Abstract
Maternal cells have recently been found in the circulation and tissues of mothers' immune-competent children, including in adult life, and is referred to as maternal microchimerism (MMc). Whether MMc confers benefits during development or later in life or sometimes has adverse effects is unknown. Type 1 diabetes (T1D) is an autoimmune disease that primarily affects children and young adults. To identify and quantify MMc, we developed a panel of quantitative PCR assays targeting nontransmitted, nonshared maternal-specific HLA alleles. MMc was assayed in peripheral blood from 172 individuals, 94 with T1D, 54 unaffected siblings, and 24 unrelated healthy subjects. MMc levels, expressed as the genome equivalent per 100,000 proband cells, were significantly higher in T1D patients than unaffected siblings and healthy subjects. Medians and ranges, respectively, were 0.09 (0–530), 0 (0–153), and 0 (0–7.9). Differences between groups were evident irrespective of HLA genotypes. However, for patients with the T1D-associated DQB1*0302-DRB1*04 haplotype, MMc was found more often when the haplotype was paternally (70%) rather than maternally transmitted (14%). In other studies, we looked for female islet β cells in four male pancreases from autopsies, one from a T1D patient, employing FISH for X and Y chromosomes with concomitant CD45 and β cell insulin staining. Female islet β cells (presumed maternal) formed 0.39–0.96% of the total, whereas female hematopoietic cells were very rare. Thus, T1D patients have higher levels of MMc in their circulation than unaffected siblings and healthy individuals, and MMc contributes to islet β cells in a mother's progeny.
Keywords: quantitative PCR, chimerism, autoimmunity, pancreas, HLA
Eleven years ago, Hall et al. (1) reported that 20% of cord blood samples from male infants contained female cells, presumed to be maternal, thus bringing into question the assumption that maternal cells rarely pass into the fetal circulation. The study used FISH with X and Y chromosome probes and visual counting of cells according to whether the cell had two X signals or one X and one Y signal. This report was followed by two others that estimated maternal-to-fetus transfer to be even more frequent, with maternal DNA detected in 40–100% of cord blood samples when PCR-based techniques were used (2, 3). Although it could be argued that these findings reflect maternal to fetus trafficking during labor, other studies indicate that transfer occurs in the course of gestation because maternal DNA has been reported in fetal blood from second and third trimester pregnancy terminations (4, 5). In experimental studies, maternal cells were found in the marrow cavities of developing bones in immune-competent mice (6).
Microchimerism refers to the harboring of a small number of cells (or DNA) by one individual that originate from another genetically distinct individual. Maternal microchimerism (MMc) was recognized in children with severe combined immunodeficiency more than 20 years ago (7) and more recently was found to persist into adult life in healthy subjects (8). These observations raise questions as to whether MMc affects growth and development or sometimes contributes to disease or to tissue repair in a mother's progeny.
Fetal cells also persist in the mother, and fetal microchimerism has been implicated in some autoimmune diseases, notably systemic sclerosis (scleroderma) (9). An important observation that arose from studies of fetal microchimerism in systemic sclerosis was that the simple presence of microchimerism is common in healthy individuals, and quantitative techniques are important in the comparison of patients to controls (10, 11). Whereas male DNA can be quantified in women who gave birth to sons as a measure of fetal microchimerism, another approach was needed for MMc. As described above, MMc can be quantified by FISH, but this approach is time and labor intensive and is limited to sex-mismatched pairs. We therefore developed a panel of HLA-specific quantitative real-time PCR (Q-PCR) assays that target nontransmitted, nonshared maternal-specific HLA alleles (12). In the current studies we used the panel of Q-PCR assays to investigate MMc in type 1 diabetes (T1D), an autoimmune disease that primarily affects children and young adults. Additionally, we examined pancreatic autopsy specimens from boys for maternal cells with concomitant phenotyping to establish whether any identified female cells were hematopoietic or, because others have described cellular plasticity (13, 14), were insulin-producing islet β cells.
Results
MMc Prevalence and Levels in Peripheral Blood.
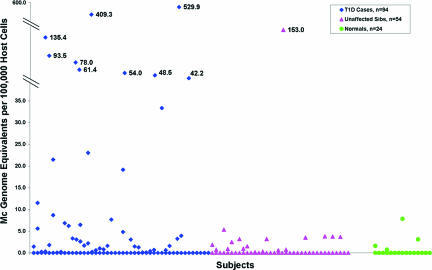
A panel of HLA-specific Q-PCR assays targeting nontransmitted, nonshared maternal-specific HLA alleles was used to identify and quantify MMc in DNA extracted from whole blood samples. Ninety-four T1D patients and 54 unaffected siblings were studied. Patient and sibling characteristics are given in Table 1. Twenty-four unrelated healthy subjects were also studied. Overall, higher levels of MMc were observed more often among T1D patients than among unaffected siblings and healthy individuals (Fig. 1). The amount of chimeric maternal DNA was expressed according to the number of genome equivalents present in 100,000 proband cells. In T1D patients, MMc ranged from 0 to 529.9 with a median level of 0.09 (mean 17.5). MMc levels in unaffected siblings ranged from 0 to 153.0, median 0 (mean 3.5). In healthy subjects the MMc levels ranged from 0 to 7.9, median 0 (mean 0.6). MMc levels were significantly higher in T1D patients than in unaffected siblings (P = 0.012) and healthy individuals (P < 0.001). In a multivariable model, adjusting for variables that might confound results, including age at blood draw, sex, birth order, and total number of cells tested, gave similar results (P = 0.028 and <0.001, respectively) and indicated that none of these variables was a significant confounding factor. The quantity of DNA tested was similar among the groups (medians were 99,517, 88,735, and 104,204 and means were 97,964, 91,435, and 97,335 in T1D, unaffected siblings, and healthy subjects, respectively).
Table 1.
Characteristics of type 1 diabetics and unaffected siblings
| Characteristic | T1D (n = 94) | Unaffected siblings (n = 54) |
|---|---|---|
| Age at draw, range (median) | 2–25 (13) | 4–29 (14) |
| Birth order* | ||
| First | 37% | 39% |
| Second | 45% | 33% |
| Third or later | 18% | 28% |
| Sex* | ||
| Male | 70% | 41% |
| Female | 30% | 59% |
| Affected relative* | 12% | 15% |
| Disease duration (yr)* | ||
| ≤1 | 53% | n.a. |
| 1 to ≤5 | 13% | n.a. |
| 5 to ≤10 | 18% | n.a. |
| >10 | 16% | n.a. |
| Age at diagnosis* | ||
| ≤5 | 19% | n.a. |
| 5 to ≤10 | 34% | n.a. |
| 10 to 16 | 47% | n.a. |
n.a., not applicable.
*Percentage of all T1D patients or unaffected siblings
Fig. 1.
MMc in T1D patients, unaffected siblings, and healthy subjects. A total of 172 probands were studied, 94 with T1D, 54 unaffected siblings, and 24 healthy controls. A panel of HLA-specific Q-PCR assays targeting nontransmitted, nonshared maternal HLA alleles was used (12) to test DNA extracted from whole peripheral blood. The amount of chimeric maternal DNA was expressed according to the number of genome equivalents present in 100,000 proband cells.
The prevalence of MMc (any positive result) among T1D patients was 51%, among unaffected siblings was 33%, and among healthy subjects was 17%. Although unaffected siblings often had MMc, levels were lower than among T1D patients, as seen in Fig. 1. Twenty-one percent of T1D patients had strikingly high levels of MMc, which exceeded levels seen in all subjects other than one unaffected sibling. If analysis considered subjects with MMc of 50 or more genome equivalents present in 100,000 proband cells, the difference between T1D patients and unaffected siblings became more evident, and the difference between unaffected siblings and unrelated healthy subjects became less evident (T1D vs. unaffected siblings, P = 0.009; unaffected siblings vs. unrelated subjects, P = 0.92; multivariable analysis).
HLA Class II Genotypes and MMc.
The DQB1*0201-DRB1*03 and DQB1*0302-DRB1*04 haplotypes are strongly associated with susceptibility to T1D (15), and it may be asked whether MMc in patients was due to enrichment for these two haplotypes. Overall, MMc was similarly prevalent among T1D patients with the DQB1*0201-DRB1*03 haplotype and/or DQB1*0302-DRB1*04 haplotypes (n = 86, 51% with MMc) and among the few patients who lacked both haplotypes (n = 8, 50% with MMc). Risk of T1D also varies with the number of copies of a T1D-associated haplotype and particular haplotype combinations (greatest risk is observed with compound heterozygosity for the two different haplotypes) (15). To further address the possibility that increased MMc among patients might be due to enrichment of particular HLA genotypes in patients, MMc results were analyzed with patients and unaffected siblings in HLA genotype groups as follows: group A DQB1*0201-DRB1*03,DQB1*0302-DRB1*04; group B homozygous for DQB1*0201-DRB1*03; group C homozygous for DQB1*0302-DRB1*04; group D DQB1*0201-DRB1*03,X; group E DQB1*0302-DRB1*04,X; and group F X,X, where X is any HLA haplotype other than the two T1D-associated haplotypes described. The genotype groups A, B, D, E, and F generally exhibited a similar pattern (Table 2) with higher MMc levels in patients than unaffected siblings (group C had too few subjects for meaningful comparison). However, for patients with one copy of the T1D-associated DQB1*0302-DRB1*04 haplotype (group E), MMc was found more often when the haplotype was paternally transmitted. Thus 70% (7/10) vs. 14% (2/14) of patients had MMc when DQB1*0302-DRB1*04 was paternally vs. maternally transmitted (P = 0.01). Among patients heterozygous for the other T1D-associated haplotype, DQB1*0201-DRB1*03, there was no difference in MMc according to the parent who transmitted the haplotype (group D).
Table 2.
HLA genotypes of type 1 diabetics and unaffected siblings and MMc
| Group A |
Group B |
Group C |
Group D |
Group E |
Group F |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Sibs | Cases | Sibs | Cases | Sibs | Cases | Sibs | Cases | Sibs | Cases | Sibs |
| n = 17 | n = 3 | n = 17 | n = 4 | n = 5 | n = 3 | n = 23 | n = 18 | n = 24 | n = 9 | n = 8 | n = 17 |
| 48.6 | 0 | 93.5 | 1.0 | 61.4 | 153 | 409.3 | 3.5 | 135.4 | 3.8 | 529.9 | 5.3 |
| 33.3 | 0 | 54.0 | 0.9 | 6.9 | 0 | 23.0 | 3.2 | 42.2 | 3.8 | 78.0 | 3.7 |
| 6.5 | 0 | 19.1 | 0 | 1.4 | 0 | 8.7 | 1.5 | 21.5 | 2.5 | 5.6 | 3.2 |
| 1.8 | 4.8 | 0 | 0 | 7.7 | 0.7 | 11.5 | 0 | 1.6 | 1.9 | ||
| 1.7 | 3.9 | 0 | 6.2 | 0.6 | 3.1 | 0 | 0 | 0.8 | |||
| 0.8 | 3.2 | 3.3 | 0.3 | 2.6 | 0 | 0 | 0.2 | ||||
| 0.7 | 3.1 | 2.2 | 0 | 1.5 | 0 | 0 | 0 | ||||
| 0.6 | 0.6 | 1.7 | 0 | 0.1 | 0 | 0 | 0 | ||||
| 0.2 | 0.4 | 1.3 | 0 | 0.07 | 0 | 0 | |||||
| 0 | 0.4 | 1.0 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0.2 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0.2 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0.1 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| 0 | 0 | 0 | |||||||||
| 0 | 0 | ||||||||||
| 0 | 0 | ||||||||||
| 0 | 0 | ||||||||||
| 0 | 0 | ||||||||||
| 0 | 0 | ||||||||||
| 0 | |||||||||||
Genotype of group A: DQB1*0201-DRB1*03,DQB1*0302-DRB1*04; genotype of group B: DQB1*0201-DRB1*03,DQB1*0201-DRB1*03; genotype of group C: DQB1*0302-DRB1*04,DQB1*0302-DRB1*04; genotype of group D: DQB1*0201-DRB1*03,X; genotype of group E: DQB1*0302-DRB1*04,X; genotype of group F: X,X. X is any haplotype other than DQB1*0201-DRB1*03 or DQB1*0302-DRB1*04; Cases, type 1 diabetics; Sibs, unaffected siblings.
MMc Prevalence and Levels According to Demographic and Clinical Characteristics.
Levels of MMc and prevalence did not differ between males and females or between patients who were younger compared with those who were older at disease onset. MMc levels were somewhat greater in T1D patients who were first-born (n = 35) compared with those of later birth order (n = 59) (medians 0.1 vs. 0.0 respectively). Among the T1D patients, 53% (n = 50) were studied within 1 year of diagnosis, 13% (n = 12) >1 and <5 years, 18% (n = 17) 5 to <10 years, and 16% (n = 15) >10 years from diagnosis. Levels of MMc were not significantly different according to time from disease onset (respective medians levels 0, 0, 0.24, and 1.5 genome equivalents present in 100,000 proband cells). There was no apparent correlation of MMc in patients with antibodies to GADA or IA2; however, the number of patients with autoantibody tests was limited (n = 52 and n = 51, respectively; data not shown). MMc did not differ substantially for subjects with an affected relative although the number of subjects was small. The increase of MMc in T1D patients was not explained by differences in HLA-specific Q-PCR assays used, and all assays were developed to have the same sensitivity level (see Methods). It should be noted, however, that the current studies investigated MMc among subjects for whom the mother differed for at least one DQB1, DRB1, or DQA1 allele (see Methods).
Female Cells in Male Pancreatic Autopsy Specimens.
In other studies, pancreatic autopsy specimens from four unrelated male individuals were investigated to determine whether maternal cells were present in the pancreas and if so to determine whether the cells were hematopoietic or were insulin-producing islet β cells. Pancreases were provided by a pathologist and examined by investigators blinded to clinical information. Two autopsy specimens had been chosen as a case and control because they derived from the same calendar year and from boys of a similar age. One boy had T1D and ketoacidosis (age 11) and the other had acute myeloblastic leukemia (age 14). Two other samples derived from infants: one infant had hypoplastic heart disease (8 weeks) and the other had cardiomyopathy (4 weeks), with the latter born to a mother with T1D. Female cells (presumed maternal) were identified by FISH for X and Y chromosomes in male pancreases and simultaneously studied for phenotype employing concomitant immunohistochemistry for insulin and CD45 on the same section (and same cells) (16).
Female cells were found in all four pancreases. In the T1D pancreas, slightly <1% of islet β cells were female [47 among 4,876 β cells counted (0.96%)]. Fewer female cells were identified in the control pancreas from the boy with acute myeloblastic leukemia [34 among 5,782 β cells (0.59%), odds ratio = 1.65, P = 0.026] (Table 3). Examples are shown in Fig. 2[multidimensional movies provided in the supporting information (SI)]. Results were similar if analyzed according to the area of pancreatic tissue with approximately twice as many female islet β cells in the former as in the latter pancreas (37 per 7.97 cm2 vs. 19 per 7.32 cm2, respectively). Female cells were very rarely identified among cells staining with the hematopoietic marker CD45; three female cells were found among 4,022 CD45+ cells (0.07%), and three were found among 2,863 (0.1%) in the T1D pancreas and acute myeloblastic leukemia pancreases, respectively (data not shown). In the two other pancreas samples from infants who died within weeks of birth, female islet β cells were also found and were fewer than in the T1D pancreas (Table 3).
Table 3.
Quantification of female and male cells among islet β cells in male pancreases
| Pancreas | Age at autopsy | Disease | XX cells | XY cells |
|---|---|---|---|---|
| A | 11 yr | T1D, ketoacidosis | 47 | 4,829 |
| B | 14 yr | Acute myeloblastic leukemia | 34 | 5,748 |
| C | 8 wk | Hypoplastic heart | 11 | 2,782 |
| D | 4 wk | Cardiomyopathy, maternal T1D | 17 | 2,262 |
Pancreas A vs. pancreas B, odds ratio (OR) = 1.65, P = 0.026 [95% confidence interval (CI) 1.1,2.6]; pancreas A vs. pancreas C, OR = 2.5, P = 0.006 (95% CI 1.3,4.8); pancreas A vs. pancreas D, OR = 1.3, P = 0.36 (95% CI 0.7,2.3).
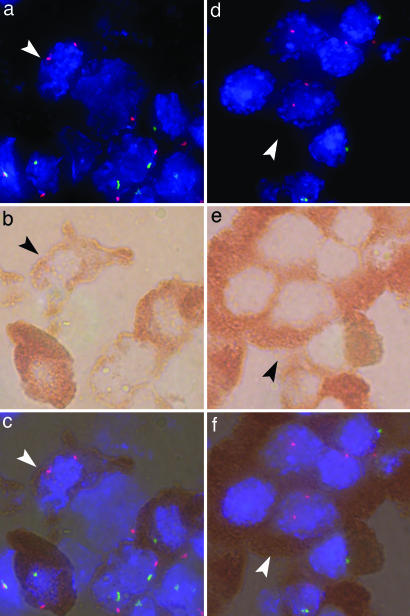
Fig. 2.
Female cells in male pancreas tissues. Immunohistochemistry for insulin and CD45 was used with concomitant FISH for X and Y chromosomes for the same cells as recently described (16). a, b, and c are from a boy with T1D and ketoacidosis; d, e, and f are from a boy with acute myeloblastic leukemia. (a) Fluorescence microscopy showing a female cell (arrowhead) with two (red) X chromosome signals. (Magnification: ×100.) Other cells contain one red and one Y chromosome (green) signal. Nuclei are identified with DAPI (blue). (b) Light microscopy of the same cells as in a. (Magnification: ×100.) The red-brown substrate identifies β cell insulin expression. (c) Overlay of a and b showing the identical cells with FISH and immunohistochemistry. (d) Fluorescence microscopy showing a female cell (arrowhead). (Magnification: ×100.) (e) Light microscopy of the same cells as in d. (Magnification: ×100.) (f) Overlay of d and e. Female cells were morphologically similar to surrounding β cells, and the cells did not express CD45.
Identification of some cells with more than two signals may be expected because of cell replication. If maternal cells are actively contributing to islet β cell regeneration, replicating maternal cells would be predicted, including cells with XXX and XXXX signals. Replicating male cells could include XXY, XYY, or XYXY signals, which could also arise by fusion with a maternal cell followed by chromosome loss (17), as could also occur for cells with two X chromosome signals. An XXXY cell could only be expected to occur as a result of fusion. We also counted all cells, including those with two as well as more than two signals, among a total of 2,424 and 2,035 islet β cells in the pancreas from the boy with T1D and the boy with acute myeloblastic leukemia, respectively. In the T1D compared with the acute myeloblastic pancreas, respectively, the number of cells found were 12 and 1 with XXX, 5 and 0 with XXXX, 29 and 19 with XXY, 4 and 3 with XYY, 14 and 3 with XYXY, and 1 and 0 with XXXY. Thus, the results supported active replication of female islet β cells in the T1D pancreas with little evidence to suggest fusion. Counting was blinded both to clinical information for the two specimens and to the reasons for counting cells with more than two signals.
Discussion
In the current studies we found MMc in the peripheral blood of subjects ranging from 3 to 28 years of age and found that levels of MMc were higher in T1D patients than unaffected siblings as well as unrelated healthy individuals. Whereas maternal cells have long been known to persist in immune-deficient children, it is only recently that long-term persistence of MMc in a mother's immune-competent children has been appreciated. MMc has been reported in infants in the thymus, liver, thyroid, skin (18), and heart (16) and in the peripheral blood of healthy adults (8). In an experimental model, maternal cells were found in the bone marrow of immune-competent mice during gestation and increased postnatally (19). Thus, accumulating evidence indicates MMc persists in the circulation and tissues of a mother's children, raising important questions as to whether MMc confers benefits during development, has beneficial or adverse effects later in life, and about the meaning of tolerance within a chimeric self.
Why levels of MMc are greater in patients with T1D is not known. A number of different potential roles of MMc may be considered. On the detrimental side, maternal cells could be effectors in “autoimmunity” as has been proposed in children with dermatomyositis (20). A second possibility is that differentiated, tissue-specific maternal cells could become targets of an “autoimmune” response as has been suggested in infants with neonatal lupus and congenital heart block (16). A third possibility is that MMc could function beneficially by providing an additional source for tissue repair as has been suggested for fetal microchimerism (21) or possibly diversity in response to infection. Of further interest, also on the beneficial side, the hypothesis has been proposed that MMc could contribute to growth and development and provide “educational” advantages (22).
Events leading to elevated MMc levels are unknown, but when results of MMc studies in peripheral blood are considered along with pancreatic autopsy studies, some conclusions are suggested. The first possibility, as outlined above, can be rejected because very few female hematopoietic cells were found in the pancreas, arguing strongly against any role of maternal cells as effectors of autoimmunity (whether due to absence of hematopoietic cells or migration failure is not known). Regarding the second possibility, maternal islet β cells were identified and thus could be targets for “autoimmunity.” However, observations are more supportive of the third possibility that MMc functions beneficially and contributes to attempts to regenerate islet β cell function. Higher levels of MMc were not observed in patients who were within a short interval from diagnosis, as might be anticipated if maternal islet β cells were targets of an “autoalloimmune” response, and increased numbers of female islet β cells suggests active replication in the T1D pancreas. Thus, although the events leading up to our observations are not known, our interpretation is that pancreatic MMc most likely contributes to efforts to restore function and regenerate diseased tissue.
The increase of MMc in T1D patients was not explained by an enrichment of particular HLA genotypes among T1D patients. MMc was more common and was present at higher levels in T1D patients compared with unaffected siblings across HLA genotype groups. Results were also not explained by the specificity of any of the HLA-specific Q-PCR assays used to identify MMc, because results were similar irrespective of the specific Q-PCR assay used. However, patients who had one copy of the T1D-associated haplotype DQB1*0302-DRB1*04 had MMc more often when the haplotype was transmitted from the father than when this haplotype was transmitted from the mother. This finding merits further exploration in larger studies and, if confirmed, suggests a differential effect on MMc of paternal and maternal genomes reminiscent of other reported epigenetic effects, such as described with imprinting (23). Of potential interest in this regard, uniparental disomy of chromosome 6 has been reported in association with islet β cell agenesis (24).
MMc was also increased in unaffected siblings relative to unrelated controls in the current studies, although less than among T1D patients. One potential explanation could be that some are prediabetic, because a 10-fold increased T1D risk has been described in first-degree relatives compared with the general population (25). Other possibilities include shared environmental factors as well as the possibility that a sibling may be more likely to share HLA and other genes with the mother than an unrelated individual. In a murine model, some histocompatibility relationships were found to correlate with increased levels of MMc (26). We were unable to explore this question because our methods were based on HLA incompatibility.
There are a number of limitations to our study. First, blood samples derived from a single time point and MMc levels could fluctuate with time and disease activity. Second, the Q-PCR assays test a relatively small amount of DNA (100,000 cell equivalent), and testing larger amounts may detect more positive results. Third, although islet β cells with two X chromosome signals within a clearly defined nucleus were found, some could nevertheless derive from overlapping cells. An additional possibility is that some were products of cell fusion (17) with chromosome loss. Although the possibility of maternal-progeny cell fusion is intriguing, in our studies there was little to support this possibility. Blood transfusion history was not known for T1D patients, unaffected siblings, or pancreatic autopsy samples in our study, and microchimerism can occur after a blood transfusion (27). However, transfusion is not common in T1D care, and reports of transfusion-associated microchimerism were in multiply transfused trauma patients given nonirradiated blood products; even for the leukemic patient from autopsy where transfusion might be expected, irradiated blood products would have been given (and any difference with the T1D pancreas would only be magnified).
Our findings bring to light an aspect of normal human biology that is relatively unexplored because it is not known how circulatory- and tissue-specific MMc is tolerated and what the consequences are for the lifetime of a mother's progeny. That maternal islet β cells were found in all pancreases studied, albeit more numerous in the T1D pancreas, suggests that MMc constitutively contributes to some cellular functions in a mother's progeny. The transfer of humoral immunity from mother to child is well recognized, but to our knowledge, a maternal contribution to endocrine function has not previously been described. Our findings also raise the possibility that naturally acquired chimerism might be exploited to therapeutic benefit. Regeneration of β cells is an area of major active investigation with recent studies reporting differentiation of pancreatic and nonpancreatic progenitors as well as replication of existing islet β cells (28–30). In conclusion, elevated levels of circulating MMc were found in patients with T1D and chimeric islet β cells were observed in pancreatic autopsy specimens. Further studies are needed to elucidate the function(s) of naturally acquired MMc in human health and disease.
Methods
Subjects.
T1D patients and unaffected siblings were recruited from the Bart's-Oxford study, a prospective, population-based study that identified >95% of families of children developing T1D before age 21 in the former Oxford Health Authority Region, U.K., since 1985 (31). The study population was 95% Caucasian European, with others mainly from the Indian subcontinent (data from Office of Population Censuses and Surveys for 1991). T1D classification was assigned by the referring clinician on the basis of World Health Organization criteria (32) and requirement for insulin treatment from diagnosis. Whole blood samples were selected from T1D patients and siblings for whom the mother was not homozygous or heterozygous identical for DQB1, DQA1, and DRB1 and thus were informative for the study. A total of 94 T1D patients, 54 unaffected siblings, and 24 healthy individuals were studied. Thirty-three families contributed 33 probands and their 40 siblings, 61 contributed a T1D patient only, and 10 contributed unaffected siblings only. Six T1D patients and five siblings had a father or mother with T1D, and seven patients and three siblings had an affected grandparent. The median age of T1D patients at the time of testing was 13 years (range 2–25), and the median age of unaffected siblings was 14 (range 4–29). In healthy controls, the median age was 14 (range 5–31). Among T1D patients, unaffected siblings, and healthy subjects, respectively, 66, 22, and 17 were male and 28, 32, and 7 female. Among the 94 T1D patients, 50 were within 1 year of diagnosis, 12 were between >1 and ≤5 years from diagnosis, 17 were between 5 and ≤10 years from diagnosis, and 15 were >10 years from diagnosis. Informed consent was obtained from study subjects. Eighteen patients were 5 or younger at diagnosis, 32 were >5 to <10 at diagnosis, and 44 were 10–16 years old at diagnosis. The study was approved by the research ethics committees in all centers involved.
HLA Genotyping.
HLA class II genotyping for DRB1, DQA1, and DQB1 was carried out on DNA extracted from whole blood or mouth swab samples with DNA-based HLA class II typing and analysis methods as reported (15).
HLA-Specific Q-PCR.
The development and validation of a panel of HLA-specific Q-PCR assays was recently described (12). Thirteen Q-PCR assays were used for the current studies (five new assays were added) targeting specific sequences of DRB1*01, DRB1*15/16, DRB1*03, DRB1*07, DRB1*08, DRB1*10, DRB4*01, DQA1*01, DQA1*03, DQA1*05, DQB1*02, DQB1*03, and DQB1*06. Six aliquots of test DNA and a calibration curve were run on each plate, using a dilution of the equivalent DNA of 500, 100, 50, 10, 5, 1, or 0.5 cells homozygous for the HLA sequence in a background of 20,000 cell equivalents negative for the HLA sequence and two additional DNA aliquots for β-globin amplified concurrently to define the total DNA of the test sample. The acceptable range for each aliquot tested DNA could not exceed 30,000 genome equivalents because larger DNA amounts can result in PCR inhibition. An initial incubation at 50°C for 2 min was followed by 95°C for 10 min, 45 cycles of 95°C for 15 sec and 60°C for 1 min (various temperatures depending on the HLA-specific assay). Data were collected with an Applied Biosystems (Foster City, CA) 7000 sequence detector and analyzed with Sequence Detection System software (Applied Biosystems). The quantity of microchimeric DNA was expressed as the genome equivalent number of microchimeric cells per 100,000 host cells (conversion factor of 6.6 pg of DNA for one cell). All assays established had similar sensitivity, detecting a single cell equivalent in a background of 20,000 host cells (0.005%). Specificity was validated by testing each HLA-specific assay against an extended panel of 47 well characterized HLA cell lines representing all HLA-DRB1, DRB3, DRB4, DRB5, DQA1, and DQB1 allele groups. As noted above, the sensitivity levels of all HLA-specific Q-PCR assays were the same, and the percent of subjects positive was not different according to the HLA-specific Q-PCR assay. Q-PCR testing was done on DNA extracted from whole blood.
FISH with Concomitant Immunohistochemistry.
To identify and characterize female cells in male pancreases as hematopoietic or islet β cells, a method was used that combines FISH and immunohistochemistry for the same section (and cells) as recently described (16). Briefly, tissue sections were first subjected to immunohistochemistry with antibodies to insulin (ICN Biomedicals, Irvine, CA) and CD45 for hematopoietic cells (DAKO, Carpinteria, CA). Stains were developed with substrates DAB (BioFX Laboratories, Owings Mills, MD) and SG (Vector Laboratories, Burlingame, CA). As negative controls, no primary antibody was added. Additionally, the CD45 signal served as an internal control, showing female cells expressing insulin were not hematopoietic cells overlying islet β cells and vice versa. The same section was then probed for X and Y chromosomes. DXS1 probe was labeled with red fluorescent Cy3-dUTP (Amersham Pharmacia Biotech U.K. Limited, Little Chalfont, Buckinghamshire, U.K.) and DYS1 probe with green fluorescein-12-dUTP (Roche Applied Science, Mannheim, Germany). Cells were required to have two signals within a clearly defined nucleus to be counted. Cells with two X signals in close proximity were excluded because of the possibility of a split single signal. Two additional independent blinded individuals reviewed each candidate female cell. After identification of cells with two X chromosome signals, coordinates were recorded so cells could also be examined throughout three dimensions by using the DeltaVision microscopy system (Applied Precision, Issaquah, WA). In secondary studies, cells with more than two signals in a clearly defined nucleus were also counted.
Statistical Analysis.
Univariable and adjusted comparisons of microchimerism levels were carried out by using linear regression models applied to ranks of the MMc values. For the analysis of MMc prevalence, each subject's outcome was dichotomozied to positive (>0) or negative (0) to compare proportions of subjects with a positive MMc result. Logistic regression was carried out to evaluate odds ratios, associated confidence intervals, and P values. Variables evaluated for confounding in both the linear model and logistic model included age at blood draw, sex, birth order, and total DNA cell equivalents tested. For both the linear and logistic regression models, generalized estimating equations were used to obtain robust standard error estimates that appropriately account for correlation between siblings.
Because HLA class II genotypes are an important factor in T1D risk, to address the possibility that differences of MMc in T1D patients and unaffected siblings was explained by enrichment for the T1D-associated HLA haplotypes in patients, analysis was also conducted with subjects grouped into genotype groups: homozygous for the T1D-associated DQB1*0201-DRB1*03 haplotype, or for the DQB1*0302-DRB1*04 haplotype, with one of each haplotype, with a single copy of either haplotype, or with no copy of either haplotype. Any two siblings can share both, one, or no HLA haplotype and, although T1D patients derived from families with at least one unaffected sibling, individual proband-unaffected sibling pairs constituted less than half the total study population because some DNA samples did not meet assay quality criteria, were not available, or the study subject was not informative for MMc. The subjects in the current studies were very similar to the overall study subject population for percentage of probands and unaffected siblings informative for MMc and for the distribution of nontransmitted maternal HLA haplotypes.
For the analysis of female cells in each of four male pancreases, cells were the unit of measurement and odds ratios, and associated confidence intervals were calculated for the T1D pancreas compared with each of the other pancreases. For example, the subject with T1D was compared with the age-similar control, and the odds of XX cells were compared between the two.
Supplementary Material
Acknowledgments
We thank Beverly Torok-Storb and Thomas Spies for discussion and critical reading of the manuscript. This work was supported by National Institutes of Health Grants AI-45659 and AI-47121 (to J.L.N.), the Juvenile Diabetes Research Foundation, the European Association for the Study of Diabetes (K.M.G.), the Iacocca Foundation (L.S.L.), and the Wellcome Trust and Diabetes U.K. (E.A.M.G.).
Abbreviations
- MMc
maternal microchimerism
- T1D
type 1 diabetes
- Q-PCR
quantitative real-time PCR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606169104/DC1.
References
- 1.Hall J, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Blood. 1995;86:2829–2832. [PubMed] [Google Scholar]
- 2.Lo DYM, Lo ES, Watson N, Noakes L, Sargent IL, Thilaganathan B, Wainscoat JS. Blood. 1996;88:4390–4395. [PubMed] [Google Scholar]
- 3.Petit T, Gluckman E, Carosella E, Brossard Y, Brison O, Socie G. Exp Hematol. 1995;23:1601–1605. [PubMed] [Google Scholar]
- 4.Petit T, Dommergues M, Socie G, Dumez Y, Gluckman E, Brison O. Br J Hematol. 1997;100:767–771. doi: 10.1046/j.1365-2141.1997.2603076.x. [DOI] [PubMed] [Google Scholar]
- 5.Lo ESF, Lo YMD, Hjelm NM, Thilaganathan B. Br J Hematol. 1998;100:605–606. doi: 10.1046/j.1365-2141.1998.0636a.x. [DOI] [PubMed] [Google Scholar]
- 6.Piotrowski P, Croy BA. Biol Reprod. 1996;54:1103–1110. doi: 10.1095/biolreprod54.5.1103. [DOI] [PubMed] [Google Scholar]
- 7.Pollack MS, Kapoor N, Sorell M, Kim SJ, Christinasen FT, Silver DM, Dupont B, O'Reilly RJ. Transplantation. 1980;30:331–334. doi: 10.1097/00007890-198011000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Maloney S, Smith A, Furst DE, Myerson D, Rupert K, Evans PC, Nelson JL. J Clin Invest. 1999;104:41–47. doi: 10.1172/JCI6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson JL, Furst DE, Maloney S, Gooley T, Evans PC, Smith A, Bean MA, Ober C, Bianchi DW. Lancet. 1998;351:559–562. doi: 10.1016/S0140-6736(97)08357-8. [DOI] [PubMed] [Google Scholar]
- 10.Adams K, Nelson JL. J Am Med Assoc. 2004;291:1127–1131. doi: 10.1001/jama.291.9.1127. [DOI] [PubMed] [Google Scholar]
- 11.Ohtsuka T, Miyamoto Y, Yamakage A, Yamazaki S. Arch Dermatol Res. 2001;293:387–391. doi: 10.1007/s004030100245. [DOI] [PubMed] [Google Scholar]
- 12.Lambert NC, Erickson TD, Yan Z, Pang JM, Guthrie KA, Furst DE, Nelson JL. Arthritis Rheum. 2004;50:906–914. doi: 10.1002/art.20200. [DOI] [PubMed] [Google Scholar]
- 13.Krause DS, Theise ND, Collector MI, Henegariu O, Hwang S, Gardner R, Neutzel S, Sharkis SJ. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 14.Brazelton T, Rossi F, Keshet G, Blau HM. Science. 2000;290:1775–1779. doi: 10.1126/science.290.5497.1775. [DOI] [PubMed] [Google Scholar]
- 15.Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EA, Bingley PJ. J Clin Endocrinol Metab. 2004;89:4037–4043. doi: 10.1210/jc.2003-032084. [DOI] [PubMed] [Google Scholar]
- 16.Stevens AM, Hermes H, Rutledge J, Buyon J, Nelson JL. Lancet. 2003;362:1617–1623. doi: 10.1016/S0140-6736(03)14795-2. [DOI] [PubMed] [Google Scholar]
- 17.Terada N, Hamazaki T, Oka M, Hoki M, Mastalerz DM, Nakano Y, Meyer EM, Morel L, Petersen BE, Scott EW. Nature. 2002;416:542–545. doi: 10.1038/nature730. [DOI] [PubMed] [Google Scholar]
- 18.Srivatsa B, Srivatsa S, Johnson KL, Bianchi DW. J Pediatr. 2003;142:31–35. doi: 10.1067/mpd.2003.mpd0327. [DOI] [PubMed] [Google Scholar]
- 19.Marleau AM, Greenwood JD, Wei Q, Singh B, Croy BA. Lab Invest. 2003;83:673–681. doi: 10.1097/01.lab.0000067500.85003.32. [DOI] [PubMed] [Google Scholar]
- 20.Reed AM, McNallan K, Wettstein P, Vehe R, Ober C. J Immunol. 2003;172:5041–5046. doi: 10.4049/jimmunol.172.8.5041. [DOI] [PubMed] [Google Scholar]
- 21.Khosrotehrani K, Johnson KL, Cha DH, Salomon RN, Bianchi DW. J Am Med Assoc. 2004;292:75–80. doi: 10.1001/jama.292.1.75. [DOI] [PubMed] [Google Scholar]
- 22.Rinkevich B. Hum Immunol. 2001;62:651–657. doi: 10.1016/s0198-8859(01)00249-x. [DOI] [PubMed] [Google Scholar]
- 23.Moore G, Abu-Amero SN, Bell G, Wakeling E, Kingsnorth A, Stanier P, Jauniaux E, Bennett ST. Diabetes. 2001;50:199–203. doi: 10.2337/diabetes.50.1.199. [DOI] [PubMed] [Google Scholar]
- 24.Abramowicz MJ, Andrien M, Dupont E, Dorchy H, Parma J, Duprez L, Ledley FD, Courtens W, Vamos E. J Clin Invest. 1994;94:418–421. doi: 10.1172/JCI117339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risch N. Am J Human Gen. 1987;40:1–14. [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan J, Land S. J Immunol. 2005;174:7123–7128. doi: 10.4049/jimmunol.174.11.7123. [DOI] [PubMed] [Google Scholar]
- 27.Lee TH, Paglieroni T, Ohto H, Holland PV, Busch MP. Blood. 1999;93:3127–3139. [PubMed] [Google Scholar]
- 28.Kodama S, Kuhtreiber W, Fujimura S, Dale EA, Faustman DL. Science. 2003;302:1223–1226. doi: 10.1126/science.1088949. [DOI] [PubMed] [Google Scholar]
- 29.Seaberg RM, Smukler SR, Kieffer TJ, Enikolopoy G, Asghar Z, Wheeler MB, Korbutt G, van der Kooy D. Nat Biotech. 2004;22:1115–1124. doi: 10.1038/nbt1004. [DOI] [PubMed] [Google Scholar]
- 30.Dor Y, Brown J, Martinez OI, Melton DA. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 31.Bingley PJ, Gale EAM. BMJ. 1989;298:558–560. doi: 10.1136/bmj.298.6673.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. Diabetes Mellitus. Report of a WHO Study Group. Geneva: WHO; 1985. Technical Report Series 727. WHO/NCD/NCS/99.2. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.