Abstract
Elucidation of the molecular mechanisms underlying carcinogenesis has benefited tremendously from the identification and characterization of oncogenes and tumor suppressor genes. One new advance in this field is the identification of PTPN11 as the first proto-oncogene that encodes a cytoplasmic tyrosine phosphatase with 2 Src-homology 2 (SH2) domains (Shp2). This tyrosine phosphatase was previously shown to play an essential role in normal hematopoiesis. More recently, somatic missense PTPN11 gain-of-function mutations have been detected in leukemias and rarely in solid tumors, and have been found to induce aberrant hyperactivation of the Ras-Erk pathway. This progress represents another milestone in the leukemia/cancer research field and provides a fresh view on the molecular mechanisms underlying cell transformation.
Introduction
Leukemia and other types of cancer continue to be a leading cause of death in the United States, and biomedical scientists sorely note that victories against cancer remain unacceptably rare. Nevertheless, due to the genetic and biochemical analyses of multiple oncogenes and tumor suppressor genes, significant progress has been made in understanding the molecular basis for transformation of a normal cell to a cancer cell. Gain-of-function mutations of normal cellular genes, termed proto-oncogenes, generate oncogenes that confer a proliferative or survival advantage to the cell. In contrast, loss-of-function mutations of tumor suppressor genes lead to dysregulated cellular proliferation and survival.
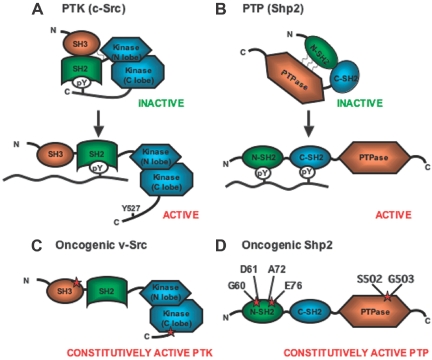
Reversible phosphorylation of protein tyrosine residues plays a critical role in signaling cascades for control of cell proliferation, differentiation, migration, and death. Tyrosyl phosphorylation levels are determined by the opposing activities of protein tyrosine kinases (PTKs) and phosphatases (PTPs). Many PTKs are known to transmit signals resulting in cell proliferation, and thus oncogenic mutations in human cancers commonly target PTKs. Typically, the mutation disrupts the structural integrity of the kinase, such that autoinhibitory mechanisms are impaired, causing constitutive activation.1 One well-illustrated example is the v-Src oncogene, first identified as an essential oncogenic component in Rous sarcoma virus. Phosphorylation of tyrosine 527 on the wild-type (WT) Src induces an intramolecular phosphotyrosine–Src-homology 2 (SH2) interaction and intramolecular contact between the SH3 domain and the SH2-kinase linker region, both of which contribute to repression of the kinase activity (Figure 1A).1 v-Src encodes a constitutively active mutant kinase lacking the C-terminal tail containing Y527 and also possessing missense mutations in the SH3 and kinase domains (Figure 1C).
Figure 1.
Schematic diagram of inactive and active forms of Src kinase (PTK) and Shp2 phosphatase (PTP). (A) c-Src is maintained in an inactive conformation by intramolecular interactions between phosphorylated tyrosine 527 on the C-terminal tail and the SH2 domain and between the SH3 domain and the kinase linker region. Dephosphorylation of tyrosine 527 and binding of a p-Tyr–containing peptide to the SH2 domain leads to a switch of c-Src to an open conformation with activation of the kinase function. (B) Deletion of sequences encoding for tyrosine 527 and missense mutations within the SH3 or kinase domains (schematically represented by red stars) converts the c-Src proto-oncogene to an oncogene encoding for mutant v-Src with constitutive kinase activity. (C) Shp2 is maintained in an inactive conformation by hydrophobic interactions between amino acid residues within the N-SH2 domain and the PTP domain. Binding of a p-Tyr–containing peptide to the N-SH2 domain causes Shp2 to assume an open conformation with activation of the phosphatase function. (D) Mutations within the N-SH2 or phosphatase domains cause disruption of the hydrophobic interactions resulting in constitutive activation of the phosphatase activity. The residues most commonly mutated in childhood leukemias are shown.
PTPs conventionally are thought to reverse PTK activities, thereby attenuating signals for cell proliferation. Although genes encoding PTPs are strong candidates for tumor suppressor genes, for example in colorectal cancers,2 few phosphatase genes have been unequivocally identified as tumor suppressors. Density-enhanced phosphatase 1 (Dep-1) is a receptor PTP that exhibits tumor suppressor activity when overexpressed in vitro, and Ptprj maps to the mouse colon cancer susceptibility locus,3 implicating Ptprj as a potential tumor suppressor gene. Germline PTEN mutations are found in Cowden syndrome, characterized by multiple hamartomas and a high proclivity for cancer development, and somatic loss-of-function PTEN mutations are detected in several human cancers; however, substantial data suggest that PTEN mainly acts as a lipid phosphatase, rather than a tyrosine phosphatase catalytically, and suppresses the phosphoinositide 3-kinase (PI3K) signaling pathway needed for survival and proliferation.4
Recent studies identified somatic gain-of-function mutations in PTPN11, which encodes the Shp2 tyrosine phosphatase, in childhood leukemias and, rarely, in adult leukemias and solid tumors.5,6 Similar to multiple mutant tyrosine kinases, the missense PTPN11 mutations destabilize noncovalent interactions between the N-SH2 and PTP domains, resulting in constitutively active phosphatase activity (Figure 1B,D). Therefore, PTPN11 was identified as the first proto-oncogene encoding a PTP.
The tyrosine phosphatase Shp2
Shp2 is a widely expressed cytoplasmic PTP containing 2 tandemly arranged SH2 domains at its amino terminal end, a central phosphatase domain, and a carboxy terminal tail.7,8 Mice homozygous for a targeted deletion of exon 3, encoding residues 46 to 110 in the N-SH2 domain of Shp2, are embryonic lethal due to abnormal gastrulation.9 More recently, Yang et al10 showed that homozygous Shp2 null mutants displayed peri-implantation lethality and that Shp2 had a critical role in trophoblast stem cell survival. Shp2 associates with activated receptor PTKs or cytokine receptors, which lack intrinsic kinase activity, either directly by docking to phosphorylated tyrosine residues on the receptors or indirectly via adaptor/scaffolding proteins, such as Grb2-associated binder 1-3 (GAB1-3), fibroblast growth factor receptor substrate-2 (FRS-2), and insulin receptor substrate 1-4 (IRS1-4), all of which possess 2 conserved tyrosine sites for engagement of the 2 SH2 domains of Shp2.7,8 Genetic and biochemical analyses in Caenorhabditis elegans, Drosophila, Xenopus, and mammals support the notion that Shp2 promotes Ras activation by growth factors and cytokines.11–18 Although different models have been proposed involving phosphatase-dependent and -independent mechanisms, most functional analyses suggest that the catalytic activity of Shp2 is required for promotion of signaling through the Ras-Erk pathway. Several studies suggest that Shp2 dephosphorylates, and thus inhibits, RasGAP and sprouty proteins, both negative regulators of Ras activation.8,19 Alternatively, Shp2 has been proposed to lead to the dephosphorylation of Src kinase, either directly or indirectly, leading to Src activation with subsequent Ras activation.8,20 However, the unequivocal identification of Shp2 substrates continues to be sought.
Shp2 enzymatic studies revealed low basal phosphatase activity with rapid induction upon occupancy of the SH2 domains.21–24 Consistently, the crystal structure of Shp2 revealed direct contact between the N-SH2 domain and the catalytic domain, constituting an autoinhibitory effect (Figure 1B).25 Occupation of the N-SH2 domain by a phosphotyrosine (p-Tyr)–containing ligand alleviates the autoinhibition, and therefore, association of Shp2 with a regulatory protein is directly coupled to phosphatase activation (Figure 1B). Importantly, the N-SH2 residues that interact with the PTP domain are not part of the p-Tyr–binding pocket. Therefore, mutation of catalytic domain-interacting residues within the N-SH2 domain (Figure 1D) will generate a molecule with enhanced phosphatase activity and retained capacity to bind p-Tyr–containing proteins. Indeed, Neel's group26 has demonstrated this principle in Xenopus embryo animal cap elongation studies.
Role of Shp2 in hematopoiesis
In contrast to Shp1, which has a negative regulatory role in myeloid/lymphoid cell development and functions, Shp2 has been found to be essential for normal hematopoietic cell development.27–30 In an in vitro hematopoietic differentiation assay, homozygous mutant embryonic stem (ES) cells for the Shp2Δ46-110 deletion mutation exhibited severely decreased differentiation capacity to erythroid and myeloid progenitors.27 This in vitro result was supported by the in vivo chimeric animal analysis, in which neither erythroid nor myeloid progenitor cells of Shp2Δ46-110 rigin were detected in the fetal liver or bone marrow of chimeric animals that were derived from mutant ES cells and wild-type embryos.28 Notably, Shp2 mutant ES cells did have significant contributions to many other organs or tissues of the chimeras, suggesting a more stringent requirement for Shp2 in hematopoiesis than development of many other cell types in mammals.28 A RAG-2–deficient blastocyst complementation assay further demonstrated a critical role of Shp2 for lymphopoiesis in a cell-autonomous manner, as development of lymphoid cell lineages in Shp2−/−/Rag-2−/− chimeric mice was blocked before pro-T- and pro-B-cell stages.29 Thus, Shp2 is positively required for development of all hematopoietic cell lineages, suggesting a role of Shp2 in the commitment/differentiation of hematopoietic stem cells.
Indeed, experimental data suggest that lack of normal Shp2 function leads to decreased differentiation from ES cells to hemangioblasts (BL-CFC),30 a multipotential precursor that has the capacity to differentiate into primitive and definitive erythroid cells as well as endothelial cells.31,32 Consistently, Shp2 mutant ES cells displayed decreased and delayed expression of brachyury and flk-1, markers of mesoderm and endothelial cells, respectively, upon differentiation in vitro.30 Shp2 appears to play a positive role in promoting the Erk pathway and a negative role in the Jak/Stat3 pathway in control of stem cell self-renewal and differentiation. In factor-dependent hematopoietic cell lines, Shp2 has been shown to participate in the relay of signals elicited by interleukin-6 (IL-6), leukemia inhibitory factor (LIF), IL-3/granulocyte macrophage–colony-stimulating factor (GM-CSF), erythropoietin (Epo), or stem cell factor (SCF).33–36 Functional analysis suggests that Shp2 may act in both a catalytic-dependent and -independent manner in mediating IL-3–stimulated proliferation and survival of hematopoietic cells.37
Germline PTPN11 mutations in Noonan syndrome and related congenital disorders
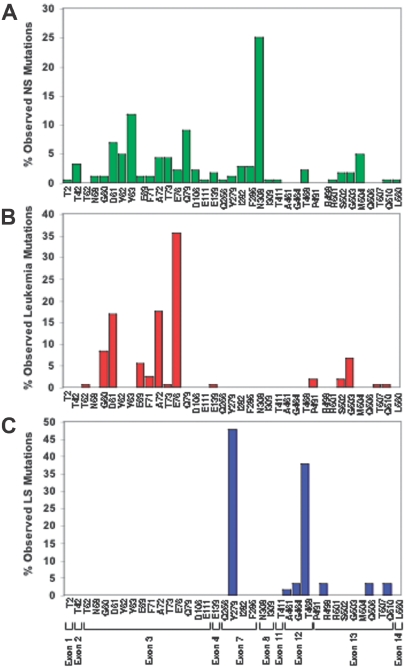
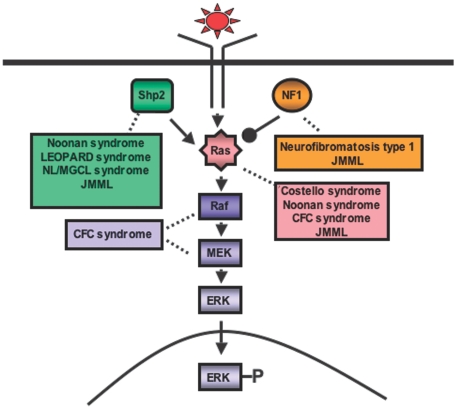
The significance of Shp2 in human disease became evident when PTPN11 germ line mutations were identified in individuals with Noonan syndrome (NS), an autosomal dominant disorder with an estimated incidence of 1 in 2500 live births.38,39 The most common abnormalities are dysmorphic facial features, heart defects, skeletal abnormalities, and growth retardation.39 Hematologic abnormalities, including hepatosplenomegaly unexplained by cardiac failure (25%-50%) and, rarely, aggressive juvenile myelomonocytic leukemia (JMML), are also observed in patients with NS.40 Based on genotyping data, the most commonly mutated residue in NS is asparagine 308 (26%) followed by tyrosine 63 (12%) and glutamine 79 (8.5%; Figure 2A).39,41,42 Individuals with generalized lentigines are classified into a Noonanlike syndrome called LEOPARD (multiple lentigines, electrocardiographic-conduction abnormalities, ocular hypertelorism, pulmonary stenosis, abnormal genitalia, retardation of growth, sensorineural deafness) syndrome (LS). PTPN11 mutations are observed in approximately 90% of patients with LS, with 85% of the mutations involving residues tyrosine 279 or threonine 468 (Figure 2C).39,43,44 PTPN11 mutations are also found in Noonanlike/multiple giant cell lesion syndrome, which clinically can be confused with cherubism.41,45,46 Notably, PTPN11 mutations have not been identified in the phenotypically overlapping cardio-facio-cutaneous (CFC) or Costello syndromes39; however, recent reports reveal HRAS mutations in Costello syndrome,47 KRAS, BRAF, MEK1, or MEK2 mutations in CFC syndrome,48,49 and KRAS mutations in a small percentage of patients with NS,50,51 implicating the common theme of aberrant Ras-Erk signaling in these overlapping human developmental disorders (Figure 3). Consistent with this, neurofibromatosis type 1 (NF1), which results from loss-of-function mutations in neurofibromin, a GTPase activating protein (GAP) that accelerates the conversion of active Ras-GTP to inactive Ras-GDP, shares clinical similarities with NS, including neurofibromas, neural crest–derived malignant tumors, cardiac anomalies, and JMML.52
Figure 2.
Graphic representation of mutation prevalence among individuals bearing PTPN11 mutations with Noonan syndrome (NS), childhood leukemia, and LEOPARD syndrome (LS). (A) PTPN11 mutations observed in NS are distributed widely throughout the PTPN11 gene, with the most common affected residue being glutamine (N) 308 (based on 175 patients with NS identified with PTPN11 mutations). (B) PTPN11 mutations observed in childhood leukemias are clustered within exons 3 and 13 (based on 163 patients with pediatric leukemia identified with PTPN11 mutations). (C) The vast majority of PTPN11 mutations observed in LS involve residues tyrosine (Y) 279 and threonine (T) 468 (based on 63 patients with LS identified with PTPN11 mutations).
Figure 3.
Schematic diagram showing ligand-stimulated Ras activation, the Ras-Erk pathway, and the human diseases found to date associated with mutation of multiple molecules participating in this signaling cascade. NL/MGCL indicates Noonanlike/multiple giant cell lesion; CFC, cardio-facio-cutaneous; JMML, juvenile myelomonocytic leukemia.
Somatic PTPN11 mutations in leukemia
The observation that JMML, a rare form of childhood leukemia, was observed in patients with NS, predicted that individuals with nonsyndromic JMML may bear somatic PTPN11 mutations. Indeed, after identification of germ line mutations of PTPN11 in patients with NS,38 Tartaglia and colleagues5 also pioneered the search of somatic PTPN11 mutations in individuals with nonsyndromic JMML. Collectively, recent studies by several groups indicate that somatic mutations within PTPN11 occur in 35% of JMML cases, as well as in childhood acute myeloid leukemia (4%), myelodysplastic syndrome (10%), and acute lymphoid leukemia (7%).5,53–59 While germ line mutations found in NS occur in multiple exons throughout PTPN11 (Figure 2A and Table S1, which is available on the Blood website; see the Supplemental Table link at the top of the online article), pediatric leukemia-associated mutations (both syndromic and nonsyndromic) are concentrated in exons 3 and 13 (Figure 2B and Table S1). Mutation of residues glycine 60, aspartate 61, glutamate 69, alanine 72, glutamate 76, all encoded by exon 3, accounts for approximately 83% of the PTPN11 mutations observed in all pediatric leukemias combined. The most common mutation found in individuals with NS and associated myeloproliferative disease (MPD) is T73I (42.1% of reported cases).5,53,55,56 T73I, along with D61G, are the only mutations found in NS,39 NS in association with MPD,5,53,55,56 and de novo, nonsyndromic leukemia.53,54
Functional and phenotypic implications of PTPN11 mutations
Comparison of PTPN11 germ line mutations observed in patients with NS to somatic mutations identified in individuals with nonsyndromic leukemia reveals that the 2 groups are largely nonoverlapping (exceptions include G60A, D61G, D61N, F71L, T73I, E139D, and R498W; Table S1). This observation prompted the notion that increasingly higher levels of phosphatase activity induce more severe phenotypes and that the most functionally severe mutations may induce in utero lethality if sustained in the germ line.60 This hypothesis is supported by the finding that the leukemia-associated PTPN11 mutants D61Y and E76K encode for mutant Shp2 with very high basal and unregulated phosphatase activity.5,56,60,61 Consistently, mutations observed in NS, syndromic leukemia, or nonsyndromic leukemia (D61G and T73I) encode for mutant Shp2 with high basal, yet p-Tyr-peptide–inducible phosphatase activity.61 Germ line mutations observed only in patients with NS (N308D) encode mutant Shp2 with only modestly high basal phosphatase activity.5,56,61 However, this correlation does not hold true for all the mutant Shp2 proteins examined. Keilhack et al61 determined that p-Tyr-peptide affinity, in contrast to absolute phosphatase activity, also contributes to the gain-of-function effect of some Shp2 mutants. Moreover, Niihori et al56 observed that 2 mutations found only in NS (D61N and F71I) had higher relative phosphatase activity than 2 mutations found in nonsyndromic leukemia (E76A and G503V).
Recent findings further highlight the difficulty in predicting a disease phenotype based merely on in vitro phosphatase activity assay. Surprisingly, PTPN11 mutations observed in LEOPARD syndrome (Y279C and T468M) reduce Shp2 phosphatase activity in vitro and inhibit ligand-stimulated phospho-Erk activation in cells, in contrast to the gain-of-function mutations observed in NS.60,62 However, the Shp2 crystal structure predicts that the Y279C and T468M mutant proteins reside preferentially in the open conformation, suggesting that these mutant proteins may exert defective signaling in vivo due to aberrant adapter function in assembling multiprotein signaling complexes or due to competition with WT Shp2 for substrates.
Functional and signal transduction alternations induced by PTPN11 mutations
JMML is a rare clonal MPD of children younger than 5 years of age and is characterized clinically by hepatosplenomegaly, increased circulating monocytes and erythroblasts, and increased production of fetal hemoglobin (HbF), implying a regression to fetallike hematopoiesis.63 In vitro, hematopoietic progenitors from patients with JMML demonstrate hypersensitivity to GM-CSF.64 Hyperactivation of Ras is implicated in the pathogenesis of JMML based on the identification of activating NRAS and KRAS mutations65,66 and loss-of-heterozygosity of NF1 in patient samples (Figure 3).67 Well-studied molecular mechanisms leading to JMML in humans with NF1 and in animal models bearing Nf1 loss-of-function or activating Kras mutations are instructive when considering the potential molecular aberrations induced by activating PTPN11 mutations. Biochemical analysis of primary leukemic cells from patients with NF1 demonstrates increased levels of Ras-GTP and Erk hyperactivation.68 Likewise, hematopoietic progenitors from Nf1−/− and KrasG12D mice demonstrate elevated Ras-GTP, hypersensitivity to GM-CSF, and induce MPD in vivo.69–71 Conditional inactivation of Nf1 in hematopoietic cells induces MPD with 100% penetrance, hypersensitivity to GM-CSF, and resistance to apoptosis.72 Likewise, wild-type mice reconstituted with fetal liver progenitors from Nf1−/− animals develop MPD similar to JMML, which is attenuated in mice lacking GM-CSF; however, despite a longer latency, some mice lacking GM-CSF eventually succumb to MPD.73 These findings highlight that although crucial, dysregulated GM-CSF signaling alone likely is not sufficient to induce JMML. Indeed, hematopoietic progenitors from KrasG12D and Nf1−/−; mice also demonstrate hypersensitivity to IL-369 and IL-3 plus stem cell factor,74 respectively. Recognizing and understanding the relevance of additional hematopoietic growth factor pathways in JMML pathogenesis is imperative for the successful development of novel therapeutic strategies in this disease.
Retroviral transduction of 3 commonly observed somatic PTPN11 mutants (E76K, D61V, and D61Y) into murine bone marrow– or fetal liver–derived mononuclear cells induces hematopoietic progenitor hypersensitivity to GM-CSF, similar to that observed in patients with JMML, as well as to IL-3.75–77 Likewise, transduction of bone marrow– or fetal liver–derived mononuclear cells with mutant E76K results in a significant increase in erythroid burst-forming unit (BFU-E) colonies, consistent with increased circulating erythroblasts observed in patients with JMML.77 Macrophage progenitors bearing the E76K, D61Y, or D61V mutants hyperproliferate in response to GM-CSF, and each mutation induced elevated constitutive and prolonged GM-CSF–stimulated phospho-Erk activation.75 Additionally, IL-3 stimulation of mutant-bearing mast cells promoted hyperproliferation and induced hyperactivation of phospho-Akt and phospho-Stat5.76 Structure-function studies defined that ablation of the phosphatase function (by both C463S or R465M mutations), impairment of binding activity of the N-SH2 domain or C-SH2 domains, or mutation of tyrosine 542, severely abrogated the transforming ability of the E76K mutant,76,77 suggesting that both the enzymatic function as well as the adapter function of Shp2 substantially contribute to the transforming ability of the Shp2 mutants.
Approximately 50% of mice reconstituted with hematopoietic progenitors transduced with PTPN11 mutants E76K or D61Y succumbed to malignant hematologic disease by 7 months after transplantation. At death, the majority of mice were found to bear a severe MPD, including hepatosplenomegaly and bone marrow hyperplasia with increased immature myelomonocytic cells.76 This progressive, lethal form of MPD is in contrast to the later-onset, nonlethal, chronic myelomonocytic hyperplasia observed in a mouse model bearing the common NS germ line PTPN11 mutation, D61G.78 This variable transforming ability of the different PTPN11 mutants in part rationalizes the unpredictable clinical course of JMML in patients with NS bearing germ line PTPN11 mutations, which spans from mild disease ending in spontaneous remission to progressive cases ending in death.40
One peculiar observation is that the incidence of somatic PTPN11 mutations is higher in JMML compared with acute leukemias and solid tumors. A contributing factor to this phenomenon may be the cell population targeted by PTPN11 mutations leading to JMML. A recent study examining a GATA1 mutation found in individuals with Down syndrome and transient MPD or acute megakaryoblastic leukemia revealed a novel embryonic progenitor population targeted by this mutation.79 Similarly, fetal myeloid progenitors may be exceptionally sensitive to the effects of activating PTPN11 mutations compared with those of adult myeloid progenitors. Interaction with a second, cooperating mutation or modifier loci may be required for clonal outgrowth progressing to frank JMML, while the absence of these functional genetic interactions may allow for the spontaneous remission of JMML, as observed in some patients with NS with germ line PTPN11 mutations.40 Although speculative, this hypothesis is consistent with the finding that murine transplantation of PTPN11 mutant–bearing cells produced robust MPD on the Balb/c background and only subtle abnormalities on the C57Bl/6 background.76,77
Although PTPN11 mutations are found only rarely in adult leukemias,80–82 recent studies demonstrate elevated Shp2 expression in primary leukemia cell specimens from multiple adult acute leukemias, compared with Shp2 levels in bone marrow mononuclear cells from healthy controls.82 Upon terminal differentiation, Shp2 expression diminished, suggesting that persistently elevated Shp2 expression may lead to perturbations in hematopoietic cell differentiation.82 Consistently, overexpression of WT Shp2 inhibited macrophage progenitor differentiation in response to macrophage colony-stimulating factor (M-CSF) based on F4/80 expression,75 suggesting that excessive Shp2 may inhibit or delay the maturation of myeloid progenitors, contributing to leukemogenesis.
Conclusion
Based on genotyping studies, in vitro and in vivo functional studies, and enzymatic studies, PTPN11 is the first identified proto-oncogene that encodes a PTP. Strikingly, a similar molecular mechanism is employed in oncogenic activation of an intracellular PTK (c-Src) or a PTP (Shp2), which involves disruption of an autoinhibitory intramolecular interaction leading to constitutive activation. Thus, the studies on mutant Shp2 not only expand the scope of oncogenes, but also refresh our views on the critical difference between proto-oncogenes and oncogenes mediating either physiologic regulation or pathologic dysregulation in normal cells and malignant cells, respectively. One important functional consequence of activating PTPN11 or KRAS mutations or loss-of-function NF1 mutations is Ras hyperactivation; therefore, Ras effectors, such as Mek and PI3K, are rational targets for novel therapeutics in JMML. Indeed, in vitro treatment of hematopoietic progenitors with the Mek inhibitor, UO126, or with rapamycin, an inhibitor of the PI3K-mTOR pathway, reduces the transforming ability of PTPN11 mutant E76K.76 The current challenge entails building on this broad fund of knowledge to define novel molecular targets with the objective of developing improved therapeutics for pediatric and adult myeloid leukemias.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the tenacious genotyping work of multiple investigators reviewed by Tartaglia and Gelb.39 Additionally, the authors thank Dr Mignon Loh for helpful discussion in the presentation of the genotyping data. Work in authors' laboratories was supported by the March of Dimes (Basil O'Connor Starter Scholar Award, R.J.C.) and the National Institutes of Health (RO1HL082 981, R.J.C.; CA078 606 and CA102 583, G.-S.F.).
Footnotes
The online version of this article contains a data supplement.
Authorship
Contribution: R.J.C. and G.-S.F. wrote and revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gen-Sheng Feng, Burnham Institute for Medical Research, 10901 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: gfeng@burnham.org.
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 3.Ruivenkamp CA, van Wezel T, Zanon C, et al. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat Genet. 2002;31:295–300. doi: 10.1038/ng903. [DOI] [PubMed] [Google Scholar]
- 4.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tartaglia M, Niemeyer CM, Fragale A, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 6.Bentires-Alj M, Paez JG, David FS, et al. Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 7.Lai LA, Zhao C, Zhang EE, Feng GS. The Shp-2 tyrosine phosphatase. In: Arino J, Alexander D, editors. Protein Phosphatases. Vol 5. Berlin, Heidelberg, Germany: Springer-Verlag; 2004. pp. 275–299. [Google Scholar]
- 8.Neel BG, Gu H, Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 9.Saxton TM, Henkemeyer M, Gasca S, et al. Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J. 1997;16:2352–2364. doi: 10.1093/emboj/16.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Klaman LD, Chen B, et al. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev Cell. 2006;10:317–327. doi: 10.1016/j.devcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Perkins LA, Larsen I, Perrimon N. Corkscrew encodes a putative protein tyrosine phosphatase that functions to transduce the terminal signal from the receptor tyrosine kinase torso. Cell. 1992;70:225–236. doi: 10.1016/0092-8674(92)90098-w. [DOI] [PubMed] [Google Scholar]
- 12.Gutch MJ, Flint AJ, Keller J, Tonks NK, Hengartner MO. The Caenorhabditis elegans SH2 domain-containing protein tyrosine phosphatase PTP-2 participates in signal transduction during oogenesis and vulval development. Genes Dev. 1998;12:571–585. doi: 10.1101/gad.12.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milarski KL, Saltiel AR. Expression of catalytically inactive Syp phosphatase in 3T3 cells blocks stimulation of mitogen-activated protein kinase by insulin. J Biol Chem. 1994;269:21239–21243. [PubMed] [Google Scholar]
- 15.Tang TL, Freeman R, Jr, O'Reilly AM, Neel BG, Sokol SY. The SH2-containing protein-tyrosine phosphatase SH-PTP2 is required upstream of MAP kinase for early Xenopus development. Cell. 1995;80:473–483. doi: 10.1016/0092-8674(95)90498-0. [DOI] [PubMed] [Google Scholar]
- 16.Feng GS. Shp-2 tyrosine phosphatase: signaling one cell or many. Exp Cell Res. 1999;253:47–54. doi: 10.1006/excr.1999.4668. [DOI] [PubMed] [Google Scholar]
- 17.Shi ZQ, Lu W, Feng GS. The Shp-2 tyrosine phosphatase has opposite effects in mediating the activation of extracellular signal-regulated and c-Jun NH2-terminal mitogen-activated protein kinases. J Biol Chem. 1998;273:4904–4908. doi: 10.1074/jbc.273.9.4904. [DOI] [PubMed] [Google Scholar]
- 18.Qu CK, Yu WM, Azzarelli B, Feng GS. Genetic evidence that shp-2 tyrosine phosphatase is a signal enhancer of the epidermal growth factor receptor in mammals. Proc Natl Acad Sci U S A. 1999;96:8528–8533. doi: 10.1073/pnas.96.15.8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanafusa H, Torii S, Yasunaga T, Matsumoto K, Nishida E. Shp2, an SH2-containing protein-tyrosine phosphatase, positively regulates receptor tyrosine kinase signaling by dephosphorylating and inactivating the inhibitor Sprouty. J Biol Chem. 2004;279:22992–22995. doi: 10.1074/jbc.M312498200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SQ, Yang W, Kontaridis MI, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–355. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 21.Lechleider RJ, Sugimoto S, Bennett AM, et al. Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding site, phosphotyrosine 1009, on the human platelet-derived growth factor receptor. J Biol Chem. 1993;268:21478–21481. [PubMed] [Google Scholar]
- 22.Sugimoto S, Wandless TJ, Shoelson SE, Neel BG, Walsh CT. Activation of the SH2-containing protein tyrosine phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin receptor substrate-1. J Biol Chem. 1994;269:13614–13622. [PubMed] [Google Scholar]
- 23.Dechert U, Adam M, Harder KW, Clark-Lewis I, Jirik F. Characterization of protein tyrosine phosphatase SH-PTP2: study of phosphopeptide substrates and possible regulatory role of SH2 domains. J Biol Chem. 1994;269:5602–5611. [PubMed] [Google Scholar]
- 24.Pluskey S, Wandless TJ, Walsh CT, Shoelson SE. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J Biol Chem. 1995;270:2897–2900. doi: 10.1074/jbc.270.7.2897. [DOI] [PubMed] [Google Scholar]
- 25.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 26.O'Reilly AM, Pluskey S, Shoelson SE, Neel BG. Activated mutants of SHP-2 preferentially induce elongation of Xenopus animal caps. Mol Cell Biol. 2000;20:299–311. doi: 10.1128/mcb.20.1.299-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu CK, Shi ZQ, Shen R, Tsai FY, Orkin SH, Feng GS. A deletion mutation in the SH2-N domain of Shp-2 severely suppresses hematopoietic cell development. Mol Cell Biol. 1997;17:5499–5507. doi: 10.1128/mcb.17.9.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu CK, Yu WM, Azzarelli B, Cooper S, Broxmeyer HE, Feng GS. Biased suppression of hematopoiesis and multiple developmental defects in chimeric mice containing Shp-2 mutant cells. Mol Cell Biol. 1998;18:6075–6082. doi: 10.1128/mcb.18.10.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu CK, Nguyen S, Chen J, Feng GS. Requirement of Shp-2 tyrosine phosphatase in lymphoid and hematopoietic cell development. Blood. 2001;97:911–914. doi: 10.1182/blood.v97.4.911. [DOI] [PubMed] [Google Scholar]
- 30.Chan RJ, Johnson SA, Li Y, Yoder MC, Feng GS. A definitive role of Shp-2 tyrosine phosphatase in mediating embryonic stem cell differentiation and hematopoiesis. Blood. 2003;102:2074–2080. doi: 10.1182/blood-2003-04-1171. [DOI] [PubMed] [Google Scholar]
- 31.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy M, Firpo M, Choi K, et al. A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature. 1997;386:488–493. doi: 10.1038/386488a0. [DOI] [PubMed] [Google Scholar]
- 33.Boulton TG, Stahl N, Yancopoulos GD. Ciliary neurotrophic factor/leukemia inhibitory factor/interleukin 6/oncostatin M family of cytokines induces tyrosine phosphorylation of a common set of proteins overlapping those induced by other cytokines and growth factors. J Biol Chem. 1994;269:11648–11655. [PubMed] [Google Scholar]
- 34.Welham MJ, Dechert U, Leslie KB, Jirik F, Schrader JW. Interleukin (IL)-3 and granulocyte/macrophage colony-stimulating factor, but not IL-4, induce tyrosine phosphorylation, activation, and association of SHPTP2 with Grb2 and phosphatidylinositol 3′-kinase. J Biol Chem. 1994;269:23764–23768. [PubMed] [Google Scholar]
- 35.Tauchi T, Feng GS, Marshall MS, et al. The ubiquitously expressed Syp phosphatase interacts with c-kit and Grb2 in hematopoietic cells. J Biol Chem. 1994;269:25206–25211. [PubMed] [Google Scholar]
- 36.Tauchi T, Feng GS, Shen R, et al. Involvement of SH2-containing phosphotyrosine phosphatase Syp in erythropoietin receptor signal transduction pathways. J Biol Chem. 1995;270:5631–5635. doi: 10.1074/jbc.270.10.5631. [DOI] [PubMed] [Google Scholar]
- 37.Yu WM, Hawley TS, Hawley RG, Qu CK. Catalytic-dependent and -independent roles of SHP-2 tyrosine phosphatase in interleukin-3 signaling. Oncogene. 2003;22:5995–6004. doi: 10.1038/sj.onc.1206846. [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia M, Mehler EL, Goldberg R, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 39.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 40.Bader-Meunier B, Tchernia G, Mielot F, et al. Occurrence of myeloproliferative disorder in patients with Noonan syndrome. J Pediatr. 1997;130:885–889. doi: 10.1016/s0022-3476(97)70273-7. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Tartaglia M, Gelb BD, et al. Phenotypic and genotypic characterisation of Noonan-like/multiple giant cell lesion syndrome. J Med Genet. 2005;42:e11. doi: 10.1136/jmg.2004.024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Kogaki S, Kurotobi S, et al. A novel mutation in the PTPN11 gene in a patient with Noonan syndrome and rapidly progressive hypertrophic cardiomyopathy. Eur J Pediatr. 2005;164:497–500. doi: 10.1007/s00431-005-1679-y. [DOI] [PubMed] [Google Scholar]
- 43.Keren B, Hadchouel A, Saba S, et al. PTPN11 mutations in patients with LEOPARD syndrome: a French multicentric experience. J Med Genet. 2004;41:e117. doi: 10.1136/jmg.2004.021451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkozy A, Conti E, Seripa D, et al. Correlation between PTPN11 gene mutations and congenital heart defects in Noonan and LEOPARD syndromes. J Med Genet. 2003;40:704–708. doi: 10.1136/jmg.40.9.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jafarov T, Ferimazova N, Reichenberger E. Noonan-like syndrome mutations in PTPN11 in patients diagnosed with cherubism. Clin Genet. 2005;68:190–191. doi: 10.1111/j.1399-0004.2005.00475.x. [DOI] [PubMed] [Google Scholar]
- 46.Tartaglia M, Kalidas K, Shaw A, et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aoki Y, Niihori T, Kawame H, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 48.Niihori T, Aoki Y, Narumi Y, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Viciana P, Tetsu O, Tidyman WE, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 50.Schubbert S, Zenker M, Rowe SL, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 51.Carta C, Pantaleoni F, Bocchinfuso G, et al. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Luca A, Bottillo I, Sarkozy A, et al. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am J Hum Genet. 2005;77:1092–1101. doi: 10.1086/498454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kratz CP, Niemeyer CM, Castleberry RP, et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 2005;106:2183–2185. doi: 10.1182/blood-2005-02-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loh ML, Reynolds MG, Vattikuti S, et al. PTPN11 mutations in pediatric patients with acute myeloid leukemia: results from the Children's Cancer Group. Leukemia. 2004;18:1831–1834. doi: 10.1038/sj.leu.2403492. [DOI] [PubMed] [Google Scholar]
- 55.Loh ML, Vattikuti S, Schubbert S, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103:2325–2331. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 56.Niihori T, Aoki Y, Ohashi H, et al. Functional analysis of PTPN11/SHP-2 mutants identified in Noonan syndrome and childhood leukemia. J Hum Genet. 2005;50:192–202. doi: 10.1007/s10038-005-0239-7. [DOI] [PubMed] [Google Scholar]
- 57.Tartaglia M, Gelb BD. Germ-line and somatic PTPN11 mutations in human disease. Eur J Med Genet. 2005;48:81–96. doi: 10.1016/j.ejmg.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 58.Tartaglia M, Martinelli S, Iavarone I, et al. Somatic PTPN11 mutations in childhood acute myeloid leukaemia. Br J Haematol. 2005;129:333–339. doi: 10.1111/j.1365-2141.2005.05457.x. [DOI] [PubMed] [Google Scholar]
- 59.Tartaglia M, Martinelli S, Cazzaniga G, et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104:307–313. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- 60.Tartaglia M, Martinelli S, Stella L, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants: implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 62.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 63.Arico M, Biondi A, Pui CH. Juvenile myelomonocytic leukemia. Blood. 1997;90:479–488. [PubMed] [Google Scholar]
- 64.Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–929. [PubMed] [Google Scholar]
- 65.Kalra R, Paderanga DC, Olson K, Shannon KM. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood. 1994;84:3435–3439. [PubMed] [Google Scholar]
- 66.Miyauchi J, Asada M, Sasaki M, Tsunematsu Y, Kojima S, Mizutani S. Mutations of the N-ras gene in juvenile chronic myelogenous leukemia. Blood. 1994;83:2248–2254. [PubMed] [Google Scholar]
- 67.Shannon KM, O'Connell P, Martin GA, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 68.Bollag G, Clapp DW, Shih S, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 69.Braun BS, Tuveson DA, Kong N, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat Genet. 1996;12:137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 71.Chan IT, Kutok JL, Williams IR, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le DT, Kong N, Zhu Y, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103:4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- 73.Birnbaum RA, O'Marcaigh A, Wardak Z, et al. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol Cell. 2000;5:189–195. doi: 10.1016/s1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- 74.Zhang YY, Vik TA, Ryder JW, et al. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J Exp Med. 1998;187:1893–1902. doi: 10.1084/jem.187.11.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan RJ, Leedy MB, Munugalavadla V, et al. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood. 2005;105:3737–3742. doi: 10.1182/blood-2004-10-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mohi MG, Williams IR, Dearolf CR, et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7:179–191. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 77.Schubbert S, Lieuw K, Rowe SL, et al. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 2005;106:311–317. doi: 10.1182/blood-2004-11-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Araki T, Mohi MG, Ismat FA, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 79.Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 80.Johan MF, Bowen DT, Frew ME, et al. Mutations in PTPN11 are uncommon in adult myelodysplastic syndromes and acute myeloid leukaemia. Br J Haematol. 2004;124:843–844. doi: 10.1111/j.1365-2141.2004.04862.x. [DOI] [PubMed] [Google Scholar]
- 81.Watkins F, Fidler C, Boultwood J, Wainscoat JS. Mutations in PTPN11 are rare in adult myelodysplastic syndromes and acute myeloid leukemia. Am J Hematol. 2004;76:417. doi: 10.1002/ajh.20134. [DOI] [PubMed] [Google Scholar]
- 82.Xu R, Yu Y, Zheng S, et al. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood. 2005;106:3142–3149. doi: 10.1182/blood-2004-10-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.