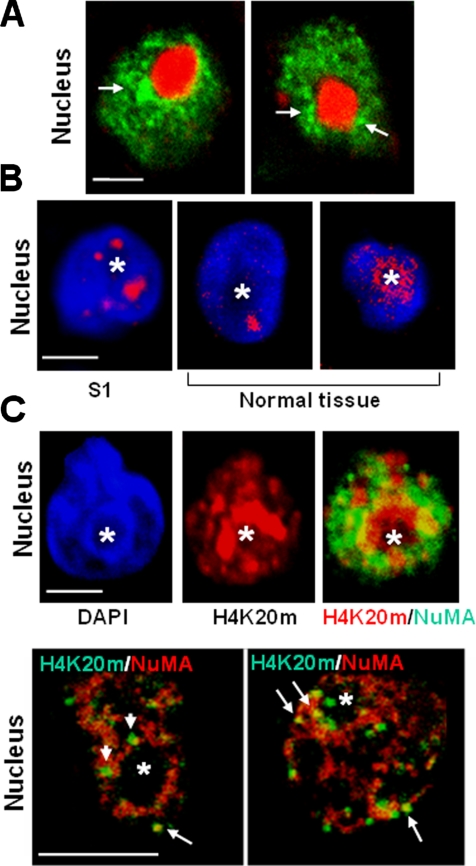
The quantitative analysis of the distribution of NuMA in differentiated S1 cells that have not undergone DNase I treatment reveals that NuMA staining foci are abundant in the midnuclear region (Figure 1D; Knowles et al., 2006). Using DAPI staining as a landmark for DNA and, hence, chromatin, we observed that the midnuclear accumulation of NuMA foci usually corresponded to the perinucleolar region (nucleoli occur as spherical regions almost devoid of DAPI staining). The preeminence of NuMA around nucleoli in acinar cells was confirmed by dual staining for NuMA and nucleophosmin, a marker of nucleoli (Biggiogera et al., 1989) (Figure 4A). Interestingly, the perinucleolar region is a major area of higher order chromatin organization; this type of organization corresponds to the concentration of euchromatin and heterochromatin domains to specific nuclear locations. Classically, heterochromatin domains have been observed to concentrate at the nuclear periphery and around the nucleolus upon cellular differentiation (Chaly and Munro, 1996; Dillon and Festenstein, 2002; Olson et al., 2002; Garagna et al., 2004). Examination of the immunostaining for heterochromatin marker H4K20m (Schotta et al., 2004) in acinar HMECs, both in 3D culture and on sections of normal breast tissue, indicated that the perinucleolar concentration of heterochromatin is also a trait of the differentiation of HMECs (Figure 4B). In addition, in acinar cells costained for NuMA and H4K20m, H4K20m foci often intercalated and sometimes overlapped with aggregates of NuMA that located to the periphery of the nucleolus (Figure 4C). Thus, NuMA is present in several nuclear compartments, and part of NuMA staining is found in regions of higher order chromatin organization in differentiated cells.
Figure 4.
In acinar cells, NuMA locates to regions enriched in heterochromatin. (A and C) S1 cells were cultured in 3D for 10 d to induce acinar differentiation. (B) S1 acini and paraffin sections of archival biopsies of normal breast tissue were used for immunostaining. (A) Immunostaining for NuMA (green) and nucleophosmin (red) in S1 acinar cells. Arrows indicate areas where NuMA concentrates around the nucleolus. (B) Immunostaining for H4K20m (red) in S1 acinar cells (S1) and luminal cells from normal breast tissue (normal tissue). Nuclei are counterstained with DAPI (blue). (C) Top, dual staining for H4K20m (red) and NuMA (green) in S1 acinar cells shows the concentration of H4K20m and NuMA domains around the nucleolus and some staining overlap (yellow). The nucleus is identified by DAPI staining (blue). The same nucleus is shown in each of the three images. Bottom, higher magnification of a dual staining for NuMA (red) and H4K20m (green) reveals that domains formed by the two proteins often intercalate around the nucleolus (see arrowheads). A few H4K20m domains colocalize with NuMA staining (as shown by yellow at arrows). One nucleus is shown per image. Bar, 2.5 μm. Asterisk indicates nucleolus.
NuMA CT Is Critical for Proper Acinar Differentiation
Acinar differentiation corresponds to the formation of polarized multicellular structures characterized by the presence of a basement membrane and a lumen at the basal and apical side of cells, respectively (Petersen et al., 1992). We have shown that introduction of antibodies directed against the C terminus of NuMA in acini modifies the nuclear distribution of NuMA and induces the loss of acinar differentiation by altering basal polarity, as evidenced by the lack of continuous staining for collagen IV, a major component of the basement membrane (Lelièvre et al., 1998). Interestingly, the alteration of NuMA organization was accompanied by changes in the distribution of the chromatin marker acetyl-H4 (Lelièvre et al., 1998), suggesting that altering NuMA function at its C terminus may affect chromatin organization.
NuMA-CT spans amino acids 1701–2101 and the distal portion (starting at amino acid 1915) of the C terminus of NuMA (NuMA-CTDP) is highly conserved in vertebrates (Abad et al., 2004), suggesting a potentially important role for this region in NuMA function. We used residues 1965–2101 of NuMA (NP_006176)—a sequence downstream of known binding sites for NuMA ligands involved in mitosis—as query for a BLASTP search against the National Center for Biotechnology Information nonredundant database of 2,315,908 sequences. We found a statistically significant similarity (E-value 0.006) between NuMA residues 1988–2068 and the central portion (residues 327–407) of the histone promoter control 2 protein (HPC2; NP_009774). HPC2 is a yeast regulator of histone expression and chromatin structure (Xu et al., 1992). When using residues 1965–2101 of NuMA (NP_006176) in a BLASTP search against the SwissProt nonredundant database of 173,195 human protein sequences, the majority of known proteins that produced significant alignment with the selected sequence of NuMA were chromatin-associated proteins. The first two hits were chromodomain-helicase-DNA-binding protein CHD-6 (residues 154–237; NP_115597) and CCAAT displacement protein CUTL1 (residues 413–496; NP_852477). Interestingly, CHD-6, CUTL1, and HPC2 aligned with similar regions of NuMA (1979–2069, 1982–2062, and 1988–2068, respectively). Thus, the ∼80-amino acid HPC2-like region of NuMA-CTDP could reflect a function of NuMA related to chromatin structure.
We engineered a cDNA construct coding for a FLAG-tagged peptide, corresponding to NuMA-CT residues 1965–2101, that would compete with the C terminus of endogenous NuMA for binding. This peptide contains the NLS, located in the 1984–2005 region of the protein (Gueth-Hallonet et al., 1996). We engineered another construct coding for a FLAG-tagged peptide (residues 2002–2101) that lacked the NLS (Figure 5). Expression of these NuMA-CT peptides was verified by Western blot analysis for FLAG from stable S1 HMEC transfectants cultured under 2D conditions and/or FLAG immunostaining from stable S1 HMEC transfectants cultured under 3D conditions (Figure 6, A and B).
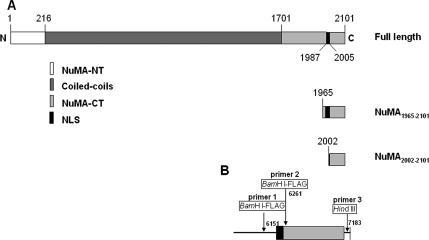
Figure 5.
cDNA inserts for NuMA1965–2101 and NuMA2002–2101. (A) Schematic of NuMA peptides expressed in S1 cells; numbers indicate the position of amino acids. (B) Schematic of primers used to synthesize the cDNAs for NuMA1965–2101 (primers 1 and 3) and NuMA2002–2101 (primers 2 and 3). Numbers indicate the position of nucleotides. NT, N terminus.
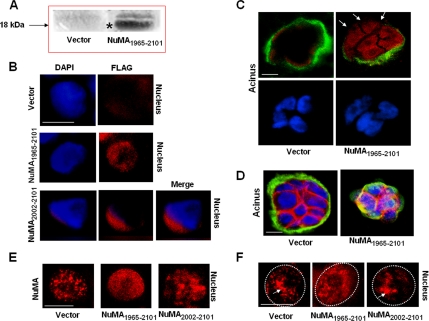
Figure 6.
Cells expressing NuMA1965–2101 display a different organization of endogenous NuMA and an altered acinar differentiation. (A) Western blot for 18-kDa FLAG-NuMA1965–2101 peptide (*) in control (Vector) and NuMA1965–2101-S1 cells cultured under 2D conditions. (B–E) NuMA1965–2101-S1, NuMA2002–2101-S1, and insertless vector-S1 cells were cultured for 10 d under 3D conditions to induce acinar morphogenesis. (B) Immunostaining for FLAG (red). Nuclei were counterstained with DAPI (blue). The images for NuMA2002–2101-S1 nucleus have been merged to show that FLAG staining is cytoplasmic. (C) Dual immunostaining for FLAG (red) and collagen IV (green) shows alterations of basement membrane integrity (see arrows) in cells expressing NuMA1965–2101 peptide. Nuclei were counterstained with DAPI (blue). (D) Dual immunostaining for beta-catenin (red) and α6-integrin (green) shows alterations in the internal organization of acinar cells in NuMA1965–2101-S1 transfectants. In differentiated acini, the distribution of β-catenin is characterized by its location only at plasma membrane sites involved in cell–cell adhesion, and the distribution of α6-integrin is characterized by continuous staining at plasma membranes only involved in cell-extracellualr matrix adhesion (compare distribution in [vector] with altered distribution in [NuMA1965–2101]). Nuclei were counterstained with DAPI (blue). (E) Immunostaining for endogenous NuMA (red). The nuclei of NuMA1965–2101-S1 cells typically display a diffuse distribution pattern for endogenous NuMA. (F) NuMA1965–2101-S1, NuMA2002–2101-S1, and insertless vector-S1 cells were cultured for 10 d in 3D to induce acinar morphogenesis and incubated with or without DNase I before fixation. Shown is immunostaining for NuMA following DNase I treatment. The arrow indicates NuMA domains remaining near the center of the cell nucleus. A dotted white circle indicates approximate nuclear boundary. Bar, 5 μm.
Nine clones of NuMA1965–2101-S1 stable transfectants and three clones of NuMA2002–2101-S1 stable transfectants were cultured under 3D conditions to induce the formation of acini. Five clones of insertless pcDNA3.1 vector-S1 stable transfectants were cultured under 3D conditions. Immunostaining for FLAG in multicellular structures obtained under 3D culture conditions revealed that the FLAG-NuMA1965–2101 peptide was present in the cell nucleus, whereas the location of FLAG-NuMA2002–2101 peptide was cytoplasmic (Figure 6B). Expression of the peptide was observed in 41–74% of S1 acini, depending on the clone (S1 cells easily shut down transgene expression; Table 1). Only S1 cells transfected with insertless vector or NuMA2002–2101 formed well-differentiated acini. In contrast, there was a decrease of well-formed acini in NuMA1965–2101-transfectants, as shown by altered organization of α-6 integrin, β-catenin, and collagen IV—all markers of the polarity-axis—compared with the organization observed in phenotypically normal epithelium (Figure 6, C and D). The percentage of acini presenting loss of differentiation in the different clones was in accordance with the extent of expression of FLAG-NuMA1965–2101 peptide in each clone (Table 1). Antibodies that do not recognize the distal C-terminal region of NuMA were used to assess the distribution of endogenous NuMA. Results show that NuMA was diffusely distributed in 37–62% (corresponding to the extent of expression of FLAG-NuMA1965–2101 peptide in each clone; Table 1) of the nuclei of acini formed by NuMA1965–2101-S1 cells. Whereas it was distributed as the foci-like pattern characteristic of acinar differentiation in insertless vector-S1 cells as well as in NuMA2002–2101-S1 cells (Figure 6E). To further investigate how expression of NuMA1965–2101 peptide altered the organization of NuMA, we assessed whether the distribution of endogenous NuMA was sensitive to DNA degradation in the transfectants. NuMA1965–2101-S1, NuMA2002–2101-S1, and insertless vector-S1 cells were cultured under 3D conditions for 10 d and subjected to DNase I digestion in situ. Immunostaining for endogenous NuMA revealed that, in contrast to NuMA2002–2101-S1 cells and insertless vector-S1 cells, NuMA distribution was not affected by DNase I treatment in NuMA1965–2101-S1 cells (Figure 6F). This observation suggests that the relationship between endogenous NuMA and chromatin may differ upon expression of NuMA1965–2101 peptide.
Table 1.
Expression and distribution of specific proteins in S1 cells stably transfected with insertless vector (pcDNA3.1), NuMA1965–2101 (containing the NLS), NuMA2002–2101(without the NLS), TIN2-15, and hTERT
| Clone | % of acini showing FLAG expression |
% of acini showing altered collagen IV |
% of nuclei showing diffuse NuMA |
% of nuclei showing altered H4K20m |
|---|---|---|---|---|
| NuMA1965–2101 CL 1-5 | 61.5 | 41 | 62 | 55 |
| NuMA1965–2101 CL 1-6 | 74 | 75 | 59.3 | 52.4 |
| NuMA1965–2101 CL 4-2 | 41 | 37.7 | 36.9 | 39.6 |
| NuMA1965–2101 CL 4-4 | 66.6 | 68.6 | 51.5 | 51 |
| NuMA2002–2101 (T+10) | 64.6 | 34.6 | 9.8 | 13.8* |
| NuMA2002–2101 CL 1 | 72 | 17.33 | 3.3 | 7* |
| NuMA2002–2101 CL 2 | 76.9 | 15.38 | 0.75 | 15.2* |
| pcDNA3.1 CL 2 | 34.3 | 3.6 | 17.2* | |
| pcDNA3.1 CL 7 | 20 | 3.9 | 14* | |
| TIN2-15 | 15.8* | |||
| hTERT | 17.8* |
Frozen sections of 10-day-old 3D cultures of different clones were stained for collagen IV, FLAG, NuMA, or H4K20m. At least 75 acini were scored for the presence of collagen IV and FLAG, and at least 100 cells were scored for the distribution of NuMA and H4K20m in each replicate. In the clones marked with an asterisk (*), all cells showing some concentration of H4K20m at the nuclear periphery had nuclei with unusual angular or folded shape. This may be an artifact of cryosectioning in these 3D cultures. In contrast, the majority of NuMA1965–2101cells with heavy concentration of H4K20m at the nuclear periphery had nuclei with round and smooth shape. Only NuMA1965–2101 and NuMA2002–2101clones with >20% expression of FLAG-tagged peptides are shown.
Peptide- and Antibody-induced Alteration of NuMA Distribution Is Associated with Changes in Higher Order Chromatin Organization Characteristic of Acinar Differentiation
To assess whether expression of NuMA1965–2101 was associated with changes in chromatin organization as previously observed upon introduction of antibodies against the C terminus of NuMA (Lelièvre et al., 1998), insertless vector-S1, NuMA1965–2101-S1, and NuMA2002–2101-S1 cells were immunostained for chromatin markers acetyl-H4 and H4K20m. Depending on the extent of expression of the peptide in the different clones, 40–55% of the nuclei in acini formed by NuMA1965–2101-S1 cells had altered higher order chromatin organization, as shown by the accumulation of enlarged domains of acetyl-H4 and H4K20m at the nuclear periphery (Figure 7, A and B, and Table 1). In contrast, the distributions of acetyl-H4 and H4K20m in NuMA2002–2101-S1 and insertless vector-S1 acinar cells were similar to those seen in nontransfected S1 cells (scoring of 300 nuclei per sample showed that <6 and 18% of cells had peripheral accumulation of acetyl-H4 and H4K20m, respectively). Because this visual scoring of acetyl-H4 and H4K20m staining pattern does not give a measure of the amount or density of staining at the nuclear periphery, we have used two independent approaches to quantify the presence of nuclear peripheral accumulation of staining foci for these two proteins, as described in Materials and Methods. The first approach (method 1) made use of the radial-LBF analysis (Knowles et al., 2006) and was applied to calculate the density of staining foci in a nuclear peripheral shell corresponding to 50% of the nuclear cross-sectional area as measured on the optical section. The second approach (method 2) was developed to give a measure of the portion of staining in the periphery of the cell nucleus (Figure 7C). Analyses applied to the images shown in Figure 7A reveal that NuMA1965–2101-S1 cells have, on average, 50 and 33% more nuclear peripheral density of bright foci for acetyl-H4 and H4K20m, respectively, compared with all control cells (method 1), and 45% more staining for acetyl-H4 and H4K20m in the peripheral area of the cell nucleus compared with all control cells (method 2). To confirm that the observed alteration of higher order chromatin organization was not triggered by the presence of just any peptide in the cell nucleus, acini formed by S1 cells that expressed the catalytic subunit of telomerase (hTERT), or a truncated form of the telomeric protein TIN2 (Kaminker et al., 2005), were immunostained for acetyl-H4 and H4K20m. The percentage of hTERT-S1 cells and TIN2-15-S1 cells showing accumulation of these proteins at the nuclear periphery was not different from that observed for other controls (<18% for H4K20m, Table 1; 4.3 and 3.5%, respectively, for acetyl-H4).
We also silenced NuMA in differentiating S1 cells to verify that changes in chromatin structure were truly dependent on NuMA. Cells growing as a monolayer (2D culture) were transfected with siRNAs. Twenty hours posttransfection, these cells were plated in 3D culture and incubated for 8 d, a period of time long enough to foster differentiation, and short enough to still permit the detection of gene silencing in a significant amount of cells. For each biological replicate, immunostaining revealed that the percentage of cells without detectable NuMA staining, after 8 d of 3D culture, increased with the concentration of siRNA (on average the lack of NuMA staining occurred in 14% of cells treated with 10 nM NuMA siRNA, 28% of cells treated with 50 nM NuMA siRNA, and 37% of cells treated with 100 nM NuMA siRNA) (Figure 7D). In the control samples that included cells treated with 50 and 100 nM scrambled siRNA, 50 nM GAPDH siRNA, Lipofectamine only, and nontransfected cells, the absence of NuMA staining occurred in <4.3% of the cell population. The percentage of cells with peripheral concentration of heterochromatin marker H4K20m increased 1.4-fold in the population of cells treated with 10 nM NuMA siRNA and tripled in the population of cells treated with 50 and 100 nM NuMA siRNA compared with controls. The alteration of H4K20m distribution was essentially found in cells that showed greatly decreased or no NuMA staining (Figure 7D) and occurred on average in 36% of these cells. Interestingly, the peripheral distribution of acetyl-H4 was not a characteristic of the cells that lacked NuMA staining. Instead, in these cells, acetyl-H4 was barely visible, whereas it was strongly stained in cells in which NuMA was expressed (Figure 7D). We conclude that like with the treatment with NuMA antibody and the expression of NuMA- CT peptide, the absence of full-length NuMA triggers changes in chromatin organization.
Another means to analyze gross alterations in chromatin organization is to reveal the location of regions highly sensitive to DNase I digestion by in situ nick translation. Using this technique it has been shown that DNase I-sensitive chromatin densely locates to the periphery of the cell nucleus in differentiated and reverse-transformed cells but not in transformed cells (Krystosek and Puck, 1990; Linares-Cruz et al., 1998). In situ nick translation revealed that 83% of NuMA2002–2101-S1, 68.7% of insertless vector-S1, and 89.6% of control S1 acinar cells had a marked nuclear peripheral location of DNase I-sensitive chromatin. Whereas sensitive sites were mainly found throughout the cell nucleus in NuMA1965–2101-S1 cells (depending on the clone, 12.9 to 41.1% of nuclei had a nuclear peripheral location of DNase I-sensitive chromatin) (Figure 7E). These data suggest that preventing the differentiation-specific distribution of NuMA, by expressing the NuMA1965–2101 peptide, influences higher order chromatin organization.
HMECs expressing NuMA1965–2101 show marked alterations in acinar differentiation due to the loss of basement membrane integrity. Therefore, the changes observed in chromatin organization in these cells compared with control cells could be the result of signaling induced by the loss of basement membrane integrity originally triggered by the alteration of NuMA (Figure 6C). Alternatively, the changes in chromatin organization could be a direct effect of the alteration of NuMA and, in turn, prevent acinar differentiation. To evaluate whether altering NuMA may influence higher order chromatin organization before impairing acinar differentiation, acini were briefly permeabilized with digitonin and treated with function-blocking antibody against NuMA-CT as reported previously (Lelièvre et al., 1998). Cultures were stopped after 30 min of incubation with the antibody. On such a short incubation time, there were no remarkable alterations in acinar morphogenesis and the NuMA antibody did not induce an increase in the number of acini without a complete staining for collagen IV (77.7% of acini with intact collagen IV staining in nonspecific IgGs-treated cells versus 79.2% of acini with intact collagen IV staining in NuMA antibody-treated cells) (Figure 8A). Nevertheless, introduction of antibodies against NuMA-CT in the cell nucleus induced a peripheral concentration of acetyl-H4 domains (23.1% of nuclei with accumulation of acetyl-H4 domains at the nuclear periphery in NuMA antibody-treated cells compared with 5.3% in IgGs-treated cells; data not shown) and H4K20m domains (31.9% of nuclei with accumulation of H4K20m domains at the nuclear periphery in NuMA antibody-treated cells compared with 5.2% in IgGs-treated cells) (Figure 8B).
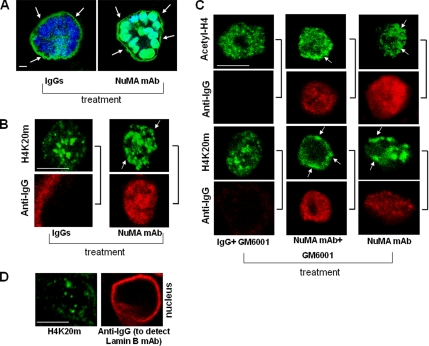
Figure 8.
Effects of altering NuMA on higher order chromatin organization are independent of the loss of basement membrane integrity. (A–C) S1 cells were cultured for 10 d in 3D to induce acinar morphogenesis and incubated with an antibody against NuMA-CT (NuMA mAb) or nonspecific IgGs for 30 min (A and B) or 3 d (C) in the presence or absence of metalloproteinase inhibitor GM6001. (A) Acini were immunostained with an antibody against collagen IV to estimate the percentage of acini with basement membrane containing intact collagen IV (green) after incubation with nonspecific IgGs and NuMA antibody. Because function blocking NuMA antibody, nonspecific IgGs, and the antibody against collagen IV are all mouse immunoglobulins, images show staining (green) for collagen IV around the acini (see arrows) and also either staining for IgGs in the cytoplasm or for nuclear NuMA antibody used for treatment of live acini. Nuclei are counterstained with DAPI (blue). (B and C) Dual immunostaining with secondary antibodies against mouse IgGs (anti-IgG) to detect nonspecific IgGs and NuMA antibodies in the cells (red) and for acetyl-H4 (green) or H4K20m (green). Arrows indicate domains of acetyl-H4 and H4K20m concentrated at the nuclear periphery. Sidebars indicate images obtained from the same nucleus. Bar, 5 μm. Note: Incubation of NuMA antibody-treated cells with GM6001 maintained collagen IV integrity in 59% of acini compared with 37% in NuMA antibody-treated cells without GM6001 and 78% in IgGs- or (IgG + GM6001)-treated cells. (D) Dual immunostaining against H4K20m (green) and lamin B (red). For lamin B, during the staining procedure we only used secondary antibodies against mouse IgGs (anti-IgG) to detect lamin B antibodies previously introduced in the cells upon digitonin permeabilization.
We also incubated S1 acinar cells with NuMA antibodies or IgGs for 3 d, an interval sufficient to induce changes in chromatin organization and loss of basement membrane integrity (Lelièvre et al., 1998). During this time, GM6001, an inhibitor of metalloproteinases that prevents degradation of the basement membrane in NuMA antibody-treated cells (Lelièvre et al., 1998; Figure 8C), was added to the culture medium of half of the samples. Acini cultured with or without GM6001 in the presence of NuMA antibodies had alterations in the distribution of acetyl-H4 (35% of nuclei in NuMA antibody-treated cells and 26.6% of nuclei in NuMA antibody + GM6001-treated cells versus 3.2% of nuclei in IgGs-treated cells and 5.6% of nuclei in IgGs + GM6001-treated cells) and H4K20m (26.7% of nuclei in NuMA antibody-treated cells and 31.1% of nuclei in NuMA antibody + GM6001-treated cells versus 7.7% of nuclei in IgGs-treated cells and 3.6% of nuclei in IgGs + GM6001-treated cells) (Figure 8C). The two methods described in the preceding paragraphs were used to measure the accumulation of staining foci at the nuclear periphery for the images shown in Figure 8, B and C. By comparison to all control cells, S1 cells incubated with NuMA antibody had, on average, 28 and 25% more nuclear peripheral density of bright foci for acetyl-H4 and H4K20m, respectively (method 1) and 38 and 31% more staining for acetyl-H4 and H4K20m, respectively, in the peripheral area of the cell nucleus (method 2).
To confirm that the induced alteration of higher order chromatin organization is not due to the introduction of merely any antibody into the cell nucleus, in a separate set of experiments, we also treated digitonin-permeabilized S1 acinar cells with antibodies against the nuclear matrix protein lamin B. After 3 days, we verified that the antibodies had reached their target by staining with fluorescence-tagged secondary antibodies only (Figure 8D). On average, peripheral accumulation of H4K20m was detected in 1.1% of the cell population in lamin B antibody-treated acinar cells versus 29.5% of the cell population in NuMA antibody-treated acinar cells. Similarly, on average, peripheral accumulation of acetyl-H4 was detected in 6% of the cell population in lamin B antibody-treated acinar cells versus 14.6% of the cell population in NuMA antibody-treated acinar cells. Collectively, these data confirm that the observed changes in higher order chromatin organization are linked to the alteration of NuMA and occur before changes in acinar differentiation resulting from the alteration of NuMA.
DISCUSSION
To date, NuMA has been considered a nuclear matrix protein because it remains in the cell nucleus upon detergent extraction and DNA removal. In this work, we demonstrate that NuMA is abundant also in the chromatin fraction. The presence of NuMA in different compartments of the cell nucleus may explain the great variation in its distribution reported for different cell and tissue phenotypes (Sodja et al., 1997; Merdes and Cleveland, 1998; Lelièvre et al., 1998; Gribbon et al., 2002; Plachot and Lelièvre, 2004; Knowles et al., 2006). The compartmentalization of NuMA is particularly interesting to observe upon DNase I treatment in situ. The staining pattern for NuMA changed dramatically upon DNase I treatment. Specifically, we observed residual staining for NuMA next to nondigested DNA. Under these conditions, heterochromatin marker H4K20m was mostly absent, suggesting that the remaining DNA was particularly inaccessible. Other nuclear matrix proteins, for example PML, remained in the insoluble nuclear fraction, but they were not consistently associated with nondigested DNA (Figure 3). A role for NuMA in DNA anchoring has been proposed based on an in vitro interaction with MAR sequences (Ludérus et al., 1994). Such sequences anchor DNA onto the nuclear matrix and, in some cases, coincide with the boundaries of a DNase I-sensitive domain (Blasquez et al., 1989; Levy-Wilson and Fortier, 1989). Whether the specific delineation of nondigested DNA by NuMA signifies an interaction between NuMA and MAR sequences remains to be determined.
We find evidence for a novel role for NuMA in chromatin organization. Impairing endogenous NuMA during or upon acinar differentiation using NuMA-CT peptide or anti-NuMA-CT antibodies modifies the distribution of the chromatin markers acetyl-H4 and H4K20m. Changes in higher order chromatin organization are confirmed by the alteration of the distribution of DNase I sensitive regions, indicative of modifications in genome exposure (Linares-Cruz et al., 1998). The induced alteration of H4K20m and acetyl-H4 distribution by NuMA antibodies, which occurs before the loss of acinar differentiation, suggests a direct effect of NuMA on chromatin organization. In addition, silencing NuMA also triggers modifications in chromatin markers H4K20m and acetyl-H4, indicating that the effect on chromatin is indeed linked to an altered function of NuMA. One means by which NuMA could influence chromatin structure is via the displacement of NuMA from MAR sequences during differentiation, because certain MAR associated-proteins may control tissue-specific gene expression by influencing the chromatin environment (Cai et al., 2003). Another explanation for the effect of NuMA on chromatin structure may be linked to the location of NuMA in the chromatin compartment. In this compartment, NuMA could interact with the putative transcription factor GAS41 (Harborth et al., 2000), a protein shown to bind the chromatin remodeling complex component INI1 (Debernardi et al., 2002). In addition, chromatin-remodeling complexes have been shown to contain actin-related proteins. An interaction between NuMA and chromatin-remodeling complexes mediated by actin-related proteins might also exist, because a putative actin-binding domain has been identified at the N terminus of NuMA (Novatchkova and Eisenhaber, 2002).
Our data indicate that the distal portion of NuMA-CT is important for the effect of NuMA on chromatin organization. These findings are in agreement with previous data obtained with HeLa cells, which showed that expression of NuMA with a deletion of the distal portion of the C terminus induced the relocation of DNA and histone H1 within the nucleus (Gueth-Hallonet et al., 1998). How the NuMA1965–2101 peptide exerts its effects remains to be understood. Based on very preliminary studies involving protein–protein binding experiments, we are inclined to conclude that the peptide does not bind to NuMA. Rather, it may compete with endogenous NuMA to bind ligand(s) of the distal C terminus. By doing so, it could alter the relationship between endogenous NuMA and chromatin, as suggested by the lack of removal of NuMA staining upon DNase I treatment of NuMA1965–2101 cells (Figure 6F). The conservation of the distal C-terminal sequence of NuMA among vertebrate species reinforces the idea that this sequence may play an essential role in this phylum (Abad et al., 2004); hence, it will be important to identify critical features or domains within this region. The ligands of the distal portion of NuMA remain to be found, and as discussed in the preceding paragraph it is possible that other regions of NuMA may have a role in chromatin organization. Indeed, sequence analysis suggests that regions at both C and N termini of NuMA could bind structural elements and chromatin components (Harborth et al., 2000; Novatchkova and Eisenhaber, 2002; Abad et al., 2004).
The role of NuMA in differentiated cells remains the subject of an ongoing debate (Lelièvre et al., 1998; Merdes and Cleveland, 1998; Gribbon et al., 2002; Abad et al., 2004; Taimen et al., 2004). Although some data point to the absence of NuMA in differentiated tissues, the examples reported include primarily nonepithelial tissues (Merdes and Cleveland, 1998; Taimen et al., 2004). Differentiation processes are generally accompanied with growth arrest. Although it has been argued that disappearance of NuMA expression could be associated with growth arrest (Taimen et al., 2000), our immunostaining of biopsy sections from growth-arrested epithelial tissues and previous results showing NuMA abundantly present in growth-arrested mammary epithelial cells (Lelièvre et al., 1998; Knowles et al., 2006) suggest otherwise. In this study, we confirmed the expression of NuMA in epithelial tissues from lung, colon, stomach, and mammary gland as reported previously (Taimen et al., 2000). However, our immunofluorescence-based analysis indicates that the distribution patterns of NuMA in these epithelia seem somewhat different, ranging from a few large foci (in colon and stomach) to abundant foci (in breast) concentrated in specific nuclear regions. These differences may result from the fact that differentiation-linked nuclear organization can be cell type specific (Kaminker et al., 2005). As a consequence, NuMA itself may be differently localized, or alternatively, changes in nuclear organization may modify the accessibility of the epitope recognized by the antibody used for NuMA.
Differentiation processes are accompanied with dramatic changes in the gene expression profile. These changes are the consequences of global as well as gene-specific modifications in transcriptional activity. Thus, a link between NuMA and transcriptional activity might explain differences that exist in the distribution of NuMA depending on the type of differentiated tissue. This possibility is suggested by the correlation between loss of NuMA and low transcriptional activity noticed in spermatozoa and nonactivated granulocytes (Taimen et al., 2004). In addition, a link between transcriptional activity and NuMA distribution is supported by a study of lens epithelial differentiation, showing the location of NuMA to Cajal bodies and splicing speckles upon decrease in the levels of transcriptional activity (Gribbon et al., 2002). Interestingly, NuMA is one of the proteins that remain intact as mouse erythrocytes undergo differentiation, and it was suggested that NuMA integrity is important to maintain the gene expression profile necessary for the differentiation process in these cells (Krauss et al., 2005). All epithelial tissues that reveal nuclei with moderately high euchromatin/heterochromatin ratio are involved in high transcriptional activity, even if this activity supports one major function (e.g., lactation for breast epithelium) that can be switched on or off. Thus, qualitative rather than quantitative changes in transcriptional activity upon differentiation of certain epithelial tissues could also be accompanied with the redistribution of NuMA, as seen in mammary epithelial differentiation.
In this study, we have demonstrated that NuMA remains abundantly expressed in the mammary epithelium along different stages of differentiation and that NuMA influences the organization and maintenance of higher order chromatin structure associated with the differentiation of mammary epithelium. It will be important to determine whether, by influencing chromatin organization, NuMA participates in transcriptional regulation.
ACKNOWLEDGMENTS
We thank Scott Briggs for critical reading of the manuscript, Cedric Plachot for critical reading of the manuscript and for developing the siRNA transfection technique for 3D culture studies in the Lelièvre laboratory, Ali Ravanpay for helpful discussion, and Tushendan Rasiah and Lesley Chaboub for technical assistance. Special thanks also to Dr. Ronald Hullinger for helping with the interpretation of NuMA distribution in the various tissues analyzed and critical reading of the manuscript. This work was supported by the Department of Defense/Breast Cancer Research Program (DAMD17-00-1-0226 and W81XWH-04-1-0670 to S.A.L.), the Walther Cancer Institute (WCI-110-114 to S.A.L.), the Lawrence Berkeley National Laboratory (subcontract 6806563 to S.A.L.), the California Breast Cancer Research Program and Department of Energy-Office of Health and Environmental Research (to I.S.M.), a Purdue Doctoral Fellowship and a Bilsland Dissertation Fellowship (to P.C.A.), the Purdue Research Foundation (to S.A.L.), the Purdue Cancer Center, and the Purdue School of Veterinary Medicine.
Abbreviations used:
- acetyl-H4
acetylated histone 4
- CSK
cytoskeleton buffer, CSK
- CT
C terminus
- DNase
deoxyribonuclease
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride:hydrate
- H4K20m
histone 4 methylated on lysine 20
- HMEC
human mammary epithelial cells
- LBF
local bright feature
- MAR
matrix attachment region
- NLS
nuclear localization signal
- NuMA
nuclear mitotic apparatus protein
- PI
protein and phosphatase inhibitors
- 3D
three-dimensional
- TBS
Tris-buffered saline
- TBST
Tris-buffered saline with Tween
- 2D
two-dimensional
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0551) on November 15, 2006.
REFERENCES
- Abad P. C., Mian I. S., Plachot C., Nelpurackal A., Bator-Kelly C., Lelièvre S. A. The C terminus of the nuclear protein NuMA: phylogenetic distribution and structure. Protein Sci. 2004;13:2573–2577. doi: 10.1110/ps.04906804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggiogera M., Fakan S., Kaufmann S. H., Black A., Shaper J. H., Busch H. Simultaneous immunoelectron microscopic visualization of protein B23 and C23 distribution in the HeLa cell nucleolus. J. Histochem. Cytochem. 1989;37:1371–1374. doi: 10.1177/37.9.2768807. [DOI] [PubMed] [Google Scholar]
- Blaschke R. J., Howlett A. R., Desprez P. Y., Petersen O. W., Bissell M. J. Cell differentiation by extracellular matrix components. Methods Enzymol. 1994;245:535–556. doi: 10.1016/0076-6879(94)45027-7. [DOI] [PubMed] [Google Scholar]
- Blasquez V. C., Sperry A. O., Cockerill P. N., Garrard W. T. Protein:DNA interactions at chromosomal loop attachment sites. Genome. 1989;31:503–509. doi: 10.1139/g89-098. [DOI] [PubMed] [Google Scholar]
- Briand P., Petersen O. W., Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev. Biol. 1987;23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- Cai S., Han H. J., Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Chaly N., Munro S. B. Centromeres reposition to the nuclear periphery during L6E9 myogenesis in vitro. Exp. Cell Res. 1996;223:274–278. doi: 10.1006/excr.1996.0082. [DOI] [PubMed] [Google Scholar]
- Compton D. A., Cleveland D. W. NuMA is required for the proper completion of mitosis. J. Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A., Cleveland D. W. NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 1994;6:343–346. doi: 10.1016/0955-0674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Compton D. A., Szilak I., Cleveland D. W. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J. Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A., Yen T. J., Cleveland D. W. Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J. Cell Biol. 1991;112:1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi S., Bassini A., Jones L. K., Chaplin T., Linder B., de Bruijn D. R., Meese E., Young B. D. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99:275–281. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- Dillon N., Festenstein R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 2002;18:252–258. doi: 10.1016/s0168-9525(02)02648-3. [DOI] [PubMed] [Google Scholar]
- Du Q., Stukenberg P. T., Macara I. G. A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Allis C. D. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Compton D. A. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagna S., Merico V., Sebastiano V., Monti M., Orlandini G., Gatti R., Scandroglio R., Redi C. A., Zuccotti M. Three-dimensional localization and dynamics of centromeres in mouse oocytes during folliculogenesis. J. Mol. Histol. 2004;35:631–638. doi: 10.1007/s10735-004-2190-x. [DOI] [PubMed] [Google Scholar]
- Gehmlich K., Haren L., Merdes A. Cyclin B degradation leads to NuMA release from dynein/dynactin and from spindle poles. EMBO Rep. 2004;5:97–103. doi: 10.1038/sj.embor.7400046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribbon C., Dahm R., Prescott A. R., Quinlan R. A. Association of the nuclear matrix component NuMA with the Cajal body and nuclear speckle compartments during transitions in transcriptional activity in lens cell differentiation. Eur. J. Cell Biol. 2002;81:557–566. doi: 10.1078/0171-9335-00275. [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet C., Wang J., Harborth J., Weber K., Osborn M. Induction of a regular nuclear lattice by overexpression of NuMA. Exp. Cell Res. 1998;243:434–452. doi: 10.1006/excr.1998.4178. [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet C., Weber K., Osborn M. NuMA: a bipartite nuclear location signal and other functional properties of the tail domain. Exp. Cell Res. 1996;225:207–218. doi: 10.1006/excr.1996.0171. [DOI] [PubMed] [Google Scholar]
- Harborth J., Weber K., Osborn M. GAS41, a highly conserved protein in eukaryotic nuclei, binds to NuMA. J. Biol. Chem. 2000;275:31979–31985. doi: 10.1074/jbc.M000994200. [DOI] [PubMed] [Google Scholar]
- Kallajoki M., Harborth J., Weber K., Osborn M. Microinjection of a monoclonal antibody against SPN antigen, now identified by peptide sequences as the NuMA protein, induces micronuclei in PtK2 cells. J. Cell Sci. 1993;104:139–150. doi: 10.1242/jcs.104.1.139. [DOI] [PubMed] [Google Scholar]
- Kallajoki M., Weber K., Osborn M. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle; a microinjection study using SPN monoclonal antibodies. EMBO J. 1991;10:3351–3362. doi: 10.1002/j.1460-2075.1991.tb04899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker P., Plachot C., Kim S. H., Chung P., Crippen D., Petersen O. W., Bissell M. J., Campisi J., Lelievre S. A. Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J. Cell Sci. 2005;118:1321–1330. doi: 10.1242/jcs.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles D. W., Sudar D., Bator-Kelly C., Bissell M. J., Lelievre S. A. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proc. Natl. Acad. Sci. USA. 2006;103:4445–4450. doi: 10.1073/pnas.0509944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S. W., Lo A. J., Short S. A., Koury M. J., Mohandas N., Chasis J. A. Nuclear substructure reorganization during late-stage erythropoiesis is selective and does not involve caspase cleavage of major nuclear substructural proteins. Blood. 2005;106:2200–2205. doi: 10.1182/blood-2005-04-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystosek A., Puck T. T. The spatial distribution of exposed nuclear DNA in normal, cancer, and reverse-transformed cells. Proc. Natl. Acad. Sci. USA. 1990;87:6560–6564. doi: 10.1073/pnas.87.17.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelièvre S. A., Bissell M. J. Encyclopedia of Molecular and Cellular Biology and Molecular Medicine. Vol. 14. Weinheim, Germany: Wiley-VCH; 2005. Three dimensional cell culture: the importance of context in regulation of function; pp. 383–420. [Google Scholar]
- Lelièvre S. A., Weaver V. M., Nickerson J. A., Larabell C. A., Bhaumik A., Petersen O. W., Bissell M. J. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc. Natl. Acad. Sci. USA. 1998;95:14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B., Fortier C. The limits of the DNase I-sensitive domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5′ and 3′ boundaries of the gene. J. Biol. Chem. 1989;264:21196–21204. [PubMed] [Google Scholar]
- Linares-Cruz G., et al. p21WAF-1 reorganizes the nucleus in tumor suppression. Proc. Natl. Acad. Sci. USA. 1998;95:1131–1135. doi: 10.1073/pnas.95.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lögdberg L. E., Åkerström B., Badve S. Tissue distribution of the lipocalin alpha-1 microglobulin in the developing human fetus. J Histochem. Cytochem. 2000;48:1545–1552. doi: 10.1177/002215540004801111. [DOI] [PubMed] [Google Scholar]
- Ludérus M. E., den Blaauwen J. L., de Smit O. J., Compton D. A., van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 1994;14:6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydersen B. K., Pettijohn D. E. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kuriyama R. Primary structure and microtubule-interacting domain of the SP-H antigen: a mitotic MAP located at the spindle pole and characterized as a homologous protein to NuMA. J. Cell Sci. 1993;105:589–600. doi: 10.1242/jcs.105.2.589. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Leslie R., Kuriyama R. Identification of a minus end-specific microtubule-associated protein located at the mitotic poles in cultured mammalian cells. Eur. J. Cell Biol. 1991;54:255–267. [PubMed] [Google Scholar]
- Merdes A., Cleveland D. W. The role of NuMA in the interphase nucleus. J. Cell Sci. 1998;111:71–79. doi: 10.1242/jcs.111.1.71. [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W. C., Cleveland D. W. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson J. A., Krockmalnic G., Wan K. M., Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc. Natl. Acad. Sci. USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M., Eisenhaber F. A CH domain-containing N terminus in NuMA? Protein Sci. 2002;11:2281–2284. doi: 10.1110/ps.0221002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. O., Hingorani K., Szebeni A. Conventional and nonconventional roles of the nucleolus. Int. Rev. Cytol. 2002;219:199–266. doi: 10.1016/S0074-7696(02)19014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen O. W., Ronnov-Jessen L., Howlett A. R., Bissell M. J. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachot C., Lelièvre S. A. DNA methylation control of tissue polarity and cellular differentiation in the mammary epithelium. Exp. Cell Res. 2004;298:122–132. doi: 10.1016/j.yexcr.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodja C., Walker P. R., Brown D. L., Chaly N. Unique behaviour of NuMA during heat-induced apoptosis of lymphocytes. Biochem. Cell Biol. 1997;75:399–414. [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sukhai M. A., et al. Myeloid leukemia with promyelocytic features in transgenic mice expressing hCG-NuMA-RARalpha. Oncogene. 2004;23:665–678. doi: 10.1038/sj.onc.1207073. [DOI] [PubMed] [Google Scholar]
- Szekely L., Kiss C., Mattsson K., Kashuba E., Pokrovskaja K., Juhasz A., Holmvall P., Klein G. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin- associated nuclear bodies. J. Gen. Virol. 1999;80:2889–2900. doi: 10.1099/0022-1317-80-11-2889. [DOI] [PubMed] [Google Scholar]
- Tabellini G., Riccio M., Baldini G., Bareggi R., Billi A. M., Grill V., Narducci P., Martelli A. M. Further considerations on the intranuclear distribution of HMGI/Y proteins. Ital. J. Anat. Embryol. 2001;106:251–260. [PubMed] [Google Scholar]
- Taimen P., Parvinen M., Osborn M., Kallajoki M. NuMA in rat testis–evidence for roles in proliferative activity and meiotic cell division. Exp. Cell Res. 2004;298:512–520. doi: 10.1016/j.yexcr.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Taimen P., Viljamaa M., Kallajoki M. Preferential expression of NuMA in the nuclei of proliferating cells. Exp. Cell Res. 2000;256:140–149. doi: 10.1006/excr.2000.4799. [DOI] [PubMed] [Google Scholar]
- Takei Y., Swietlik M., Tanoue A., Tsujimoto G., Kouzarides T., Laskey R. MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep. 2001;2:119–123. doi: 10.1093/embo-reports/kve026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T. K., Tang C. J., Chen Y. L., Wu C. W. Nuclear proteins of the bovine esophageal epithelium. II. The NuMA gene gives rise to multiple mRNAs and gene products reactive with monoclonal antibody W1. J. Cell Sci. 1993;104:249–260. doi: 10.1242/jcs.104.2.249. [DOI] [PubMed] [Google Scholar]
- Tousson A., Zeng C., Brinkley B. R., Valdivia M. M. Centrophilin: a novel mitotic spindle protein involved in microtubule nucleation. J. Cell Biol. 1991;112:427–440. doi: 10.1083/jcb.112.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlcek S., Dechat T., Foisner R. Nuclear envelope and nuclear matrix: interactions and dynamics. Cell Mol. Life Sci. 2001;58:1758–1765. doi: 10.1007/PL00000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver V. M., Carson C. E., Walker P. R., Chaly N., Lach B., Raymond Y., Brown D. L., Sikorska M. Degradation of nuclear matrix and DNA cleavage in apoptotic thymocytes. J. Cell Sci. 1996;109:45–56. doi: 10.1242/jcs.109.1.45. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Reilly P. T., Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Kim U. J., Schuster T., Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Lambie E. J., Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., He D., Berget S. M., Brinkley B. R. Nuclear-mitotic apparatus protein: a structural protein interface between the nucleoskeleton and RNA splicing. Proc. Natl. Acad. Sci. USA. 1994a;91:1505–1509. doi: 10.1073/pnas.91.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., He D., Brinkley B. R. Localization of NuMA protein isoforms in the nuclear matrix of mammalian cells. Cell Motil. Cytoskeleton. 1994b;29:167–176. doi: 10.1002/cm.970290208. [DOI] [PubMed] [Google Scholar]