Abstract
The data in the present study show that DNA polymerase γ and DNA ligase III interact in mitochondrial protein extracts from cultured HT1080 cells. An interaction was also observed between the two recombinant proteins in vitro. Expression of catalytically inert versions of DNA ligase III that bind DNA polymerase γ was associated with reduced mitochondrial DNA copy number and integrity. In contrast, overexpression of wild-type DNA ligase III had no effect on mitochondrial DNA copy number or integrity. Experiments revealed that wild-type DNA ligase III facilitates the interaction of DNA polymerase γ with a nicked DNA substrate in vitro, and that the zinc finger domain of DNA ligase III is required for this activity. Mitochondrial protein extracts prepared from cells overexpressing a DNA ligase III protein that lacked the zinc finger domain had reduced base excision repair activity compared with extracts from cells overexpressing the wild-type protein. These data support the interpretation that the interaction of DNA ligase III and DNA polymerase γ is required for proper maintenance of the mammalian mitochondrial genome.
Keywords: DNA integrity, DNA ligase III, DNA polymerase γ, HT1080, mitochondria, zinc finger domain
Abbreviations: APE, AP (apurinic/apyrimidinic) endonuclease; BER, base excision repair; DTT, dithiothreitol; HA, haemagglutinin; mtDNA, mitochondrial DNA; OGG1, 8-oxoguanine-DNA glycosylase; PolG, DNA polymerase γ; qRT-PCR, quantitative real time PCR; SSC, standard saline citrate; TBS, Tris-buffered saline; Xrcc1, X-ray repair cross-complementing factor 1
INTRODUCTION
Due to the existence of separate nuclear and mitochondrial genomes, mammalian cells must produce two separate sets of DNA replication and repair machinery. This requirement has resulted in several instances of nuclear genes that produce both nucleus-targeted and mitochondria-targeted versions of the same DNA repair protein. A striking example of this involves enzymes that catalyse the short-patch BER (base excision repair) process. Examples of nuclear genes that encode protein products essential for both nuclear and mtDNA (mitochondrial DNA) BER include APE1 and APE2 [AP (apurinic/apyrimidinic) endonuclease 1 and 2] [1,2], several DNA glycosylases, including OGG1 (8-oxoguanine-DNA glycosylase), UDG (uracil DNA glycosylase), hMYH (human MutY homologue) and MPG (N-methylpurine-DNA glycosylase) [3–6], and DNA ligase III [7]. In all cases, alternative splicing or alternative sites of translation initiation are responsible for the generation of distinct protein products that possess nuclear or mitochondrial targeting sequences.
These observations highlight that the BER reactions in the respective organelles are quite similar. However, critical differences must exist because some BER proteins that play essential roles in the nucleus are absent from the mitochondria. Examples include the repair enzyme DNA polymerase β, poly(ADP-ribose) polymerase, and the DNA ligase III-binding protein Xrcc1 (X-ray repair cross-complementing factor 1) [8]. PolG (DNA polymerase γ), which is the only known form of DNA polymerase present in the mitochondrial compartment, functions in place of DNA polymerase β during mitochondrial BER [9,10].
The observation that DNA ligase III can function in mitochondrial BER in the absence of its nuclear binding partner Xrcc1 is particularly intriguing. In addition to its role in stabilizing DNA ligase III [11], the Xrcc1 protein plays a central role in nuclear BER. Studies have revealed that Xrcc I binds to both DNA polymerase β [12,13], and poly(ADP-ribose) polymerase [13,14], and also binds, via its zinc-finger domain, to nicked DNA itself [15,16].
It thus seems reasonable to postulate that mtDNA ligase III function could be critically dependent on its ability to interact with a mitochondrial protein with functions analogous to Xrcc1. This hypothesis is consistent with the observation that DNA ligase III in Xenopus laevis oocytes extracts exists as part of a multi-protein complex [17]. The finding that BER can be reconstituted in vitro using purified mtDNA ligase III, PolG and APE 1 [9], led us to test the hypothesis that PolG functionally interacts with DNA ligase III during mitochondrial BER.
In the present paper we document a specific interaction between mtDNA ligase III and PolG, and identify the regions of the two respective proteins that are responsible for this interaction. We demonstrate that overexpressed wild-type and inactive versions of mtDNA ligase III bind to PolG, and that transgenic cells overexpressing non-functional mtDNA ligase III display reduced mtDNA copy number and integrity. Experiments conducted using recombinant proteins indicate that the zinc-finger domain of mtDNA ligase III facilitates the binding of PolG to nicked DNA repair substrates. Finally, we show that mitochondrial protein extracts prepared from cells expressing the version of mtDNA ligase III lacking the zinc-finger domain have significantly diminished levels of in vitro BER. These data support the hypothesis that a novel interaction between mtDNA ligase III and PolG plays an essential role in BER by facilitating the loading of the latter protein on to DNA repair substrates.
EXPERIMENTAL PROCEDURES
Materials and reagents
Human HT1080 fibrosarcoma cells (American Type Culture Collection) were grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Cellgro), penicillin (100 units/ml)/streptomycin (100 μg/ml) (Invitrogen), sodium pyruvate (1 mM) and uridine (50 μg/ml). Unless otherwise stated all reagents were from Sigma.
Plasmid constructs
A mitochondria-specific full-length DNA ligase III construct (comprising nucleotides 73–3102 of human DNA ligase III cDNA, GenBank® accession number X84740) with a 3′ terminal HA (haemagglutinin) tag sequence (5′-GGCGTAGTCGGGGACGTCGTAGGGGTA-3′) flanked by BamH1 sites in the vector pEGFP-N1 (Clontech) was used for the transgenic expression of mtDNA ligase III. Site-directed mutagenesis [18] was used to modify the portions of the DNA ligase III cDNA encoding two critical amino acid residues of the enzyme active site sequence (KYDGER, single letter amino acid codes are used). Mutagenic oligonucleotides 5′-TCTGAGATCGTATACGATGGAG-3′ and 5′-GATGGAGAGCATGTCCAGGTG-3′ were used individually to alter the DNA sequences encoding the active site lysine and arginine residues (indicated in bold above) to encode valine and histidine residues respectively. The resulting mutations were confirmed using DNA sequence analysis. The DNA ligase III construct was excised by BamH1 digestion and introduced into the BamH1 site of the episomal vector pREP4 (Invitrogen). Correct orientation was determined by restriction digest and DNA sequence analysis. The wild-type DNA ligase III construct was named pREP4-lig and the constructs encoding the mutant proteins were called pREP4-lig(K-V) and pREP4-lig(R-H). A mtDNA ligase III construct lacking 39 base pairs of the zinc-finger-encoding sequence was prepared by following a protocol similar to that described earlier [13]. Briefly, a mitochondria-specific full-length DNA ligase III construct (comprising nucleotides 73–3102 of human DNA ligase III cDNA, GenBank® accession number X84740) with a 3′ terminal HA tag sequence (5′-GGCGTAGTCGGGGACGTCGTAGGGGTA-3′) flanked by BamH1 sites was cloned into the vector pET15b (Novagen). Digestion of this with XmaI and KpnI restriction enzymes (New England BioLabs) led to the elimination of 39 base pairs. Subsequent treatment with T4 DNA polymerase and T4 DNA ligase led to rejoining of the linear construct. The modified mtDNA ligase III sequence was confirmed by DNA sequencing. Finally the mutant mtDNA ligase III sequence was excised by BamH1 restriction digestion and recloned into the episomal vector pREP4 and the correct orientation was confirmed by restriction digestion analysis. This construct was named pREP4-ΔZf-lig.
Creation of the transgenic cells
DNA samples of episomal vector pREP4, pREP4-lig, pREP4-lig(K-V), pREP4-lig(R-H) and pREP4-ΔZf-lig were individually electroporated [19] into the human fibrosarcoma cell-line HT1080 [20]. The transfectants are referred to as REP, WT, K-V, R-H and ΔZf-lig respectively. After electroporation, one million cells were plated in 10-cm dishes, allowed to recover for 1 day and placed in selection medium containing hygromycin. Colonies were obtained 11–14 days later. Since the pREP4 vector is maintained as a low-copy episome in human cells, all drug-resistant clones will harbour identical numbers of the pREP4-based vector (approx. 100 copies/cell) maintained as an autonomously replicating extrachromosomal genetic element. To minimize potential inter-clone variations (conceivably resulting from chromosome-encoded mutations unrelated to the presence of the episomal plasmid DNA) subsequent analysis was performed on populations of cells obtained by co-culturing cells obtained from 4–5 colonies. In all cases, these cell populations were maintained at low passage numbers (≤4 passages) under selection pressure.
Preparation of mitochondrial extracts
Mitochondria were isolated by differential centrifugation as described previously [21]. The mitochondrial pellet was washed twice and then lysed with buffer [10 mM Tris/HCl (pH 7.4), 10 mM MgCl2, 10 mM KCl, 1 mM DTT (dithiothreitol) and 350 mM NaCl]. The mitochondrial matrix proteins were separated from the insoluble membrane fraction by centrifugation at 70000 rev./min for 30 min at 4 °C in a TL100.3 rotor (Beckman). The protein extract was dialysed overnight at 4 °C in buffer containing 25 mM Tris/HCl (pH 7.5), 1 mM EDTA, 1 mM PMSF, 1 mM DTT and 10% glycerol. The protein concentration in the extract was determined using the Bradford assay [22].
Western blot analysis
Protein extracts in loading buffer containing 50 mM Tris/HCl (pH 6.8), 100 mM DTT, 2% SDS, 0.1% Bromophenol Blue and 10% glycerol were resolved by SDS/PAGE. For Western blots, the protein was transferred to nitrocellulose membrane (Bio-Rad). Subsequently the membrane was placed in blocking solution [TBS (Tris-buffered saline; 10 mM Tris/HCl and 150 mM NaCl, pH 7.5) containing 5% BSA] for 1 h at room temperature (25 °C). After the blocking solution was removed, a rat monoclonal anti-HA antibody (clone 3F10; Roche Diagnostics) at a dilution of 1:250 was added and incubated with the membrane overnight at 4 °C. For DNA ligase III Western blots, a mouse monoclonal anti-DNA ligase III antibody (Novus) at a 1:1000 dilution was used as the primary antibody. For PolG Western blots, a rabbit polyclonal anti-PolG antibody (Labvision) was used at a 1:1000 dilution. The membrane was subsequently washed three times in 0.1% BSA in TBS for 5 min each time. Anti-rat, anti-mouse or anti-rabbit IgG conjugated with alkaline phosphatase (1:5000 dilution; Sigma) was added and incubated with the membrane for 1 h at room temperature. The membrane was then washed as above. After the final wash the membrane was incubated with water containing alkaline phosphatase substrate (SigmaFast, 5-bromo-4-chloro-3-indolyl phosphate/Nitro Blue Tetrazolium; Sigma). The colour reaction was stopped by rinsing with water. The membrane was dried and scanned using the Molecular Analyst program.
Co-immunoprecipitation
Mitochondrial protein extracts (130 μg) were incubated in the presence or absence of 10 μg of mouse monoclonal PolG antibody (Neomarkers) or mouse monoclonal DNA ligase III antibody (Novus) at 4 °C for 2 h. Subsequently, 25 μl of a 50% slurry of Protein G–agarose beads was added and the mixture incubated overnight at 4 °C. Immune complexes were sedimented and washed 4 times with lysis buffer containing 20 mM Tris/HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.1% Nonidet P40, 1 mM DTT, 1 mM PMSF, 10% glycerol and a protease inhibitor cocktail.
DNA nick seal assay
The substrate for this reaction was prepared as previously described [23]. Briefly, a 15-mer oligodeoxynucleotide (5′-CCCAGTCACGACGTT-3′) and a 17-mer (5′-GTAAAACGACGGCCAGT-3′) were synthesized by the MicroChemical Facility of the University of Minnesota, Minneapolis. The 17-mer (40 pmol) was radiolabelled with polynucleotide kinase and [γ-32P]ATP (New England Biolabs) under standard conditions. The 15-mer and radiolabelled 17-mer were then annealed to the M13mp18 single-stranded DNA phage vector (New England Biolabs), resulting in the formation of a nicked DNA duplex. Mitochondrial protein extract (5 μg) was incubated with substrate (3×104 c.p.m.) at 37 °C for either 5 or 10 min. The reaction was stopped by heating at 95 °C for 5 min followed by the addition of loading dye (95% formamide, 20 mM EDTA, 0.1% Xylene Cyanol FF and 0.1% Bromophenol Blue), and this mixture was resolved on a 22.5% polyacrylamide/7 M urea SDS/PAGE gel. The gel was dried and radioactivity detected using a PhosphorImaging scanner (Molecular Dynamics) and quantitated using IP-Lab gel software.
Adenylation reaction
Protein adenylation reactions were performed as previously described [24]. Briefly, samples (either 20 μg of whole mitochondrial protein extract or pellets resuspended from immunoprecipitation reactions) were incubated in a reaction containing 0.5 μCi [α-32P]ATP, 60 mM Tris/HCl (pH 8.0), 10 mM MgCl2, 5 mM DTT and 50 μg/ml BSA at room temperature for 15 min. The reaction was stopped with the addition of loading buffer and resolved by SDS/PAGE. After electrophoresis the gel was fixed in 10% acetic acid and developed using a phosphorimager screen.
Isolation of cellular genomic DNA and Southern blot analysis
Cells were lysed using 10 mM Tris/HCl (pH 8.0), 1 mM EDTA and 0.5% SDS. The lysate was incubated with proteinase K (at a final concentration of 7 mg/ml) overnight at 37 °C. The following day, genomic DNA was isolated using phenol extraction followed by phenol/chloroform (1:1, v/v) extraction followed by chloroform extraction. The genomic DNA was precipitated by addition of an equal volume of isopropanol. DNA was resuspended in water and subjected to restriction enzyme digestion with EcoRI. The digested DNA was then precipitated by the addition of 2 vol. of ethanol, resuspended in water and the concentration of DNA measured using the diphenylalanine assay [25]. DNA (1 μg) was resolved on a 0.6% agarose gel. The DNA was transferred to a nylon membrane by capillary action overnight using buffer 5× SSC [standard saline citrate; 0.3 M sodium citrate and 2 M sodium chloride (pH 7.0)], prehybridized with Church buffer [26] for 2 h and then hybridized with a radiolabelled probe, specific for mitochondrial DNA, at 55 °C overnight. The probe DNA was obtained from a mouse mitochondrial genome cloned in pSP6 (a gift from Dr S. P. Ledoux, Department of Structural and Cellular Biology, University of South Alabama, Mobile, AL, U.S.A. [27]). Upon digestion with XhoI and BglII a 1778 base pair fragment is released which corresponds to nucleotides 13551–15329 of the mouse mitochondrial genome. The corresponding co-ordinates of the human mitochondrial genome to which this fragment anneals are nucleotides 14127–15905. This fragment was gel purified and labelled with [α-32P]dATP using the RadPrime DNA Labelling System (Invitrogen). After an overnight hybridization, the blot was washed and radioactivity detected using a PhosphorImaging scanner.
Real-time PCR of genomic DNA
The primers used for qRT-PCR (quantitative real-time PCR) of mtDNA were 5′-CACAAAATAGACTACGAAAGTGGC-3′ (forward) and 5′-ACTTACCATGTTACGACTGG-3′ (reverse), expected to amplify a 547 bp fragment of the human mtDNA between nucleotides 1556–1009. The primers used to amplify the nuclear β-actin gene were 5′-CAAGAGATGGCCACGGCTGCT-3′ (forward) and 5′-TCCTTCTGCATCCTGTCGGCA-3′ (reverse), yielding a 274 bp fragment. PCR reactions were set up in a 20 μl of total volume containing 0.2–2 ng of total genomic DNA, 1 μM of each primer and the Light Cycler FastStart DNA Master SYBR® Green I mix (Roche). PCR conditions used were 10 min at 95 °C followed by 35 cycles of 5 s at 95 °C (denaturation), 5 s at 60 °C (annealing) and 20 s at 72 °C (extension). The specificity of the amplified PCR product was accessed by performing a melting curve analysis and further by visualizing the products after resolving them on a 0.8% agarose gel. Standard curves were constructed for both the mtDNA product and β-actin using serial dilutions of total genomic DNA. Using the standard curves the relative amounts of nuclear DNA and mtDNA were obtained using three independent genomic DNA samples. A Light Cycler instrument (Roche) was used for the qRT-PCR analysis.
Co-immunoprecipitation of in vitro transcription/translation products
Full-length mtDNA ligase III cDNA encoding a 3′-terminal HA tag and PolG cDNA encoding a 3′-terminal myc tag were PCR amplified using Ready-To-Go-PCR beads (Amersham). PCR conditions were denaturation at 98 °C for 2 min followed by 35 cycles of 40 s at 98 °C (denaturation), 40 s at 55 °C (annealing) and 2 min at 72 °C (elongation). The forward and reverse primers for mtDNA ligase III were: 5′-TAATACGACTCACTATAGGGAGAGCCACCATGTCTTTGGCTTTCAAG-3′ and 5′-TTAAGCGTAATCTGGAACATCGTATGGGTACTGGTGGGTTTGGGAGAAG-3′. The forward and reverse primers for PolG were 5′-TAATACGACTCAGTATAGGGAGAGCCACCATGAGCCGCCTGCTCTGGAGG-3′ and 5′-TAACAGATCCTCTTCTGAGATGAGTTTTTGTTCGTGCTATGGTCCAGGCTGGCTTCG-3′. For both of the forward primers a T7 promoter sequence was present upstream of the ATG start site. The template DNA for mtDNA ligase III PCR was the mitochondria-specific DNA ligase III α-isoform cDNA sequence cloned into a pEGFP-N1 vector. The template DNA for PolG PCR was the plasmid HuGpVL102/103 [10] that was obtained as a gift from Dr William Copeland (National Institute of Environmental Health Services, Research Triangle Park, NC, U.S.A.). The primers used for PCR amplification of N-terminal, middle and C-terminal fragments of mtDNA ligase III were as follows: FORLigNterm 5′-TAATACGACTCACTATAGGGAGAGCCACTTGGCTTTCAAG-3′ and REVLigNterm 5′-TTAAGCGTAATCTGGAACATCGTATGGGTAATCATCTGGGTTGCAGTT-3′; FORLigmiddle 5′-TAATACGACTCACTATAGGGAGAGCCACCATGGCACGGGACCTAGAG-3′ and REVLigmiddle 5′-TTAAGCGTAATCTGGAACATCGTATGGGTAGGCCCCCTCGTTCAAATAG-3′; FORLigCterm 5′-TAATACGACTCACTATAGGGAGAGCCACCATGGCCGACACAGCTGAC-3′ and REVLigCterm 5′-TTAAGCGTAATCTGGAACATCGTATGGGTAGCAGGGAGCTACCAGTCTC-3′. The PCR primers used for smaller fragments of PolG were as follows: FORExo1 5′-TAATACGACTCACTATAGGGAGAGCCACCATGAGCCGCCTGCTCTGGAGG-3′ and REVExo1 5′-TTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGTACCGGGTCCAGCCCTCCGC-3′; FORExo2 5′-TAATACGACTCACTATAGGGAGAGCCACCATGGGCCCCGAGGGGGAGGCC-3′ and REVExo2 5′-TTACAGATCCTCTTCTGAGATGAGTTTTTGTTCCTTCACCTTCTTAGCTTTCTTCTGCTTAAATTC-3′; FORPol1 5′-TAATACGACTCACTATAGGGAGAGCCACCATGAAGGAACCAGCCACAGCCAGC-3′ and REVPol1 5′-TTACAGATCCTCTTCTGAGATGAGTTTTTGTTCCTCACTGCCTACTCGGTCAGG-3′; FORPol2 5′-TAATACGACTCACTATAGGGAGAGCCACCATGTTGAAAGCCATGGTGCAGGCC-3′ and REVPol2 5′-TTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGTGCTATGGTCCAGGCTGGCTTCG-3′. The PCR products were TA cloned into a pCR2.1 vector (Invitrogen). The resulting constructs were confirmed by DNA sequencing. Each construct was linearized following restriction digest with NotI. Following restriction digestion, 1–2 μg of each was used separately in T7-coupled rabbit reticulocyte lysate in vitro transcription/translation system mix (Promega) performed in the presence of 35S-radiolabelled methionine (Amersham) for 90 min at 30 °C. Co-immunoprecipitation reactions were set up by incubating 20 μl of each of the DNA ligase III and PolG transcription/translation products in a final volume of 500 μl with co-immunoprecipitation buffer [20 mM Tris/HCl (pH 8.0), 100mM NaCl, 1mM EDTA, 0.1% Nonidet P40, 1 mM DTT, 1 mM PMSF and 10% glycerol]. The above mixture was rocked at 4 °C for 1–2 h and subsequently 5 μg of an anti-HA monoclonal rat antibody (Roche) or an anti-myc monoclonal antibody (Roche) was added. After another 1 h incubation, 10 μl of a 50% slurry of Protein G–agarose beads were added and the mix incubated overnight at 4 °C on a rocker. The following day, the beads were sedimented and washed three times with co-immunoprecipitation buffer. The washed beads were boiled for 5 min in the presence of SDS/PAGE loading dye containing 2-mercaptoethanol and resolved on a 4–20% SDS/PAGE gradient gel (Bio-Rad). The gel was fixed in 10% acetic acid, dried and exposed to a phosphorimager screen.
In vitro BER assay
This was performed by incubating 2 pmol of a uracil-containing 30-mer oligonucleotide duplex with 30 μg of mitochondrial protein extracts in the presence of buffer containing 100 mM Tris/HCl (pH 7.5), 75 mM KCl, 1 mM DTT, 5 mM MgCl2, 0.1 mM EDTA, 0.2 mg/ml BSA, 20 μM each of dATP, dGTP and dTTP, 2 mM ATP, 25 mM phosphocreatine and 50 μg/ml phosphocreatine kinase (10 units). The radiolabelled nucleotide for the reaction was 10 μCi of [α-32P]dCTP. The sequences of the two strands of the 30 mer oligonucleotide duplex were: 5′-CCCAGTCACGAUGTTGTAAAACGACGGCAA-3′ and 5′-TTGCCGTCGTTTTACAACGTCGTGACTGGG-3′. The reaction was incubated for 4 h at 32 °C and terminated by addition of an equal volume of phenol/chloroform and the mixture was then centrifuged. The aqueous layer was taken and DNA was precipitated using 2.5 vol. of ethanol. The DNA was then loaded on to a 22.5% polyacrylamide/7 M urea SDS/PAGE gel with loading dye (95% formamide, 20 mM EDTA, 0.1% Xylene cyanol FF and 0.1% Bromophenol Blue). The radioactivity on the gel was detected using a PhosphorImaging scanner and quantitated using IP-Lab gel software.
DNA pull-down assay
The nicked DNA substrate (1 pmol) on M13mp18 single-stranded DNA was incubated with 5 μl each of the in vitro transcription/translation product of PolG and either WT-mtDNA ligase III or ΔZf-mtDNA ligase III with constant shaking for 1 h at 4 °C in co-immunoprecipitation buffer (a final volume of 500 μl). Next, 5 μg of an anti-single-stranded DNA monoclonal antibody (Chemicon) was added to the mixture and incubated for another 1 h at 4 °C. Later, 10 μl of a 50% Protein G–agarose beads slurry was added and the immunoprecipitation carried out for 4 h with constant shaking at 4 °C. Beads were washed 3 times with co-immunoprecipitation buffer, boiled and loaded on to a SDS/PAGE gel. Radioactivity was detected using phosphor scanner imaging.
RESULTS
Co-immunoprecipitation of mtDNA ligase III and PolG from mitochondrial protein extracts
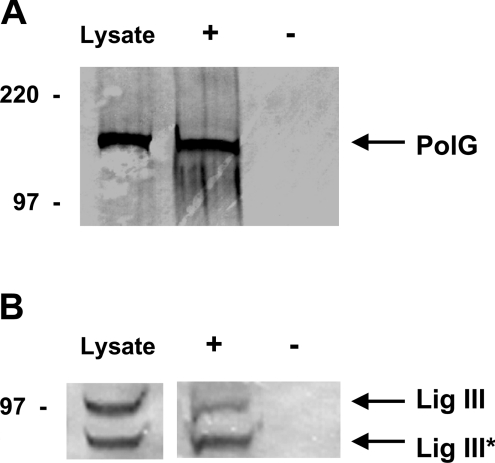
We hypothesized that PolG and mtDNA ligase III proteins associate in mammalian mitochondria. To test this hypothesis, monoclonal DNA ligase III antibody was used to precipitate DNA ligase III from mitochondrial protein extracts and the resulting pellet was resolved by SDS/PAGE and subjected to Western blot analysis using an anti-PolG antibody. Western blot analysis of mitochondrial protein extracts using the anti-PolG antibody revealed the presence of the 140 kDa protein corresponding to PolG (Figure 1A). The PolG protein was also detected in the immunoprecipitate produced using an anti-DNA ligase III antibody (Figure 1A). When a parallel immunoprecipitation was performed in the absence of a specific antibody, PolG was not detected in the pellet (Figure 1A). Additional experiments performed using a rabbit anti-Rad50 polyclonal antibody (Novus Biologicals) also failed to precipitate PolG (results not shown). Taken together, these results indicate that mtDNA ligase III and PolG associate in mitochondrial protein extracts. To confirm this finding, an analogous experiment was performed in which an anti-PolG antibody was used in the immunoprecipitation step, and an anti-DNA ligase III antibody was used in the Western blot reaction. The anti-DNA ligase III antibody recognizes two proteins in mitochondrial protein extracts (Figure 1B). The upper band, labelled Lig III in Figure 1(B), corresponds to full-length mitochondrial DNA ligase III, and the lower band, labelled Lig III* in Figure 1(B), corresponds to a C-terminal-truncated proteolytic fragment of DNA ligase III. Both forms of DNA ligase III were co-precipitated by the anti-PolG antibody (Figure 1B). When the anti-PolG antibody was omitted from the precipitation reaction, no anti-DNA ligase III immunoreactive proteins were detected in the pellet (Figure 1B). Taken together, the findings presented in Figure 1 demonstrate that the DNA ligase III and PolG proteins associate in mitochondrial protein extracts.
Figure 1. Co-immunoprecipitation of DNA ligase III and DNA polymerase γ from mitochondrial protein extracts.
Immunoprecipitation reactions were performed on mitochondrial protein extracts from HT1080 cells, as described in the Experimental procedures section, in the presence (+) or absence (−) of anti-DNA ligase III (A) or anti-PolG (B) antibodies. Precipitated material, as well as 30 μg of the untreated extract (Lysate), were resolved by SDS/PAGE and Western blots performed using anti-PolG (A) or anti-DNA ligase III (B) antibodies. Arrows indicate mobility of the 140 kDa PolG (A) and the full-length (100 kDa, Lig III) and C-terminal-truncated (80 kDa, Lig III*) DNA ligase III proteins (B). The relative mobilities of protein size standards are shown in kDa.
Co-immunoprecipitation of human recombinant DNA ligase III and PolG in vitro
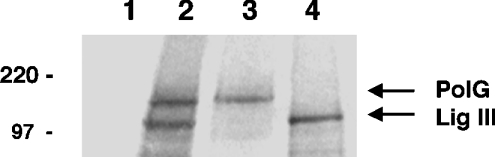
While the mtDNA ligase III and PolG proteins form a complex, the data in Figure 1 failed to address whether the interaction of the two proteins is dependent on the presence of additional mitochondrial proteins. To address this question, recombinant [35S]methionine-labelled PolG and DNA ligase III proteins were produced in vitro using a linked transcription/translation system. The two radioactive proteins, resolved by SDS/PAGE, are depicted in Figure 2, lanes 3 and 4 respectively. Immunoprecipitation experiments were performed on these proteins using an anti-HA antibody, which recognizes an HA epitope present on the C-terminus of the recombinant DNA ligase III protein. The anti-HA antibody precipitated both the DNA ligase III and PolG proteins when they were mixed prior to addition of the antibody (Figure 2, lane 2). In contrast, the anti-HA antibody did not precipitate PolG when DNA ligase III was omitted from the reaction (Figure 2, lane 1). This result shows that the recombinant human DNA ligase III and PolG proteins are capable of interacting in vitro, suggesting that the association of the two proteins in mitochondrial protein extracts involves a direct interaction. However, it is conceivable that the interaction between the two proteins may be bridged or mediated through DNA or another protein found in the protein extracts and in the in vitro translation system.
Figure 2. Co-immunoprecipitation of in vitro transcribed/translated PolG with HA-tagged DNA ligase III.
Immunoprecipitation reactions were performed using an anti-HA antibody, and the precipitated materials resolved by SDS/PAGE. Lane 1, PolG protein alone; lane 2, PolG protein plus HA-tagged DNA ligase III. The in vitro transcribed/translated PolG (lane 3) and DNA ligase III (lane 4) proteins were separately resolved by SDS/PAGE to serve as molecular mass markers. The relative mobilities of protein size standards are shown in kDa.
A central domain of DNA ligase III interacts with PolG in vitro
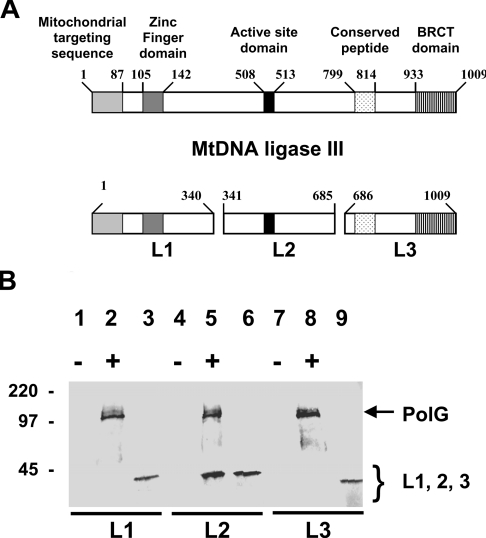
A series of experiments were performed to determine the domains of the two proteins that are responsible for their ability to interact in vitro. The in vitro transcription/translation system was used to prepare radioactive recombinant full-length PolG as well as a number of labelled sub-fragments of DNA ligase III, referred to as L1 (Figure 3B, lane 3; residues 1–340), L2 (Figure 3B, lane 6; residues 341–685) and L3 (Figure 3B, lane 9; residues 686–1009). The structure of the respective fragments of DNA ligase III are depicted schematically in Figure 3(A). Previously identified regions of the DNA ligase III protein corresponding to (from N-terminal to C-terminal ends of the protein) the mitochondrial targeting sequence, the zinc-finger domain, the active-site domain, the conserved peptide and the BRCT (BRCA1 protein C-terminal) domain are indicated by shading. In these experiments, the full-length PolG recombinant protein included a C-terminal myc epitope tag. Incubation of the full-length PolG with the DNA ligase III fragments individually followed by immunoprecipitation with an anti-myc monoclonal antibody was used to determine which fragment(s) of DNA ligase III were capable of interacting with PolG. These studies revealed that the L2 fragment of DNA ligase III co-immunoprecipitated with the full-length PolG (Figure 3B, lane 5). In contrast, we were unable to detect the L1 or L3 fragments of DNA ligase III in immmunoprecipitates formed by the anti-myc monoclonal antibody (Figure 3B, lanes 2 and 8 respectively). Taken together, these findings suggest that the PolG-interaction domain of DNA ligase III resides in the central portion of the molecule. Interestingly this central domain of DNA ligase III also harbours the KYDGER motif, the active-site consensus sequence of ATP-dependent DNA ligases.
Figure 3. The central region of DNA ligase III interacts with PolG.
(A) Schematic representation of the domain structure of the mtDNA ligase III protein. Numbers indicate amino acid residues. The shaded boxes represent previously identified domains of the DNA ligase III protein as indicated. L1, L2 and L3 refer to three fragments of mtDNA ligase III protein produced by in vitro transcription/translation reactions. (B) Immunoprecipitation reactions were performed on these proteins in the presence (+) or absence (−) of in vitro transcribed/translated myc-tagged PolG using an anti-myc antibody, and the precipitated material resolved by SDS/PAGE. Lanes 2, 5 and 8 depict the products of immunoprecipitation reactions performed on mixtures of PolG with the L1, L2 and L3 fragments respectively of DNA ligase III. Lanes 1, 4 and 7 depict control immunoprecipitation reactions performed on the L1, L2 and L3 proteins in the absence of PolG. The L1 (lane 3), L2 (lane 6) and L3 (lane 9) fragments of DNA ligase III were separately resolved by SDS/PAGE, and their respective mobilities depicted by the arrows. The relative mobilities of protein size standards are shown in kDa.
The C-terminal fragment of PolG interacts with DNA ligase III in vitro
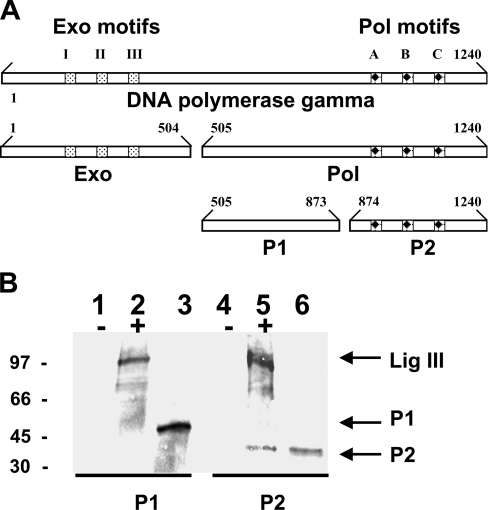
A similar approach was used to generate radiolabelled fragments of PolG (depicted schematically in Figure 4A) to determine which domain of this protein interacts with recombinant DNA ligase III in vitro. Initially, two fragments of PolG were produced. The first, referred to as Exo, contains the exonuclease domain of the protein (Figure 4A, residues 1–504), whereas the second, referred to as Pol, contains the polymerase motifs (Figure 4A, residues 505–1240). Co-immunoprecipitation experiments, analogous to those described above, demonstrated that full-length DNA ligase III associated with the Pol fragment of PolG, but not with the Exo fragment (results not shown). A subsequent series of experiments were performed to determine whether the P1 (Figure 4B, lane 3; residues 505–873) or P2 fragment (Figure 4B, lane 6; residues 874–1240) of PolG could interact with full-length recombinant DNA ligase III which harboured an HA tag at its C-terminus. To address this question radioactive sub-fragments of the Pol domain of PolG were prepared using the in vitro transcription/translation system. Subsequent immunoprecipitation experiments revealed that the P2 fragment interacted with DNA ligase III, whereas the P1 fragment did not (Figure 4B, compare lane 5 with lane 2). The data presented in Figures 3 and 4 together support the interpretation that interactions between amino acid residues present within the central portion of DNA ligase III and the C-terminal region of PolG are responsible for the association of the two proteins in vitro. Interestingly, these portions of the molecules contain the conserved catalytic DNA ligase and DNA polymerase motif domains of the two respective proteins.
Figure 4. The C-terminal fragment of PolG interacts with DNA ligase III.
(A) Schematic representation of the domain structure of the PolG protein. Numbers indicate amino acid residues. Exo, Pol, P1 and P2 refer to the fragments of PolG protein that were transcribed via in vitro transcription/translation. (B) Immunoprecipitation reactions were performed on the P1 and P2 fragments of the Pol domain of PolG in the presence (+) or absence (−) of in vitro transcribed/translated DNA ligase III protein using anti-DNA ligase III antibody, and the precipitated material resolved by SDS/PAGE. Lanes 2 and 5 depict the products of immunoprecipitation reactions performed on mixtures of DNA ligase III with the P1 and P2 fragments of PolG respectively. Lanes 1 and 4 depict control immunoprecipitation reactions performed on the P1 and P2 fragments of PolG respectively, in the absence of DNA ligase III. The P1 (lane 3) and P2 (lane 6) fragments of PolG were separately resolved by SDS/PAGE, and their respective mobilities depicted by the arrows. The relative mobilities of protein size standards are shown in kDa.
Characterization of cells expressing mutant mtDNA ligase III
A non-functional version of DNA ligase III targeted to the mitochondria in cultured human cells would presumably interact with PolG, producing an inert complex that could potentially interfere with mtDNA repair function. As an initial step towards testing this hypothesis, site-directed mutagenesis was used to create recombinant DNA ligase III minigenes encoding amino acid substitutions within the conserved domain of that molecule. One clone encoded a lysine to valine residue substitution (termed K-V) at position 421 and the other encoded an arginine to histidine residue substitution (termed R-H) at position 426 in the active-site domain of DNA ligase III. Previous reports [28–31] have indicated that these mutations destroy DNA ligase III catalytic activity. The human recombinant HA-tagged K-V and R-H variant mtDNA ligase III proteins were synthesized using the in vitro transcription/translation system and co-immunoprecipitation studies, similar to those described above, were performed to determine whether they would interact with PolG in vitro. Our results confirmed that both the K-V and R-H variants of DNA ligase III interact with PolG protein in vitro (results not shown).
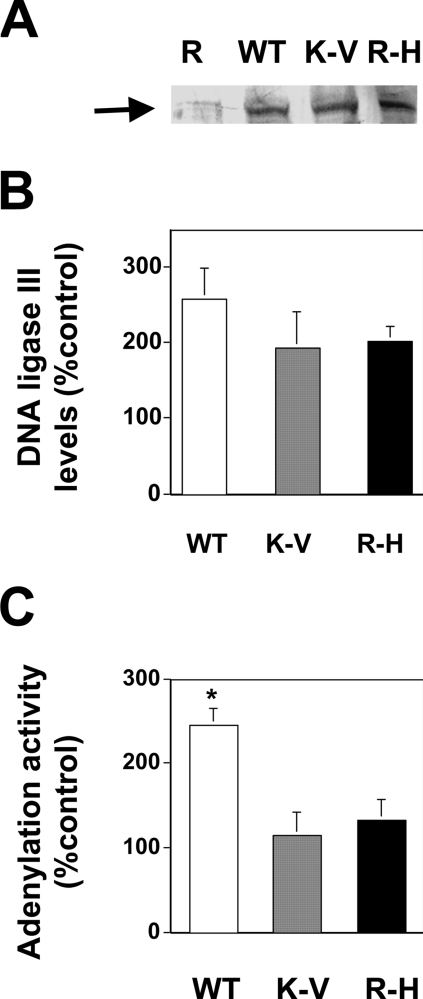
Expression constructs encoding mitochondria-targeted wild-type (WT) and catalytically inactive (K-V and R-H) variants of DNA ligase III proteins were prepared as described in the Experimental procedures section. In all three cases, the coding sequence of the DNA ligase III cDNA was mutated to destroy the ATG codon essential for nuclear DNA ligase III production. Previous studies [7] have confirmed that this modification results in mitochondria-specific DNA ligase III expression, thereby ensuring that any phenotypes observed result from the expression of these modified DNA ligase III proteins within the mitochondrial compartment. The expression constructs were introduced by electroporation into HT1080 cells, and the resulting clones are referred to hereafter as WT, K-V and R-H. In addition, cells were transfected with an unmodified pREP4 vector to create a control clone named REP. Mitochondrial protein extracts were prepared from these cells and resolved by SDS/PAGE. Figure 5(A) depicts a representative Western blot analysis using an anti-HA antibody. A band corresponding to HA-tagged mtDNA ligase III was detected in the extracts prepared from cells transfected with the recombinant mtDNA ligase III constructs. Additional Western blots were performed on multiple sets of mitochondrial protein extracts from these cell lines using an antibody that recognized both the endogenous and transgene-encoded DNA ligase III proteins. The data in Figure 5(B) indicate that the three transgenic cell lines express similar levels of mtDNA ligase III protein. These data further reveal that the levels of mtDNA ligase III expressed in the three cell lines are approximately twice that detected in control cells, consistent with the interpretation that there are roughly equal levels of endogenous and transgene-encoded mtDNA ligase III proteins in these cells. (The apparent differences in DNA ligase III protein levels in the WT, K-V, and R-H cell lines depicted in Figure 5B are not statistically significant.)
Figure 5. Transgene-encoded DNA ligase III protein detected in mitochondrial protein extracts.
(A) Western blot analysis using anti-HA antibody was performed on 10 μg of mitochondrial protein extracts from REP (R), WT, K-V, and R-H cells that had been resolved by SDS/PAGE. The arrow indicates the presence of the HA-tagged DNA ligase III trans-proteins. (B) Western blots identical with that portrayed in (A) were performed on three independently prepared sets of mitochondrial protein extracts from the REP, WT, K-V and R-H cells. The relative intensities of the anti-DNA ligase III immunoreactivity (comprised of both the endogenous and transgenic proteins) present in the WT, K-V and R-H cell extracts were determined relative to that present in the REP cell extracts using densitometric analysis. Results depict the means±S.E.M. (C) Mitochondrial protein extracts (20 μg) were incubated with [α-32P]ATP and resolved by SDS/PAGE. Scanning densitometric analysis was used to determine the amount of adenylated product formation relative to that in REP cell extracts. The graph depicts the average of two independent experiments±S.D. *P<0.05 compared with REP.
ATP-dependent DNA ligases form stable covalent radioactive adenylated products following exposure to [α-32P]ATP, and previous studies [24] have shown that mtDNA ligase III is the only protein in mitochondrial extracts that becomes adenylated following incubation with [α-32P]ATP. Thus one can quantitate the amount of catalytically active DNA ligase III present in mitochondrial protein extracts by measuring adenylation activity. This analysis revealed that adenylation activity in mitochondrial protein extracts from WT cells was roughly twice that detected in extracts from REP cells (Figure 5C). As anticipated, adenylation activity in mitochondrial protein extracts prepared from cells expressing the catalytically inactive K-V and R-H variants of DNA ligase III were indistinguishable from those present in extracts from control cells (Figure 5C).
Transgene-encoded DNA ligase III proteins bind PolG
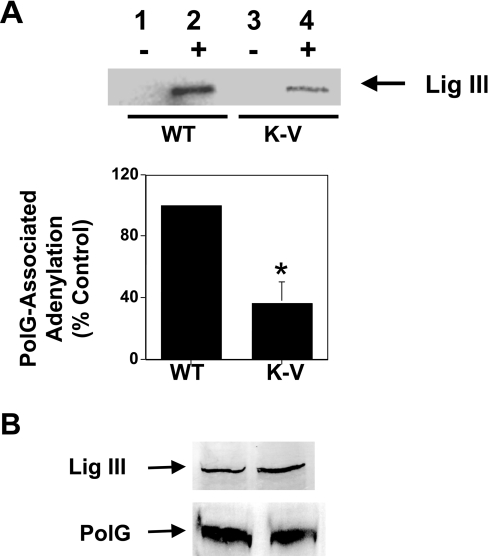
The data presented above suggest that both wild-type and catalytically inert DNA ligase III proteins would interact with PolG in vivo. Since the latter proteins lack function, one would expect that examination of adenylation activity associated with PolG would be substantially reduced in cells expressing non-functional DNA ligase III transgene, compared with that present in cells expressing a functional DNA ligase III transgene. To test this hypothesis, anti-PolG antibody was used to perform immunoprecipitation reactions on mitochondrial protein extracts from cells expressing either the WT or K-V variants of DNA ligase III, and the relative levels of adenylated DNA ligase III present in the pelleted material was determined. As the data in the upper panel of Figure 6(A) reveal, considerably more adenylated DNA ligase III protein was precipitated from mitochondrial protein extracts prepared from cells expressing WT mtDNA ligase III than was precipitated from extracts expressing the K-V variant of DNA ligase III (compare lane 2 with lane 4). As expected, no adenylation activity was precipitated when the anti-PolG antibody was omitted from the reaction (Figure 6A, upper panel, lanes 1 and 3).
Figure 6. Transgene-encoded DNA ligase III binds to PolG.
(A) Upper panel: immunoprecipitation reactions were performed on mitochondrial protein extracts from WT (lanes 1 and 2) and K-V (lanes 3 and 4) cells in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of an anti-PolG antibody. Adenylation reactions were performed on the resuspended pellets using [α-32P]ATP and the proteins subsequently resolved by SDS/PAGE. The arrow represents the mobility of the 100 kDa DNA ligase III molecule. Lower panel: a series of three immunoprecipitation/adenylation experiments were performed on mitochondrial protein extracts from WT and K-V cells. Each experiment was performed on independently prepared sets of extracts from the two cell lines, and scanning densitometry was used to calculate the relative level of PolG-associated adenylation activity present in the mitochondrial extract from K-V cells compared with that present in mitochondrial extract from WT cells. The graph depicts the means±S.E.M. (n=3), *P<0.05. (B) Immunoprecipitates formed using an anti-PolG antibody were resolved by SDS/PAGE and Western blot analysis performed using anti-DNA ligase III (upper panel) or anti-PolG (lower panel) antibodies. The mobilities of these respective proteins are indicated by the arrows.
To determine whether the difference in the amount of adenylation activity present in mitochondrial protein extracts from WT and K-V cells was statistically significant, the experiment depicted in the upper panel of Figure 6(A) was performed on three independently prepared mitochondrial protein extracts prepared from each of the two cell lines. Scanning densitometry was used to quantitate the amount of adenylation activity present in the immunoprecipitates from K-V cell-derived mitochondrial protein extracts, compared with that present in similar precipitates prepared from WT cell-derived extracts. This analysis (Figure 6A, lower panel) revealed that the amount of catalytically active DNA ligase III associated with PolG in mitochondrial protein extracts from K-V cells is approx. 40% of that observed in extracts from WT cells. These data support the interpretation that both the catalytically inert and wild-type DNA ligase III proteins are able to bind to PolG in vivo. If this interpretation is correct, whereas levels of adenylation activity vary in the immunoprecipitates prepared from the different cell lines, levels of mtDNA ligase III immunoreactivity should not vary because the anti-DNA ligase III antibody would recognize both the wild-type and mutant versions of DNA ligase III. To test this hypothesis, Western blot analysis using an anti-DNA ligase III antibody was performed on immunoprecipitation pellets generated using an anti-PolG antibody. As the results in the upper panel of Figure 6(B) illustrate, the levels of mtDNA ligase III co-precipitated by the anti-PolG antibody from mitochondrial protein extracts from WT and K-V cells were quite similar. Analogous experiments confirmed that similar levels of PolG antigen were also precipitated from the two different extracts (Figure 6B, lower panel). These results indicate that both the WT and K-V forms of mtDNA ligase III are capable of interacting with PolG.
Cells expressing non-functional DNA ligase III have reduced mtDNA copy number
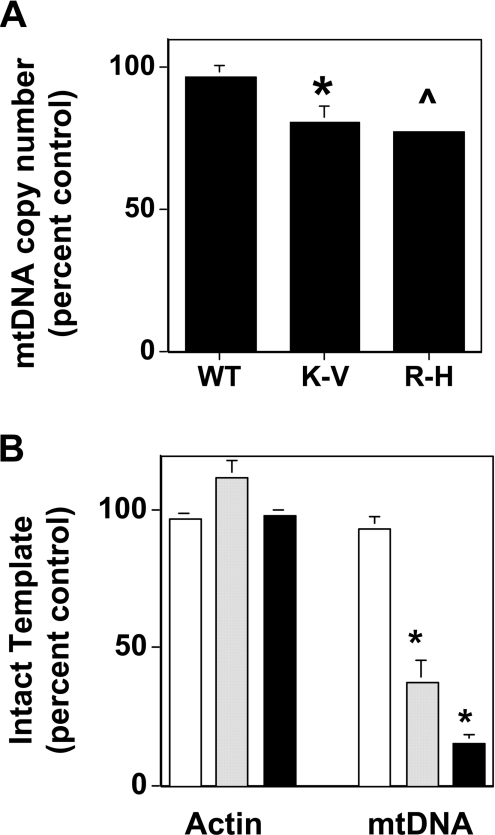
Previous studies have shown that overexpression of non-functional alleles of PolG in cultured cells exerts a dominant effect, resulting in decreased mtDNA copy number, increased accumulation of mtDNA mutations and decreased oxygen consumption [32,33]. Since the non-functional K-V variant of DNA ligase III binds to PolG in vivo, we hypothesized that these cells would have diminished PolG function, resulting in reduced mtDNA integrity. To test this hypothesis, Southern blot analysis was performed on genomic DNA samples isolated from the transgenic cells described above. The radiolabelled probe used selectively hybridizes to an approx. 9.4 kb EcoRI fragment of the mitochondrial genome, and the intensity of the hybridization signal provides a measure of the relative mtDNA copy number in transgenic cells. To ensure that each lane was loaded with DNA from an equivalent number of cells, samples were quantitated using the diphenylamine assay. In addition, scanning densitometry was performed on ethidium bromide-stained gels to confirm that each lane contained an identical amount of genomic DNA. In cultured fibroblast cell lines, mtDNA accounts for less than 1% of total genomic DNA. Thus even significant alterations in cellular mtDNA copy number would have a negligible effect on the total amount of DNA present per cell. By ensuring that each sample contained the same amount of total cellular DNA, we used the respective strength of hybridization signals to a mtDNA probe present in each lane to determine the relative mtDNA copy number per cell line.
Whereas the steady-state level of mtDNA in WT cells was indistinguishable from that in the control REP cells, the mtDNA copy number in both K-V and R-H cells was reduced by approx. 20% compared with that present in the control cells (Figure 7A). Additional experiments revealed that steady-state mtDNA levels in REP cells were indistinguishable from those detected in non-transfected cells (results not shown). While these effects are relatively modest, the reduced steady-state levels of mtDNA observed in K-V and R-H cells compared with that in REP cells were statistically significant (P<0.05 and P<0.005 respectively). Since DNA ligase III is the only known mtDNA ligase, it should play an important role in mtDNA replication. Previous studies showing that the depletion of mtDNA ligase III through an anti-sense mRNA strategy led to a similar decrease in mtDNA copy number as observed through Southern blot analysis [24]. The present study attempting to disrupt mtDNA ligase III function through a dominant negative strategy support this earlier finding.
Figure 7. Altered mtDNA copy number and integrity in K-V and R-H cells.
(A) Genomic DNA (1 μg) from transgenic cells was digested with EcoRI and resolved by electrophoresis prior to Southern blot analysis with a mtDNA specific probe that hybridizes to a 9.4 kb EcoRI fragment. Scanning densitometry was used to determine the mean relative intensity (±S.E.M.) of the respective hybridization signals compared with that obtained from the REP cell sample. n≥4, *P<0.05 compared with REP, and ˇP<0.005 compared with REP. (B) qRT-PCR was performed on three independent genomic DNA samples from REP, K-V and R-H cell lines to amplify a 547 base pair mtDNA fragment and a 274 base pair nuclear β-actin gene fragment. Serial dilutions of genomic DNA were used to construct a standard curve for quantitation, and the relative percentage of mtDNA and nuclear DNA in the cell-lines were calculated from the standard curve. Open bars, REP; grey bars, K-V; and solid bars, R-H. Bars represent means±S.E.M. for experiments performed in triplicate. *P<0.05 compared with REP.
The integrity of mtDNA in control and transgenic cell-lines was determined using qRT-PCR. Failure to amplify product during qRT-PCR can result from either diminished amounts of template DNA, or from reduced levels of intact template DNA [34,35]. Our prediction was that if the residual mtDNA from the K-V and R-H cells harboured DNA lesions, such as single-strand nicks, there would be substantially less PCR product compared with REP cells when mtDNA from these cells was amplified.
Amplifications using PCR primers specific for a 547 base pair fragment of the mitochondrial genome, revealed that the K-V and R-H cells had between one-third and one-fifth as much intact mtDNA template as did the REP cells (Figure 7B). It is noteworthy that the level of mtDNA template in WT cells was indistinguishable from that present in REP cells. Experiments performed using PCR primers specific for a fragment of the nuclear β-actin gene revealed that all of the transgenic cell lines possessed virtually identical levels of nuclear DNA template (there was no statistically significant difference). Taken together, the results presented in Figure 7 indicate that expression of the catalytically inert DNA ligase III leads to an approx. 20% decrease in mtDNA copy number, and that the residual mtDNA in these cells harbour lesions that interfere with its ability to be amplified by PCR. It is noteworthy that expression of catalytically inactive mtDNA ligase III has no effect on the copy number or integrity of a nuclear gene.
Recombinant DNA ligase III protein enhances the interaction of PolG with nicked DNA
Experiments have revealed that the relatively low affinity of free PolG protein for DNA is dramatically enhanced through interaction with its accessory subunit [36]. Interestingly, however, the accessory protein is not required to reconstitute mitochondrial BER in vitro [9]. Based on the knowledge that DNA ligase III has a zinc-finger domain that binds avidly to nicked DNA [37], we examined whether DNA ligase III could facilitate the interaction of PolG with a nicked DNA repair substrate. Recombinant human PolG protein was incubated with nicked DNA substrate and a DNA pull-down assay was performed using an anti-single-stranded DNA monoclonal antibody (see Experimental procedures section). Resolution of the immunoprecipitate by SDS/PAGE revealed that, as expected based on previous results [36], negligible amounts of PolG were precipitated (results not shown). Next we performed an experiment to determine whether inclusion of recombinant DNA ligase III protein would enhance the amount of PolG present in the immunoprecipitate. As the results in Figure 8 reveal, wild-type DNA ligase III (WT) and PolG that had been co-incubated with the nicked DNA repair substrate were efficiently precipitated during the DNA pull-down assay (PolG/WT). In contrast, when a similar experiment was performed using a modified version of DNA ligase III that lacked the zinc-finger domain (ΔZf-ligIII) in place of the wild-type DNA ligase III protein, neither that protein nor PolG were efficiently precipitated during the DNA pull-down assay (PolG/Zf). Control experiments revealed that ΔZf-ligIII interacts with PolG in vitro with an apparent affinity indistinguishable from that of the wild-type DNA ligase III (results not shown). These results support the conclusion that the association of PolG with DNA is mediated through its interaction with DNA ligase III, and also requires zinc-finger DNA-binding motif of DNA ligase III.
Figure 8. Recombinant WT-mtDNA ligase III, but not ΔZf-mtDNA ligase III, enhances the association of PolG with a nicked DNA substrate.
DNA pull-down reactions (see Experimental procedures section) were performed on mixtures containing in vitro transcribed/translated PolG in the presence of zinc-finger-deficient DNA ligase III (PolG/Zf) or wild-type DNA ligase III (PolG/WT), and the sedimented material was resolved by SDS/PAGE. Parallel samples containing recombinant PolG (PolG), recombinant zinc-finger-deficient DNA ligase III (Zf) or wild-type DNA ligase III (WT) were loaded directly on to the gel and similarly resolved. The relative mobilities of protein size standards are shown in kDa.
Mitochondrial protein extracts from ΔZf-ligIII cells have reduced in vitro BER activity compared with WT cells
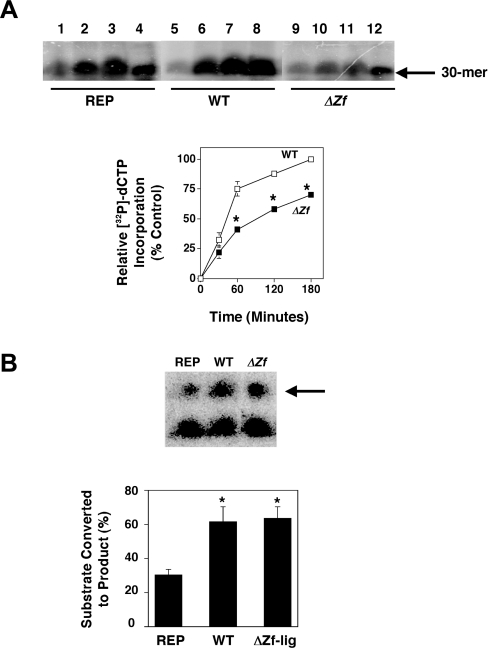
The data in Figure 8 suggest that DNA ligase III may function during BER to help PolG interact with DNA repair substrates, and that the presence of non-functional complexes of ΔZf-ligIII/PolG may inhibit BER. To test this hypothesis, we constructed a zinc-finger-deficient mtDNA ligase III minigene and overexpressed it in HT1080 cells. Mitochondrial protein extracts were prepared from transgenic cells overexpressing the ΔZf-ligIII, and WT versions of mtDNA ligase III and from control cells transfected with the empty vector, and these extracts were used to perform a series of in vitro BER assays. The upper panel in Figure 9(A) depicts the results from a representative time-course experiment in which product formation was determined at 30 min (lanes 1, 5 and 9), 60 min (lanes 2, 6 and 10), 120 min (lanes 3, 7 and 11) and 180 min (lanes 4, 8 and 12). To determine whether the reduced levels of product formation observed in the extract derived from ΔZf-ligIII cells was statistically significant, a series of BER experiments were performed using independently prepared sets of extracts from the REP, WT and ΔZf-ligIII cells. Product formation was quantified as described in the Experimental procedures section, and normalized to that obtained with the mitochondrial protein extracts from REP cells at 180 min. (This point was chosen because it represents the time at which product formation plateaus, results not shown.) The results from this analysis are presented in the lower panel of Figure 9(A). These data indicate that mitochondrial protein extracts prepared from cells expressing the ΔZf-ligIII protein had significantly less BER activity than did extracts from cells expressing the wild-type mtDNA ligase III. Analysis of the slope of the product formation curves between zero and 60 min reveals that BER activity in mitochondrial extracts prepared from the ΔZf-ligIII is reduced by approx. 50%, compared with BER activity present in extracts from control cells. Interestingly, the BER activity present in mitochondrial extracts prepared from the cells overexpressing wild-type mtDNA ligase III was not significantly different from that detected in similar extracts prepared from vector-control cells.
Figure 9. In vitro BER activity is reduced in mitochondrial protein extracts from ΔZf-lig cells, but DNA nick seal activity is not.
(A) Upper panel: in vitro BER time course experiments were performed on mitochondrial protein extracts derived from REP (lanes 1–4), WT (lanes 5–8) and ΔZf-lig (lanes 9–12) cell lines as described in the Experimental procedures section. Lanes 1, 5 and 9 represent 30 min; lanes 2, 6 and 10 represent 60 min; lanes 3, 7 and 11 represent 120 min; and lanes 4, 8 and 12 represent 180 min time points respectively. Lower panel: three independent mitochondrial protein extracts were prepared from REP, WT and ΔZf-lig cell lines and in vitro BER time course experiments performed as above. The amount of radioisotope incorporation was determined by densitometric analysis of autoradiographs of urea/polyacrylamide gels and normalized to that obtained with the mitochondrial protein extracts from vector control (REP) cells at 180 min. Results depict means±S.E.M. *P<0.05 compared with WT. (B) DNA nick-sealing assays were performed as described in the Experimental procedures section, and the reaction products were resolved on polyacrylamide/7 M urea gels. Upper panel: results from a representative experiment. Arrow indicates the formation of the nick sealed product. Lower panel: three independent mitochondrial protein extracts were prepared from REP, WT and ΔZf-lig cell lines and in vitro nick-sealing experiments performed as above. The amount of product produced (depicted as the percentage of total substrate converted into product) was determined by scanning densitometry performed on autoradiographs. Results depict the mean percentage of substrate converted into product±S.D. *P<0.05 compared with REP.
An additional analysis was performed to determine the levels of DNA nick-sealing activity in mitochondrial protein extracts from REP, WT and ΔZf-ligIII cells. Results from a representative nicksealing experiment (Figure 9B, upper panel) suggested that nick-sealing activity was elevated in both the WT and ΔZf-ligIII cell-derived mitochondrial extracts compared with that present in an extract from REP cells. A more extensive series of experiments were performed on multiple sets of independently prepared mitochondrial extracts from these cells. The results of these experiments (Figure 9B, lower panel) revealed that mitochondrial protein extracts from the WT and ΔZf-ligIII cells contained identical levels of DNA nick-sealing activity, and that the percentage of nicked substrate converted into product following exposure to extracts from these cells (approx. 60%) was approximately twice that observed in mitochondrial protein extracts cells prepared from cells transfected with the empty pREP4 vector (approx. 30%). These data are consistent with previous reports (see the Discussion section) that the zinc-finger domain of DNA ligase III is not required for efficient ligation of nicked DNA substrates. Taken together, these data support a model in which mitochondrial genome stability is dependent on the ability of DNA ligase to interact with PolG and form a functional complex. Within the mitochondria of cells expressing the ΔZf-ligIII allele, this protein presumably competes with endogenous DNA ligase III for binding to PolG. The complex formed by the ΔZf-ligIII and PolG proteins is substantially less able to catalyse BER, and possibly other functions including DNA replication, than is a complex of PolG and wild-type DNA ligase III. Consistent with this hypothesis, ΔZf-ligIII cells possess reduced steady-state levels of mtDNA (75% compared with control), and this residual DNA is inefficiently amplified by PCR (approx. 50% reduction in product formation compared with control), indicating the presence of damage to the mtDNA in these cells (results not shown). These values are similar to those obtained when similar analysis was performed on the K-V and R-H cells, which express catalytically inactive versions of mtDNA ligase III (see Figure 7).
DISCUSSION
The results from the present paper describe a novel interaction between DNA ligase III and PolG. We have shown that the proteins associate in mitochondrial protein extracts, and that the recombinant proteins are capable of interacting in vitro. Our analysis reveals that the regions of the two proteins that are required for this interaction include the DNA polymerase motifs A, B and C of PolG, and the region surrounding the conserved active site sequence KYDGER present on DNA ligase III. We have demonstrated that recombinant DNA ligase III facilitates the interaction of recombinant PolG with a gapped duplex DNA substrate in vitro, and that the DNA ligase III/PolG interaction is apparently required for efficient BER activity in vitro. Over-expressed functional and non-functional alleles of mitochondria-targeted DNA ligase III in cultured human fibroblasts were capable of interacting with PolG. Cells expressing non-functional DNA ligase III alleles had reduced mtDNA copy number and integrity, consistent with the interpretation that the DNA ligase III/PolG interaction plays an essential role in facilitating mitochondrial DNA repair and/or replication.
In recent years, considerable progress has been made in our understanding of DNA repair in the mitochondria. This has been particularly true in the case of mitochondrial BER. As was summarized in the introduction, it is now clear that many of the genes that encode nuclear BER enzymes also encode mitochondrial DNA repair enzymes. A partial list includes DNA ligase III, APE1 and APE2, and the DNA glycosylases OGG1, UDG (uracil DNA glycosylase) and hMYH (human MutY homologue) [1–7]. Interestingly, the DNA ligase III-binding partner Xrcc1 can not be detected in mammalian mitochondrial extracts [8]. The finding that there is no apparent defect in mtDNA repair in cells lacking functional Xrcc1 supports the view that this protein does not play an essential role in mitochondrial BER [8].
The absence of Xrcc1 from the mitochondria is surprising, given the critical role this protein plays in stabilizing DNA ligase III in the nucleus. It has also been postulated that Xrcc1 plays a critical role in both binding to nicked DNA substrates, and in possibly facilitating the interaction of the DNA polymerase β protein with BER substrates [13]. Others have proposed that Xrcc1 specifically binds to and stabilizes one conformation of DNA polymerase β, thereby making the complex of nicked DNA, Xrcc1 and DNA polymerase β more accessible to DNA ligase III [12].
Pinz and Bogenhagen [9] demonstrated that the purified mitochondrial proteins APE1, DNA ligase III and PolG are capable of repairing abasic sites on synthetic repair substrates. Intriguingly, Pinz and Bogenhagen showed that the accessory protein of PolG was not required for the repair reaction. This latter finding is particularly interesting because the affinity of PolG for DNA is quite low in the absence of its accessory protein [36]. Our data indicating that the presence of DNA ligase III greatly enhances the ability of recombinant PolG to bind a nicked duplex DNA substrate, suggests that the mtDNA ligase III protein functions to recruit PolG to DNA in a manner analogous to that of the accessory protein of PolG during DNA replication. Consistent with this interpretation, we found that BER activity in mitochondrial protein extracts prepared from cells overexpressing a zinc-finger-lacking DNA ligase III protein was dramatically reduced.
Our data are consistent with a model in which a complex of DNA ligase III and PolG is recruited to sites of DNA repair in the mitochondria by virtue of the DNA-binding zinc-finger motif of the former protein. Traditionally, a ‘passing the baton’ model has been proposed to explain the mechanism of BER [38–40]. According to this model, the enzymes required to catalyse the respective steps of BER sequentially interact with DNA in an assembly line fashion, with each enzyme catalysing one step in the overall reaction, and subsequently diffusing away, permitting the next repair enzyme to bind the substrate molecule and catalyse the next step in the reaction. However, our data are more consistent with a model in which the co-ordinated action of a multi-enzyme repair complex is essential for efficient BER. Our data suggest that a complex minimally comprised of the DNA ligase III and PolG proteins targets damaged DNA during BER. Our data do not indicate whether the DNA ligase III–PolG complex also contains other mitochondrial BER proteins, and our model does not address this. However, data from other laboratories suggest this may be the case. For example, it has been shown that human recombinant OGG1 had very low specific activities for excising the 8-oxoguanine damaged base and cleaving the abasic site [41]. However, the addition of stoichiometric amounts of human recombinant APE1 increased the specific activity by 5-fold. It will be of interest to examine whether the APE1 and OGG1 molecules co-immunoprecipitate with the DNA ligase III–PolG complex.
Numerous authors have noted that DNA ligase III is unique amongst eukaryotic DNA ligases in possessing a zinc-finger domain. Results from a number of papers indicate that while repair of some types of DNA damage appears to be dependent on the presence of the zinc-finger domain, repair of simple nicked substrates by the nuclear BER repair machinery does not require the zinc finger motif of the DNA ligase III molecule [42,37]. In contrast, the data contained in the present study indicate an essential role for the zinc-finger motif of DNA ligase III in mitochondrial BER.
The finding that the zinc-finger domain of DNA ligase III is essential for in vitro BER catalysed by mitochondrial protein extracts provides a possible explanation for the retention of the apparently dispensable zinc-finger domain of DNA ligase III. While this motif may play a role in facilitating the repair of clustered DNA lesions, or other complex DNA repair intermediates in the nucleus, our data suggest that retention of the zinc-finger motif of DNA ligase III is essential due to evolutionary pressure exerted by the mitochondria on the nucleus. It is therefore conceivable that loss of function of the zinc-finger motif would not exert a significantly deleterious influence on repair of the nuclear genome, but would dramatically interfere with the ability of the cell to repair the mitochondrial genome. It would be interesting to examine the phenotype of cells in which the zinc-finger-less DNA ligase III molecule was overexpressed in the nucleus, rather than, as in the present experiments, in the mitochondria. If our hypothesis were correct, one would predict that overexpressing this nucleus-targeted protein would have no effect on nuclear DNA repair in vivo, and that extracts prepared from these cells would have wild-type levels of BER activity.
Acknowledgments
This work was supported in part by a National Institutes of Health Grant AG16678. A. D. was supported by an American Heart Association Northland Affiliate Predoctoral fellowship.
References
- 1.Tell G., Crivellato E., Pines A., Paron I., Pucillo C., Manzini G., Bandiera A., Kelley M. R., Di Loreto C., Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat. Res. 2001;485:143–152. doi: 10.1016/s0921-8777(00)00068-9. [DOI] [PubMed] [Google Scholar]
- 2.Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsen H., Otterlei M., Haug T., Solum K., Nagelhus T. A., Skorpen F., Krokan H. E. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishioka K., Ohtsubo T., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohtsubo T., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izumi T., Tatsuka M., Tano K., Asano M., Mitra S. Molecular cloning and characterization of the promoter of the human N-methylpurine-DNA glycosylase (MPG) gene. Carcinogenesis. 1997;18:1837–1839. doi: 10.1093/carcin/18.9.1837. [DOI] [PubMed] [Google Scholar]
- 7.Lakshmipathy U., Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakshmipathy U., Campbell C. Mitochondrial DNA ligase III function is independent of XRCC1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinz K. G., Bogenhagen D. F. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell. Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longley M. J., Ropp P. A., Lim S. E., Copeland W. C. Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 11.Caldecott K. W., Tucker J. D., Stanker L. H., Thompson L. H. Characterization of the XRCC1–DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota Y., Nash R. A., Klungland A., Schar P., Barnes D. E., Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 13.Caldecott K. W., Aoufouchi S., Johnson P., Shall S. XRCC1 polypeptide interacts with DNA polymerase β and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marintchev A., Mullen M. A., Maciejewski M. W., Pan B., Gryk M. R., Mullen G. P. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat. Struct. Biol. 1999;6:884–893. doi: 10.1038/12347. [DOI] [PubMed] [Google Scholar]
- 16.Rice P. A. Holding damaged DNA together. Nat. Struct. Biol. 1999;6:805–806. doi: 10.1038/12257. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Jannotti R. M., Klein S. M., Bogenhagen D. F. Two forms of mitochondrial DNA ligase III are produced in Xenopus laevis oocytes. J. Biol. Chem. 2001;276:48978–48987. doi: 10.1074/jbc.M107177200. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 19.Keown W. A., Campbell C. R., Kucherlapati R. S. Methods for introducing DNA into mammalian cells. Methods Enzymol. 1990;185:527–537. doi: 10.1016/0076-6879(90)85043-n. [DOI] [PubMed] [Google Scholar]
- 20.Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Rickwood D., Wilson M. T., Darley-Usmar V. M. Isolation and characteristics of intact mitochondria. In: Darley-Usmar V. M., Rickwood D., Wilson M. T., editors. Mitochondria: a Practical Approach. IRL Press Oxford; 1987. pp. 1–16. [Google Scholar]
- 22.Bradford M. M. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Barker D. G., Johnson A. L., Johnston L. H. An improved assay for DNA ligase reveals temperature-sensitive activity in cdc9 mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 1985;200:458–462. doi: 10.1007/BF00425731. [DOI] [PubMed] [Google Scholar]
- 24.Lakshmipathy U., Campbell C. Anti-sense mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 2001;29:668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abraham G. N., Scaletta C., Vaughan J. H. Modified diphenylalanine reaction for increased sensitivity. Anal. Biochem. 1972;49:547–549. doi: 10.1016/0003-2697(72)90460-5. [DOI] [PubMed] [Google Scholar]
- 26.Church G. M., Gilbert W. Genomic sequencing. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeDoux S. P., Wilson G. L., Beecham E. J., Stevnsner T., Wassermann K., Bohr V. A. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 28.Wei Y. F., Robins P., Carter K., Caldecott K., Pappin D. J., Yu G. L., Wang R. P., Shell B. K., Nash R. A., Schar P., et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomkinson A. E., Totty N. F., Ginsburg M., Lindahl T. Location of the active-site for enzyme-adenylate formation in DNA ligases. Proc. Natl. Acad. Sci. U.S.A. 1991;88:400–404. doi: 10.1073/pnas.88.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riballo E., Critchlow S. E., Teo S. H., Doherty A. J., Priestley A., Broughton B., Kysela B., Beamish H., Plowman N., Arlett C. F., et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 31.O'Driscoll M., Cerosaletti K. M., Girard P. M., Dai Y., Stumm M., Kysela B., Hirsch B., Gennery A., Palmer S. E., Seidel J., et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 32.Spelbrink J. N., Toivonen J. M., Hakkaart G. A., Kurkela J. M., Cooper H. M., Lehtinen S. K., Lecrenier N., Back J. W., Speijer D., Foury F., Jacobs H. T. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 33.Jazayeri M., Andreyev A., Will Y., Ward M., Anderson C. M., Clevenger W. Inducible expression of a dominant negative DNA polymerase-γ depletes mitochondrial DNA and produces a rho0 phenotype. J. Biol. Chem. 2003;278:9823–9830. doi: 10.1074/jbc.m211730200. [DOI] [PubMed] [Google Scholar]
- 34.Mambo E., Gao X., Cohen Y., Guo Z., Talalay P., Sidransky D. Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1838–1843. doi: 10.1073/pnas.0437910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai R. K., Perng C. L., Hsu C. H., Wong L. J. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann. N.Y. Acad. Sci. 2004;1011:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- 36.Lim S. E., Longley M. J., Copeland W. C. The mitochondrial p55 accessory subunit of human DNA polymerase γ enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 37.Mackey Z. B., Niedergang C., Murcia J. M., Leppard J., Au K., Chen J., de Murcia G., Tomkinson A. E. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 38.Lindahl T., Wood R. D. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 39.Mol C. D., Izumi T., Mitra S., Tainer J. A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 40.Wilson S. H., Kunkel T. A. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 41.Hill J. W., Hazra T. K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor R. M., Whitehouse J., Cappelli E., Frosina G., Caldecott K. W. Role of the DNA ligase III zinc finger in polynucleotide binding and ligation. Nucleic Acids Res. 1998;26:4804–4810. doi: 10.1093/nar/26.21.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]