Abstract
Tight regulation of p53 is essential for maintaining normal cell growth. Here we report that BLIMP1 acts in an autoregulatory feedback loop that controls p53 activity through repression of p53 transcription. p53 binds to and positively regulates BLIMP1, which encodes for a known B cell transcriptional repressor. Knockdown of BLIMP1 by siRNA results in both apoptosis and growth arrest in human colon cancer cells and cell-cycle arrest in primary human fibroblasts. Interestingly, the levels of both p53 mRNA and protein are substantially increased after BLIMP1 depletion, which is accompanied by the induction of p53 target genes. Importantly, the apoptosis induced by BLIMP1 depletion in HCT116 cells is largely abrogated in cells lacking p53 or in cells depleted in p53 by siRNA. We further demonstrate that BLIMP1 binds to the p53 promoter and represses p53 transcription, and this provides a mechanistic explanation for the induction of p53 response in cells depleted of BLIMP1. Hence, suppression of p53 transcription is a crucial function of endogenous BLIMP1 and is essential for normal cell growth.
Keywords: apoptosis, growth arrest, transcription regulation
BLIMP1 is a zinc-finger-containing DNA-binding transcriptional repressor and plays essential roles in both embryonic development and adult tissues. BLIMP1 was initially identified as a postviral induction repressor of transcription of IFNB1 (1) and later was shown to recruit the histone methyltransferase G9a to the promoter of IFN-β gene to shut down transcription (2). Blimp1 plays an essential role in the terminal differentiation of B cells into Ig-secreting plasma cell through direct transcriptional silencing of several key transcription activators such as Pax5 and Bcl6 (3–5). In the differentiation of myeloid lineage, Blimp1 also is a key regulator (6). Study on Blimp1 knockout mice demonstrates that Blimp1 is a critical determinant of the germ cell lineage (7, 8), and it is crucial for consistent repression of homeobox genes that normally accompany specification of primordial germ cells (PGCs) (7). In zebrafish, Blimp1 promotes differentiation of the embryonic slow muscle lineage (9) and specifies neural crest and sensory neuron progenitors (10). Collectively, these studies indicate that Blimp1 plays a key role in the cellular differentiation process.
In addition, a number of reports suggest that Blimp1 might regulate diverse cellular processes including cell growth or survival. The PGC-like cells in Blimp1 mutant embryos failed to show the characteristic proliferation and migration (7). Blimp1 mutant embryos also display apoptosis in multiple cell types, most notably the mesenchyme cells, which express high levels of Blimp1 (8). Recent studies of Blimp1 in T cells demonstrate that mice lacking Blimp1 develop inflammatory disease and show a decrease in survival of T cells in thymocytes (11, 12). However, no studies to date have directly defined the role of Blimp1 in regulating cell proliferation and survival. Furthermore, the upstream transcription regulator of Blimp1 is also not known.
The tumor suppressor p53 responds to a variety of intrinsic and extrinsic stress signals to trigger several cellular programs, including cell-cycle arrest, apoptosis, inhibition of angiogenesis/metastasis, and DNA repair (13–16). p53 regulates the expression of downstream target genes, which serve as mediators of p53 functions (17–19). For example, p21, Bax, and Puma are direct transcriptional targets of p53, and they play critical role in the p53 pathway (20–23). Our previous study that coupled ChIP with the paired-end ditag technologies for mapping the p53 binding sites in the human genome uncovered many putative p53 target genes (24). One of these candidate genes is BLIMP1. In this study we show that BLIMP1 is a bona fide p53 target gene and, more importantly, that it acts in an autoregulatory feedback loop that controls p53 activity through repression of p53 transcription. Our study uncovers a function of BLIMP1 in regulating cell survival and demonstrates the involvement of p53 in this process.
Results
p53 Positively Regulates BLIMP1 Transcription.
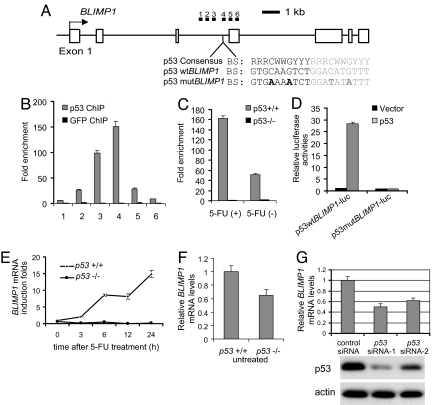
The identification of p53 binding in the BLIMP1 genomic locus suggests that BLIMP1 could be regulated by p53. The p53 binding locus was located downstream of the BLIMP1 transcription start site and within the third intron (Fig. 1A). A short sequence containing a p53 consensus site was identified within the paired-end ditag cluster (Fig. 1A). We first verified whether the p53 binding motif within the BLIMP1 genomic locus determined by ChIP paired-end ditag analysis is associated with p53 interaction in vivo. We examined a 2,200-bp region encompassing the p53 binding motif using ChIP followed by quantitative real-time PCR (ChIP-qPCR) in HCT116 cells with p53 activated by treatment with 5-fluorouracil (5-FU). ChIP-qPCR confirmed p53 binding with the highest level of occupancy coinciding with the location of the predicted p53 binding motif (Fig. 1B). Interestingly, we also observed p53 occupancy in untreated HCT116 cells, suggesting that p53 may also regulate BLIMP1 transcription even in the absence of genotoxic stress (Fig. 1C).
Fig. 1.
p53 positively regulates BLIMP1expression. (A) Genomic structure of human BLIMP1. The sequence and location of a p53 binding motif within intron 3 are indicated. The locations of the six pairs of primer sets used to detect the ChIP-enriched DNA fragments in B are indicated as filled bars. Open boxes represent exons of BLIMP1. (B) High-resolution mapping of p53 binding sites across intron 3 of BLIMP1 using the six primer sets indicated in A. HCT116 cells were treated with 5-FU for ChIP analysis. ChIP was performed by using a p53 or GFP control antibody. Fold enrichment is the relative abundance of DNA fragments at the indicated regions over a control region as quantified by real-time PCR. (C) p53 binds to BLIMP1 intron in unstressed HCT116 cells. ChIP on untreated or 5-FU-treated HCT116 cells with a p53 antibody is shown. p53−/− cells were used as control. Enrichment for fragment 4 amplicon was tested by real-time PCR. (D) p53 activates reporter containing the putative p53 binding site of BLIMP1 intron 3. Two tandem copies of wild-type or mutant p53 motif in BLIMP1 intron 3 were cloned into a pGL3 luciferase reporter construct and were cotransfected with p53 in HCT116 p53−/− cells. (E) Expression levels of BLIMP1 mRNAs in 5-FU-treated p53+/+ and p53−/− HCT116 cells. Cells were treated with 5-FU for the indicated times. RNA was isolated, reverse-transcribed, subjected to qPCR by using primers specific for BLIMP1 mRNA, and normalized with GAPDH mRNA. (F) Expression of BLIMP1 mRNA in unstressed p53+/+ and p53−/− HCT116 cells. (G) Depletion of p53 by siRNAs results in a reduction of BLIMP1 transcription in HCT116 cells. HCT116 cells were transfected with p53 siRNA or GFP siRNA as a control. Cells were harvested 48 h after transfection for mRNA analysis of BLIMP1 mRNA levels by real-time PCR (Top) and p53 protein levels by Western blotting (Middle and Bottom).
To determine whether the p53 binding motif present in BLIMP1 intron 3 verified above could mediate p53 responsiveness, two tandem copies of this binding site (p53 wtBLIMP1) or its mutants (p53 mutBLIMP1) [Fig. 1A, and see supporting information (SI) Methods] were inserted upstream of the luciferase coding sequence in pGL3 basic vector. HCT116 p53−/− cells were transfected with either p53wtBLIMP1-luc or p53mutBLIMP1-luc along with plasmids expressing wild-type p53. As shown in Fig. 1D and SI Fig. 6, p53 induced luciferase expression from p53wtBLIMP1-luc in a dose-dependent manner whereas no transcriptional activation was observed from p53mutBLIMP1-luc. This indicates that the identified p53 binding motif within intron 3 of the BLIMP1 is a bona fide p53 binding site. To determine whether BLIMP1 is positively regulated by p53 in a more physiological setting, we examined the changes in BLIMP1 mRNA levels in untreated or 5-FU-treated p53+/+ and p53−/− HCT116 cells (see SI Methods). We observed a p53-dependent induction of BLIMP1 mRNAs in HCT116 cells treated with 5-FU (Fig. 1E). Interestingly, BLIMP1 mRNA levels in unstressed p53−/− HCT116 cells were lower than in unstressed wild-type HCT116 cells (Fig. 1F). Considering p53 binding to BLIMP1 in unstressed cells (Fig. 1C), this suggests that p53 also plays a role in regulating the basal level of BLIMP1 transcription. To further substantiate this, we examined whether depletion of p53 by siRNAs would lead to a reduction of BLIMP1 transcription in HCT116 cells. As expected, BLIMP1 mRNA was reduced by 50% in HCT116 cells transfected with p53 siRNAs (Fig. 1G). This is not a dramatic change, probably because of the existence of other family members of p53 (p73 and p63), which might be able to transactivate BLIMP1. Taken together, these results indicate that p53 directly regulates BLIMP1 transcription in both stressed and unstressed conditions.
BLIMP1 Depletion Inhibits Cell Growth in HCT116 Cells and IMR90 Cells.
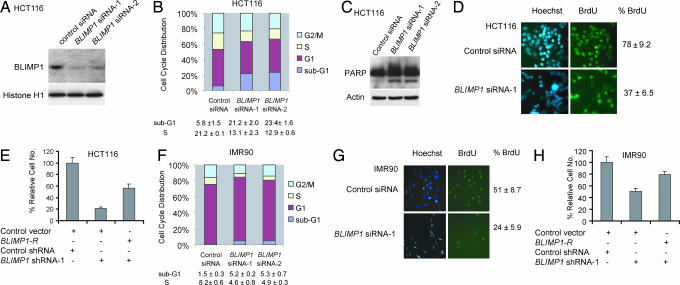
In addition to playing a key role in regulating cellular response to genotoxic stress, p53 has also been shown to be involved in the control of normal cell growth (25–28). Given that p53 regulates BLIMP1 transcription in unstressed cells, we examined whether BLIMP1 has a role in cell growth. To test this possibility, we depleted endogenous BLIMP1 with siRNAs and then monitored the growth of HCT116 cells (see SI Methods). To minimize potential off-target effects, we used two independent BLIMP1 siRNAs, and both effectively reduced endogenous BLIMP1 expression at the protein levels (Fig. 2A). In HCT116 cells, FACS analysis revealed a 3- to 4-fold increase in the sub-G1 population whereas S phase population decreased from 21% to 13% after BLIMP1 siRNA treatment as compared with control siRNA treatment (Fig. 2B and SI Fig. 7A), suggesting that BLIMP1 depletion results in both apoptosis and cell-cycle arrest. Western blotting showed that the PARP cleavage, an apoptotic marker, was increased with the depletion of BLIMP1 (Fig. 2C). By monitoring BrdU incorporation, we showed that BLIMP1 knockdown indeed inhibits proliferation of HCT116 cells (Fig. 2D). To rule out the possibility of nonspecific effects of RNAi, we showed that BLIMP1-R (an expression vector for Blimp1, which is immune to effect by the Blimp1 shRNA-1) was able to prevent the reduction in cell number mediated by BLIMP1 shRNA-1 (Fig. 2E). The functional consequence of BLIMP1 knockdown was also analyzed in the nontransformed human fibroblast IMR90 cells. Similar to the results obtained with HCT116 cells, IMR90 cells depleted of BLIMP1 also exhibited a decrease in the proportion of cells in S phase (Fig. 2F and SI Fig. 7A). The proportion of cells in sub-G1 phase after BLIMP1 depletion in IMR90 cells is less than HCT116 cells, suggesting that the apoptotic effect may be cell-type-specific. The growth arrest was substantiated by a decrease in BrdU incorporation in BLIMP1-depleted IMR90 cells (Fig. 2G). Also, we observed that the growth of IMR90 cells was strongly inhibited as a result of BLIMP1 knockdown by lentiviral transduction (SI Fig. 7B). Importantly, the BLIMP1-R was able to prevent the reduction in IMR90 cell numbers caused by BLIMP1 siRNA (Fig. 2H), confirming that the cell growth inhibition is specifically caused by the BLIMP1 depletion. Taken together, these data indicate that the constitutive expression of BLIMP1, which itself is a p53 target gene, is essential for proliferation or survival in both HCT116 and IMR90 cells.
Fig. 2.
BLIMP1 depletion by siRNA activates apoptosis and/or cell-cycle arrest. (A) The efficiency of the two independent BLIMP1 siRNAs was tested. HCT116 cells were transfected with BLIMP1 siRNA-1, BLIMP1 siRNA-2, or GFP siRNA as a control. Nuclear extracts (100 μg in each lane) were loaded to detect endogenous BLIMP1 protein, which is expressed at low levels in HCT116 cells. (B) Cell-cycle distribution of HCT116 cells transfected with control or BLIMP1 siRNAs. The cells were stained with propidium iodide (PI) for DNA content and analyzed by FACS. (C) Immunoblots of total lysates of control siRNA-transfected HCT116 cells and BLIMP1 siRNA-transfected cells with the indicated antibodies. (D) Quantification of BrdU-positive cells in HCT116 treated with BLIMP1 siRNA-1 or control siRNA. (E) Cell number count to show that the growth-inhibitory effect induced by loss of BLIMP1 in HCT116 cells could be rescued by BLIMP1-R. Transfected cells were selected with puromycin for 2 days. (F) Cell-cycle distribution of IMR90 cells transfected with control or BLIMP1 siRNAs. The cells were stained with PI for DNA content and analyzed by FACS. (G) Quantification of BrdU-positive cells in IMR90 treated with BLIMP1 siRNA-1 or control siRNA. (H) Cell number count to show that the growth defects induced by BLIMP1 depletion in IMR90 cells could be rescued by BLIMP1-R. Transfected cells were selected with puromycin for 2 days.
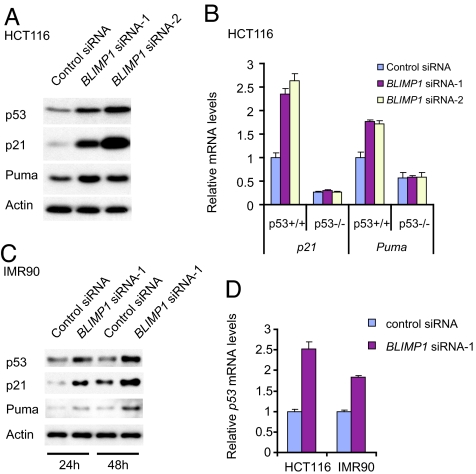
BLIMP1 depletion triggers a p53 response. Because BLIMP1 is positively regulated by p53 and depletion of BLIMP1 triggers cell apoptosis or growth arrest, which is a hallmark of p53-mediated response, we asked whether removal of BLIMP1 may affect the p53 pathway. Interestingly, the depletion of BLIMP1 in HCT116 cells resulted in an elevation of p53 expression at protein levels (Fig. 3A). More importantly, the induction of p53 was accompanied by an increase in the protein level of its target genes, p21 (20, 21) and Puma (22, 23) (Fig. 3A), which may account for the observed cell growth defects in BLIMP1-depleted HCT116 cells. Furthermore, p21 and Puma mRNAs were observed to be up-regulated in response to BLIMP1 knockdown only in p53+/+ HCT116 cells, but not in p53−/− HCT116 cells (Fig. 3B). We also observed a similar result in IMR90 cells with an increase in p53 as well as its downstream target genes, p21 and Puma, at the protein levels upon depletion of BLIMP1 by RNAi (Fig. 3C). These findings identify a role of BLIMP1 in regulating the p53 pathway. Because BLIMP1 is known to function as a transcriptional repressor, we examined whether p53 transcription is affected by BLIMP1 depletion. Indeed, qPCR indicated that p53 mRNA levels increased substantially after BLIMP1 RNAi treatment in both HCT116 and IMR90 cells (Fig. 3D). Taken together, these results demonstrate that depletion of BLIMP1 triggers a p53 response and suggest that endogenous BLIMP1 may be involved in suppressing p53 expression at the transcriptional level.
Fig. 3.
Depletion of BLIMP1 elevates p53 and induces the expression of p53 target genes. Lysates were immunoblotted with the indicated antibodies. RNA was isolated, reverse-transcribed, subjected to qPCR, and normalized with GAPDH mRNA. (A) Immunoblots of total lysates of HCT116 cells transfected with control siRNA or BLIMP1 siRNAs. (B) RT-PCR analysis of mRNA levels in HCT116 p53+/+ and p53−/− cells transfected with BLIMP1 siRNAs or control siRNA. (C) Immunoblots of total lysates of IMR90 cells transfected with control siRNA or BLIMP1 siRNAs. (D) RT-PCR analysis of p53 mRNA levels in HCT116 and IMR90 cells transfected with BLIMP1 siRNA-1 or control siRNA.
p53 Is an Important Mediator of Apoptosis Induced by BLIMP1 Knockdown in HCT116 Cells.
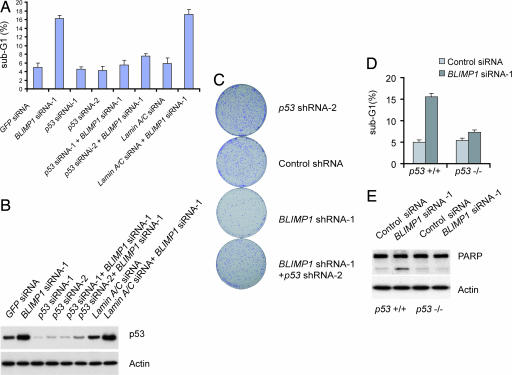
The results shown above suggest that p53 appears to be responsible for altered cell proliferation and cell survival ability in BLIMP1 knockdown cells. To determine the contribution of endogenous p53 in apoptosis that is instigated by BLIMP1 depletion in HCT116 cells, we cotransfected HCT116 cells with BLIMP1 siRNA together with either control or p53 siRNA. Control transfections included a GFP siRNA and lamin A/C siRNA, respectively. BLIMP1 reduction resulted in a 3- to 4-fold increase in the proportion of apoptotic cells; this effect was abrogated by the depletion of p53 but not by control siRNA against lamin A/C (Fig. 4A). We confirmed that p53 siRNA did not compromise the efficacy of the BLIMP1 RNAi by using qPCR to check BLIMP1 mRNA levels (data not shown). p53 siRNA-1, which was more efficient at suppressing p53 protein levels, was able to abrogate BLIMP1 RNAi-induced apoptosis more efficiently (Fig. 4 A and B). Moreover, we observed an increase in the number of surviving colonies of HCT116 cells coexpressing with p53 shRNA and BLIMP1 shRNA as compared with cells with BLIMP1 depletion alone or shRNA against lamin A/C plus Blimp1 shRNA (Fig. 4C and SI Fig. 8). To seek more evidence that p53 is an important mediator of apoptosis in HCT116 cells induced by BLIMP1 knockdown, we compared the extent of apoptosis between wild-type and p53−/− HCT116 cells after BLIMP1 depletion. Consistent with the above results, the induction of apoptosis by BLIMP1 siRNA was strikingly prohibited in p53−/− HCT116 cells relative to p53+/+ cells (Fig. 4D). We observed a slight increase of apoptosis in p53−/− cells, suggesting that a p53-independent apoptosis pathway might be involved (Fig. 4D). Moreover, we saw an increase in PARP cleavage in BLIMP1 siRNA-treated p53+/+ HCT116 cells but not in p53−/− HCT116 cells (Fig. 4E). In conclusion, p53 is, indeed, responsible for the induced apoptosis observed in HCT116 cells depleted of BLIMP1. Thus, negatively regulating p53 is a crucial function of endogenous BLIMP1 and is essential for the survival of HCT116 cells.
Fig. 4.
p53 is an important mediator of apoptosis induced by BLIMP1 knockdown in HCT116 cells. (A) Cotransfection of p53 siRNA with BLIMP1 siRNA prevented the induction in apoptotic cell population. Cells were analyzed for apoptotic cells (sub-G1) according to PI staining followed by FACS analysis. (B) Protein lysates of cells from A were immunoblotted with antibody against p53. (C) Restoration of surviving colony numbers of HCT116 cells exposed to both p53 shRNA(LV-p53 shRNA) and BLIMP1 shRNA compared with that of cells with BLIMP1 depletion alone. Cells were split at 24 h after transfection and selected with puromycin for 10 days. Plates were stained with crystal violet. (D) In the absence of p53, the apoptosis induced by BLIMP1 depletion is largely abrogated. The proportion of apoptotic cells was determined by PI staining followed by FACS analysis (sub-G1). (E) In the absence of p53, the increase in the PARP cleavage induced by BLIMP1 depletion is abrogated. Protein lysates of cells were immunoblotted with antibody against PARP.
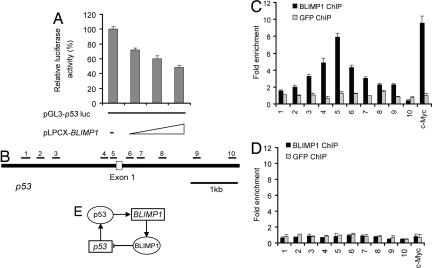
BLIMP1 Directly Suppresses p53 Transcription by Binding to Its Promoter.
In light of the significant biological consequences of BLIMP1-mediated regulation of p53 in unstressed cells, we explored the molecular mechanism. Having shown that BLIMP1 depletion results in up-regulation of p53 mRNA level (Fig. 3D), we next assessed whether BLIMP1 can suppress the activity of p53 promoter. Cotransfection of a STAT1 or IRF-7 expression vector with pGL3p53 reporter into HCT116 cells had no appreciable effect on the p53 promoter (SI Fig. 9A). In contrast, cotransfection of the BLIMP1 expression vector with the pGL3p53 reporter resulted in considerable inhibition of p53 promoter activity (Fig. 5A). Next, we addressed whether BLIMP1 binds to endogenous p53 promoter using ChIP-qPCR assays. Because of low endogenous levels of BLIMP1 in HCT116 cells, ChIP-qPCR was not sensitive enough to locate BLIMP1 binding within the p53 promoter in control vector-transfected cells (Fig. 5D). However, ectopically expressing BLIMP1 in HCT116 cells resulted in sufficient enrichment of BLIMP1–DNA complexes. Ten pairs of primers, located sequentially along the proximal promoter, first exon, and intron 1 of p53 were used to quantify the ChIP-enriched DNA by real-time PCR (Fig. 5B). A peak representing BLIMP1 binding was observed (≈8-fold above background) at a region very close to the transcriptional start site of p53 (Fig. 5C). Identical results were obtained when we used another polyclonal antibody raised against a different epitope of BLIMP1 (data not shown). ChIP using a control antibody of either GFP or E2F1 showed no significant enrichment over the entire surveyed region (Fig. 5C and SI Fig. 9B). In summary, a ChIP assay shows that BLIMP1 binds to a specific region within the p53 loci. Together with BLIMP1 suppression of p53 promoter activity, these data provide strong evidence that p53 is a direct transcription target of BLIMP1 repression.
Fig. 5.
BLIMP1 directly suppresses p53 transcription by binding to its promoter. (A) Overexpression of BLIMP1 suppressed p53 promoter activity. Vectors expressing BLIMP1 (pLPCX-BLIMP1) and pGL3-p53luc were transiently cotransfected into HCT116 cells, and luciferase activity was measured 30 h after transfection. (B) Mapping of BLIMP1 binding across the p53 promoter using 10 primer sets. The locations of the amplified products of the primer sets used to detect the ChIP-enriched DNA fragments are shown within the context of the genomic structure of human p53. Amplicons are numbered in order relative to their sites along the gene. ChIP assays were performed by using HCT116 cells transiently transfected with pLPCX-BLIMP1 (C) or vector pLPCX alone (D). Real-time PCR was performed with immunoprecipitated chromatin fragments obtained by using anti-BLIMP1 antibody or an irrelevant antibody (anti-GFP) as control. A known BLIMP1 binding site in the promoter region of c-Myc gene was amplified as a positive control for the ChIP assays. (E) A model for the negative feedback loop between p53 and BLIMP1. The loop creates a circuit composed of BLIMP1, whose mRNA synthesis is influenced by the activation of p53, and this in turn results in the alteration of p53 activity in cells.
Discussion
Our findings have uncovered a crucial role for BLIMP1 in the control of cell proliferation and survival through the negative regulation of p53 at the transcriptional level. The results presented here highlight a previously unrecognized link between BLIMP1 and p53-mediated response. The data demonstrate that the expression of p53 is suppressed by BLIMP1 in normal unstressed cells and that release of this repression leads to growth arrest or apoptosis.
A number of E3 ubiquitin ligases, such as Mdm2 (29, 30), Pirh2 (31), and Cop1 (32), have been shown to be transcriptionally regulated by p53 and form an autoregulatory loop with p53. To our knowledge, our results show for the first time that p53 expression is down-regulated by a transcriptional repressor that is itself up-regulated by p53, thereby forming a negative feedback loop that serves to maintain low p53 expression and prevent inappropriate activation of a p53 response in unstressed cells (Fig. 5E). It has been recently reported that Blimp1-deficient mice exhibit a severe reduction in the number of PGCs (7, 8). We wish to highlight an interesting correlation between Blimp1 expression and the proliferation status of PGCs. After the specification event of ≈40 founder PGCs, a rapid proliferation event is required so that a large mass of PGCs is attained. Intriguingly, the expression of Blimp1 in PGCs is first detected at embryonic day 6.5 and remains expressed throughout the phase of proliferation. This observation, together with the findings from our study, hints at the possibility that Blimp1 expression in PGCs after embryonic day 6.5 keeps PGCs in the proliferative state by maintaining a low level of p53. Another group also reported that Blimp1 knockout mice display apoptosis in branchial arch and heart tissue (8). However, it is remains unclear how Blimp1 deletion triggers apoptosis in these tissues. In line with our observations, we speculate that the apoptosis observed in branchial arch and heart tissue is due to the activation of the p53 pathway as a consequence of Blimp1 deletion.
The discovery of frequent inactivation of BLIMP1 in activated B cell-like diffuse large B-cell lymphoma (DLBCL) indicates that BLIMP1 has an important role in the pathogenesis of this lymphoma subtype (33) and possibly functions as a tumor suppressor gene. However, a recent report shows that the presence of variable percentages of BLIMP1-positive tumor cells in approximately half of DLBCL cases (34) and patients with higher BLIMP1 expression are significantly correlated with shorter failure-free survival (34). This raises the possibility that higher BLIMP1 expression could possibly promote the development of lymphoma. Deregulated BCL-6 expression contributes to lymphomagenesis in part by functional inactivation of p53 (32). Our finding that BLIMP1 represses p53 expression suggests that high expression of BLIMP1 might also promote the development of tumor by suppression of p53 activation. The exact nature of how BLIMP1 contributes to lymphomagenesis is unclear, and it would be of interest to study the relationship between BLIMP1 and p53 status along with patient survival in DLBCL.
Long-lived plasma cells are critical arm of humoral memory and are the important source of antibodies that protect us for future encounters with the pathogens. Calame and coworkers (35) have previously shown that the expression of Blimp1 in long-lived plasma cell is indispensable for their long-term maintenance; however, the molecular mechanism of Blimp1's role in long-lived plasma cells is not clear. The finding of p53 activation after Blimp1 depletion in this study suggests that one possible mechanism by which Blimp1 maintains long-lived plasma cells is by repressing p53 activation that can induce apoptosis and/or growth arrest. Generation of knockout mice deficient for both Blimp1 and p53 might rescue the loss of maintenance of long-lived plasma cells. Recently, two groups demonstrated that Blimp1 is expressed in T lymphocytes and is essential for T cell homeostasis and self-tolerance (11, 12). Interestingly, the group led by Calame (11) showed that CD4+ CD8+ (double-positive) thymocytes in mice with T cell-restricted Blimp1 deletion were severely reduced to ≈36% of the control littermates and that, furthermore, Blimp1-deficient double-positive thymocytes were more susceptible to apoptosis (11). The link between BLIMP1 and p53 presented here probably gives some insight into its mechanism. DNA double-strand break is one important process in T cell lineage maturation and is essential for T cell receptor rearrangement. BCL6 is known to suppress p53 expression in germinal-center B cells (33). Similarly, by suppressing p53 expression in thymocytes, Blimp1 may allow these cells to sustain the physiological genomic stress without eliciting a p53 response during expansion. Together with Calame's study (11), we hypothesize that double-positive thymocytes lacking Blimp1 expression are not able to reach normal numbers because an inappropriate p53 response prevents their proliferative expansion.
Materials and Methods
Cell Lines and Transfection.
The human colorectal cancer cell lines p53+/+ and p53−/− HCT116 were provided by Bert Vogelstein (The Johns Hopkins University). The human fibroblast IMR90 cells were obtained from American Type Culture Collection. siRNA duplexes and plasmids were transiently transfected by using Lipofectamine 2000 following the manufacturer's procedures (Invitrogen).
RNAi.
siRNA duplexes targeting the BLIMP1, p53, and GFP mRNAs were chemically synthesized by Dharmacon Research. Their target sequences are as follows: BLIMP1, 5′-GATCTGACCCGAATCAATG-3′ (BLIMP1 siRNA-1) and 5′-GCAACTGGATGCGCTATGT-3′ (BLIMP1 siRNA-2); p53, 5′-CAGTCTACCTCCCGCCATA-3′ (p53 siRNA-1) and 5′-GAAGAAACCACTGGATGGA-3′ (p53 siRNA-2). The control siRNAs are as follows: GFP, 5′-GGCTACGTCCAGGAGCGCACC-3′; Lamin A/C, 5′-GGTGGTGACGATCTGGGCT-3′.
For vector-based RNAi (shRNA), pSUPER.puro (Oligoengine) was used for BLIMP1 and Lamin A/C. The H1 promoter cassette in pLVTH lentivirus vector was replaced by the H1-shRNA cassette excised from pSUPER-shRNA, generating LV-shRNA (LV-control shRNA, LV-BLIMP shRNA, and LV-p53 shRNA), respectively.
BLIMP1 Expression Constructs.
The full-length BLIMP1 vector was provided by Tom Maniatis (Harvard University), cloned into pLPCX vector (Clontech), and confirmed by sequencing. For BLIMP1 shRNA-1 immune rescue construct (BLIMP1-R), there are six silent mutations (5′-gacctcactcgcattaacg-3′ as underlined) in the target sequence.
ChIP Assays.
ChIP assays were carried out as described (24, 34, 36). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and formaldehyde was then inactivated by the addition of 125 mM glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated by using anti-BLIMP1 polyclonal antibody (ab13700; Abcam), control GFP antibody (Santa Cruz Biotechnology), or control E2F1 antibody (Upstate Biotechnology). For all ChIP experiments, qPCR analyses were performed in real time by using ABI PRISM 7900 Sequence Detection System and SYBR Green Master Mix. Threshold cycles (Ct) were determined for both immunoprecipitated DNA and a known amount of DNA from the input sample for different primer pairs. Relative occupancy values (also known as fold enrichments) were calculated by determining the IP efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and normalized to the level observed at a control region, which was defined as 1.0. The control region is a 279-bp region on chromosome 22 and is amplified by using the primers 5′-GGACTCGGAAGAGGTTCACCTTCGG-3′ and 5′-GTCGCCTCCGCTTGCTGAACTCAATGC-3′. For all of the primers used, each gave a single product of the right size, as confirmed by agarose gel electrophoresis and dissociation curve analysis.
Supplementary Material
Acknowledgments
We thank Bert Vogelstein for p53+/+ and p53−/− HCT116 cells, Tom Maniatis for human BLIMP1 cDNA, Riccardo Dalla-Favera (Columbia University) for human p53 promoter, and Didier Trono (University of Geneva) for pLVTH vector. This work was supported by the Agency for Science, Technology and Research of Singapore and a Biomedical Research Council Young Investigator Award (to H.-H.N.). J.J. is supported by a National University of Singapore graduate scholarship and a National University of Singapore President graduate fellowship. C.A.L. is supported by an A*STAR graduate scholarship.
Abbreviations
- qPCR
quantitative real-time PCR
- 5-FU
5-fluorouracil
- shRNA
short hairpin RNA
- PGC
primordial germ cell.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605562104/DC1.
References
- 1.Keller AD, Maniatis T. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 2.Gyory I, Wu J, Fejer G, Seto E, Wright KL. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 3.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro-Shelef M, Calame K. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 6.Chang DH, Angelin-Duclos C, Calame K. Nat Immunol. 2000;1:169–176. doi: 10.1038/77861. [DOI] [PubMed] [Google Scholar]
- 7.Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 8.Vincent SD, Dunn NR, Sciammas R, Shapiro-Shelef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. Development (Cambridge, UK) 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 9.Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Ng T. Curr Biol. 2004;14:1772–1777. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 11.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 12.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 14.Chao C, Saito S, Kang J, Anderson CW, Appella E, Xu Y. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 16.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 17.Harris SL, Levine AJ. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 18.Jin S, Levine AJ. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 19.Lane DP. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 20.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 21.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 23.Nakano K, Vousden KH. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 24.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 25.Jiang M, Milner J. Genes Dev. 2003;17:832–837. doi: 10.1101/gad.252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 28.Agarwal ML, Agarwal A, Taylor WR, Stark GR. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 30.Kubbutat MH, Jones SN, Vousden KH. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 31.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 32.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 33.Phan RT, Dalla-Favera R. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 34.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. J Exp Med. 2005;202:1471–1476. doi: 10.1084/jem.20051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geisberg JV, Struhl K. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.