Abstract
Adaptation by natural selection proceeds most efficiently when alleles compete solely on the basis of their effects on the survival and reproduction of their carriers. A major condition for this is equal Mendelian segregation, but meiotic drive can short-circuit this process. The evolution of drive often involves multiple, interacting genetic components, together with enhancers and suppressors of drive. Chromosomal inversions that suppress crossing over are also frequently associated with drive systems. This study investigates the effects of these processes on patterns of molecular evolution in the fly Drosophila recens, which is polymorphic for a driving X chromosome (XD). Whereas standard wild-type chromosomes exhibit high levels of polymorphism at multiple loci, all of the XD chromosomes effectively carry a single multilocus haplotype that spans at least 130 cM. The XD is associated with a complex set of inversions that completely suppresses recombination between the standard wild-type chromosome and XD in heterozygous females, which maintain nonrandom associations among loci that presumably interact epistatically for the expression of drive. The long-term costs of foregoing recombination may be substantial; in combination with its low equilibrium frequency, this makes the XD chromosome susceptible to the accumulation of deleterious mutations. Consistent with this, XD chromosomes are apparently fixed for a recessive mutation that causes female sterility. Thus, the XD in D. recens appears to be in chromosome-wide linkage disequilibrium and in the early stages of mutational degradation.
Keywords: Drosophila recens, genetic conflict, inversion, population genetics, segregation distortion
Mendelian segregation ensures that alleles compete on a level playing field, thus maximizing the efficiency of natural selection (1). Although the deterministic spread of an allele under selection is generally thought to occur because of a fitness benefit to its carriers, this can also come about as a result of biased transmission through mechanisms such as meiotic drive (2). By being transmitted to more than half of the gametes, driving alleles can spread even if they adversely affect the fitness of their carriers. Furthermore, when such alleles occur on the X or Y chromosome, this can result in an unequal population sex ratio, further penalizing individuals that produce an excess of the more abundant sex (3). This can even lead to population extinction if the driver reaches a sufficiently high frequency (4).
Drive systems involve multiple, interacting components, usually with both enhancers and suppressors of drive (2). Epistatic interactions among these components favor suppression of crossing over among them (5). Close linkage among the core components of drive systems may be required for their initial evolution, because otherwise low-fitness genotypes are produced by recombination (6). It is therefore not surprising that drive loci are often found in low recombination regions, such as heterochromatin, and/or are associated with chromosomal inversions that suppress crossing over (but see ref. 7). Through an intragenomic arms race, a drive system may eventually come to involve a complex of unlinked suppressors, as well as tightly linked enhancers and core components (8).
As a consequence of selection for tight linkage, a drive system may eventually tie up a large block of genes as a single segregating unit. Accordingly, the best-studied drive systems [SD in Drosophila melanogaster (9) and t-haplotype in Mus (10)] are associated with regions of reduced recombination caused by inversions, which can extend for up to 20 cM. As a result, patterns of polymorphism and linkage disequilibrium (LD) throughout such regions may be affected. In D. melanogaster, polymorphism is reduced near SD, and very weak LD among sites extends up to 3 cM away (11). In contrast, in Drosophila pseudoobscura, four allozyme loci that span a distance of ≈50 cM on the right arm of the X (XR) are differentiated between sex ratio and standard chromosomes, which differ by a complex of three inversions (12, 13). At the DNA level, one of the two XR loci surveyed (Est-5) was in LD with sex ratio, whereas none of the three on the left arm of the X showed an association with sex ratio (14). In neither of these cases is crossing over suppressed, or LD maintained, over the entire chromosome.
Here we conducted an evolutionary genetic analysis of the X chromosome in Drosophila recens to infer the evolutionary history of X-drive and to understand its genomic consequences. D. recens is a mycophagous species in the quinaria group of the subgenus Drosophila and is found in boreal forests of eastern and central North America. In this study we surveyed six loci that span most of the X chromosome and asked whether any show an association with X-drive (the exact genes underlying X-drive in D. recens are unknown). Patterns of molecular variation of driving X chromosome (XD) and standard wild-type chromosome (XST) in D. recens indicate that the evolution of X-drive has resulted in a dramatic molecular signature: a chromosome-wide XD haplotype that harbors little polymorphism. This pattern of chromosome-wide suppression of recombination, which is associated with multiple inversions, suggests that Hill–Robertson interference among closely linked loci subject to selection (15) will eventually lead to the evolutionary degradation of this XD system.
Results
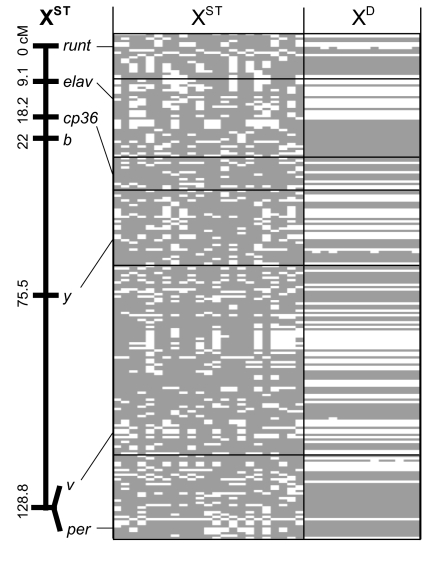
Sequences from six X-linked loci (cp36, elav, per, runt, v, and y) were obtained from D. recens. We first determined the gene order and recombination distances on the XST background among these loci and two visible markers (y and b). The recombination map in Fig. 1 indicates that the seven loci are spread over ≈129 cM. Species within the subgenus Drosophila have Muller's element A as their X chromosome, which ranges in size: 109 cM in Drosophila buzzatii (16), 116 cM in Drosophila hydei (17), 131 cM in Drosophila mojavensis (18), and 170 cM in Drosophila virilis (19). Because the cumulative runt-to-per map distance of D. recens falls within this range, it is likely that these loci span most of the X chromosome.
Fig. 1.
Kosambi-corrected map distances among loci and haplotype structure of sampled chromosomes. Each column is a sampled chromosome. Gray lines indicate a site that is identical to the consensus XST sequence, and white lines indicate a difference from the XST consensus. Only parsimony informative sites were included; sites with segregating indels and singleton polymorphisms were excluded for clarity.
To characterize X chromosomes from natural populations as XD or XST, we surveyed the offspring sex ratio of 262 wild-caught males collected throughout the range of D. recens. Seven of these males (2.6%) sired almost exclusively daughters (mean fraction female = 0.982 ± 0.009) and were classified as XD. The remaining 255 males produced ≈1:1 offspring sex ratios (mean fraction female = 0.536 ± 0.005) and were classified as XST. Offspring sex ratio is bimodal, as no males sired between 81% and 93% daughters [supporting information (SI) Fig. 5]. Populations do not vary significantly in their frequency of XD (P > 0.5, χ2 = 4.212, df = 11); thus, combining our frequency data with those of Jaenike (20), a total of 11 of 391 males were found to harbor an XD, resulting in an overall frequency among males of 0.028 with a 95% confidence interval of 0.014–0.054 (SI Table 2).
We surveyed levels of polymorphism and divergence in a total of 44 X chromosomes: 29 XST that were sampled randomly from the central part of the range of D. recens, 14 XD from populations spanning the range of the species (SI Table 2), and 1 from the closely related species Drosophila subquinaria. From each chromosome we sequenced the six loci (cp36, elav, per, runt, v, and y), for a total of ≈5 kb per chromosome. The sample of XST chromosomes shows a consistently high level of polymorphism and a low level of LD. Within the XST sample, the overall silent sites π and θ are 0.048 (SE 0.010) and 0.059 (SE 0.010), respectively. The difference between these two estimates of polymorphism indicates an excess of rare variants, and across loci the observed mean Tajima's D of −0.87 was significantly less than the simulated mean for a neutral model with constant population size [D(exp) = −0.10, P = 0.012]. Within the XST sample the standardized population recombination parameter (ρ/θ) ranged between 0.90 and 52.7 (Table 1), with a mean of 20.6. In comparison to other Drosophila species (21), this suggests that most of the loci are located in regions of at least moderate recombination. To test for nonrandom associations within and among loci, we concatenated the data from the six loci (n = 23 chromosomes), which resulted in 259 informative segregating sites and thus 10,560 pairwise comparisons among sites. We identified 22 pairs of sites with a significant nonrandom association; all 22 pairs were within loci (SI Fig. 6), indicating independence among loci on the XST background.
Table 1.
Summary of molecular population genetic analyses
| cp36 | elav | per | runt | v | y | All | |
|---|---|---|---|---|---|---|---|
| XST | |||||||
| n | 29 | 29 | 29 | 28 | 26 | 26 | 23 |
| LALL | 744 | 1140 | 648 | 375 | 759 | 1602 | 5268 |
| LSIL | 260 | 279 | 141 | 206 | 411 | 410 | 1769 |
| M | 41 | 65 | 48 | 46 | 170 | 105 | 447 |
| S | 40 | 68 | 45 | 39 | 147 | 99 | 408 |
| S1 | 26 | 34 | 15 | 18 | 73 | 61 | 220 |
| π | 2.06 | 4.67 | 8.89 | 4.71 | 7.20 | 3.68 | 4.78 |
| θW | 4.02 | 6.19 | 8.14 | 4.86 | 10.85 | 6.71 | 6.25 |
| DT | −1.75 | −0.81 | 0.35 | −0.60 | −0.90 | −1.51 | −0.87 |
| KS | 0.03 | 0.10 | 0.21 | 0.07 | 0.15 | 0.18 | 0.12 |
| ρ | 0.74 | 0.47 | 0.31 | 0.77 | 0.07 | 0.02 | — |
| ρ/θW | 52.7 | 32.2 | 13.8 | 22.9 | 1.41 | 0.90 | — |
| XD | |||||||
| n | 14 | 14 | 14 | 14 | 14 | 14 | 14 |
| LSIL | 260 | 280 | 142 | 235 | 529 | 560 | 2042 |
| S | 1 | 2 | 2 | 0 | 2 | 0 | 7 |
| S1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| π | 0.06 | 0.15 | 0.69 | 0 | 0.17 | 0 | 0.12 |
| θW | 0.12 | 0.22 | 0.44 | 0 | 0.12 | 0 | 0.11 |
| DT | — | — | — | — | — | — | 0.45 |
| XD vs. XST | |||||||
| K*ST | 0.31 | 0.31 | 0.32 | 0.14 | 0.24 | 0.28 | 0.22 |
| Snn | 1.00 | 1.00 | 1.00 | 0.96 | 0.98 | 0.98 | 1.00 |
| Fixed differences | 3 | 1 | 1 | 0 | 5 | 0 | 10 |
| Polymorphic in XST, fixed in XD | 42 | 67 | 61 | 46 | 163 | 105 | 484 |
| Polymorphic in XD, fixed in XST | 1 | 1 | 2 | 0 | 1 | 0 | 5 |
| Shared polymorphism | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
The last column is for the concatenated data set. Number of all (LALL) and silent sites (LSIL), silent segregating mutations (M), sites (S), and singletons (S1) are excluding alignment gaps. π and θW estimate silent-site nucleotide diversity, also excluding gaps, with values per site and multiplied by 10−2. KS is the Jukes–Cantor corrected silent-site divergence to D. subquinaria, and ρ estimates the per-site population recombination parameter. Values in bold are significant at the P< 0.0001 level.
In contrast to the XST sample, the XD chromosomes show a consistently low level of polymorphism. Summed over all loci, we identified seven segregating sites, of which two were singletons, resulting in silent site polymorphism estimates of π = 0.0012 and θ = 0.0011. None of these seven segregating sites results in an amino acid change, and two of these are also polymorphic within XST. Because of the abundance of multiple hits and high recombination in the XST samples it is difficult to determine with certainty whether these two shared polymorphisms are identical by descent. We also identified two indels within noncoding regions of the XD sample: a polymorphic (A)n in y that differs by 1 bp and is polymorphic within both XD (in 2 of 14 XD chromosomes) and XST, and a 17-bp deletion in v that is polymorphic only within XD (in 4 of the 14 XD chromosomes). The overall Tajima's D for the XD sample is 0.45, which is not significantly different from the neutral expectation. Within the XD chromosomes there is some evidence for recombination: excluding indels we find evidence for a single recombination event, and including indels there is evidence for three recombination events. To test for LD between XST and XD we used the concatenated sequences of both XST and XD haplotypes (n = 37), which resulted in 284 informative segregating sites and 14,290 valid comparisons among sites. We identified 561 pairs of sites with a significant nonrandom association: of these, 325 pairs (58%) occurred among loci, including all pairwise combinations (SI Figs. 7 and 8), indicating that LD between chromosome types (XST vs. XD) is maintained throughout the chromosomal region we sampled.
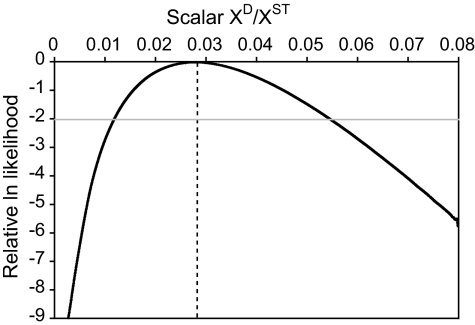
Comparisons of the levels of silent nucleotide polymorphism between XD and XST chromosomes yield ratios π[ST]/π[D] = 0.0478/0.0012 ≈ 40 and θ[ST]/θ[D] = 0.0589/0.011 ≈ 53. Using coalescent simulations assuming a constant population size, we determined the maximum likelihood estimate (MLE) of this difference in polymorphism, and hence effective population size (Ne), between the XST and XD subpopulations. As shown in Fig. 2, the XD has an Ne of 2.8% of the XST, with a 2-unit support interval of 1.2–5.4%.
Fig. 2.
MLE of the ratio of the Ne of XD relative to XST. The MLE of this ratio of 0.0276 is indicated by the dotted line, and the 2-unit support interval of 0.0118–0.054 is indicated by the gray line.
The phylogenetic tree made from the concatenation of the six loci shows several important aspects of the evolutionary history of the XD (Fig. 3). First, the XD samples are monophyletic, both for the concatenated data set and for each of the individual loci (SI Fig. 9), indicating that the XD chromosome in D. recens has a single origin. For each individual locus, as well as for the concatenated data, there is a perfect association between the XD haplotype group and a biased offspring sex ratio for the males that carry these chromosomes (Fisher's exact test, P < 0.0001). This perfect association indicates that suppressors of drive are rare or absent in D. recens. Thus, the XD forms a single and unique haplogroup that encompasses all of the loci we surveyed and that must, therefore, span at least 129 cM of the X chromosome.
Fig. 3.
Phylogenetic tree made from the concatenated sequences of the 14 XD and 23 XST chromosomes of D. recens, using D. subquinaria to root the tree. The tree was constructed by using the neighbor-joining algorithm with Kimura two-parameter adjusted branch lengths. Support values were generated from 10,000 bootstrap replicates. Sites with gaps were excluded from the analyses, resulting in a total of 3,757 bp.
Second, the phylogenetic tree clearly shows that the XD chromosome is derived from XST and does not represent an ancient polymorphism (Fig. 3). Nevertheless, all of the loci are differentiated between the XD and XST chromosomes (Table 1). Comparing the XD with all XST samples, there are 10 fixed nucleotide differences, including four synonymous changes and one nonsynonymous change in coding regions and five changes in noncoding regions. To estimate the evolutionary age of the current XD haplotype, we used the nine silent fixed differences between XD and all XST and assume a molecular clock of 1.5 × 10−9 silent substitutions per site per year (22, 23). This gives an expected XD age of 300,000 years, with a range of 100,000–400,000 years. To determine the minimum age of the XD (or time to most recent common ancestor) we used the method of Andolfatto et al. (24). This method assumes, first, that the XD inversion has a unique origin from a XST ancestor, and, second, that the XD inversion brought about complete genetic isolation from the XST. If we assume the same molecular clock as above and three generations per year, the minimum age of the XD haplotype is 250,000 years. Note that these methods estimate the age of the current XD haplotype, not the age of the underlying drive system (which may be much older).
To estimate recombination rates in XD/XST females we used the visible X-linked mutants y and b. In XST/XST females there is nearly free recombination between y and b: among 1,936 F2 progeny of ++/y b females, 40.7% were recombinants, resulting in a distance of 60 cM using Kosambi's (25) mapping function. To estimate recombination between y, b, and the loci responsible for drive in XD/XST females, we scored 5,402 progeny of y b/XD females and recovered no recombinant y + or + b genotypes. We determined the offspring sex ratio of 281 F2 males and found that all y b males produced normal 1:1 sex ratios, whereas all ++ males sired only daughters. Thus, whereas y and b segregate nearly independently in XST/XST females, recombination is completely suppressed between y, b, and drive in XD/XST females.
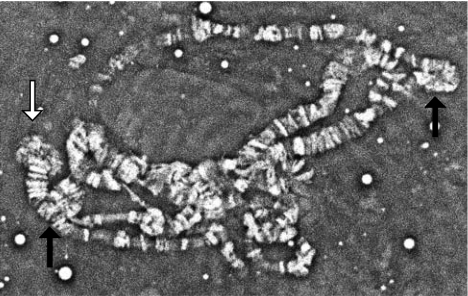
Chromosomal inversions are common in Diptera (26) and are known to be associated with driving chromosomes in other systems. Fig. 4 shows the polytene X chromosomes of an XD/XST(y b) female. As evidenced by the two inversion shifts noted in the figure, which are indicative of overlapping inversions, this complex probably contains at least four different and overlapping inversions. It is likely that these inversions suppress recombination and thus maintain linkage among the multiple loci required for full drive expression.
Fig. 4.
Polytene X chromosomes of XD/XST female. Inversion shifts, which indicate overlapping inversions, are noted by the black arrows, and the tip of the chromosome is indicated by the white arrow.
Laboratory culturing revealed that eight of eight independently sampled XDs from across the range of D. recens carried a recessive female sterile mutation. To determine whether the different XD isolates carry the same sterility mutation, we conducted complementation tests. By crossing different strains we produced XD i/XDBemidji females, where XDBemidji represents an XD from Bemidji, MN, and XDi represents one of six other XD isolates. In no case did an XDi/XDBemidji female produce any offspring (n > 20 tested females per cross type). Thus, the sterility mutations carried by the different XDs are allelic and most likely share a single origin.
Discussion
X chromosome drive has had a dramatic impact on the genome of D. recens. Our data show that drive is associated with a unique X chromosome haplotype that harbors little polymorphism in comparison to the nondriving XST chromosome. Furthermore, all six of the X-linked loci we examined are strongly differentiated between the XD and XST chromosomes. These loci span ≈130 cM, which is likely to encompass most of the X chromosome. Thus, the evolution of this drive system has tied up the entire XD chromosome as a single nonrecombining unit. This pattern is probably the result of the accumulation of inversions that now suppress recombination between XD and XST chromosomes and have been selected to maintain associations among the loci that control drive (6).
Our sample of D. recens XST chromosomes indicates that these are highly polymorphic and freely recombining chromosomes. However, compared with XST, the XD sample harbors 20- to 80-fold less polymorphism, with perfect association with X-drive across all six loci. Males carrying an XD were found across the range of D. recens, but there was no geographic differentiation among the XD samples (HST =−0.03, P =0.60, using populations with at least two XD alleles). Such patterns could have arisen if the XD chromosome has gone through a recent bottleneck as the result of the spread of a new sex ratio inversion or enhancer-of-drive allele. Although such sweeps probably have occurred several times during the history of the XD (because it has apparently accumulated multiple inversions), three lines of evidence suggest that the XD has experienced neither a very recent sweep nor a recent dramatic reduction in frequency. First, a sweep of a nonrecombining chromosome will eliminate standing polymorphism, yet we find several polymorphic sites among extant XD chromosomes, almost all of which are mutations absent from the XST class. Second, during the initial recovery phase after a sweep, most polymorphisms will be unique, resulting in a star-like phylogeny and an excess of low-frequency mutations. However, only 2 of the 11 polymorphisms (including indels) are singletons, resulting in a frequency spectrum of mutations consistent with neutral expectations, or possibly with a gradual decline in frequency of the XD, because there is a (nonsignificant) deficit of low-frequency mutations. Finally, after accounting for the difference in recombination between the two types of chromosomes, the polymorphism-based MLE of the ratio of Ne between XD and XST of 0.0276 is nearly identical to the current XD/XST frequency ratio of 0.028 (Fig. 2). In other words, the XD chromosome harbors a level of variation consistent with equilibrium expectations. Thus, although the XD may have experienced selective sweeps in the past, we see no evidence of this; if anything, our data suggest that XD has been at a consistently low or declining frequency in the recent evolutionary past.
The inversions that suppress recombination between XD and XST in D. recens probably maintain linkage among the genes that interact epistatically for the expression of drive (6). There is some evidence for historical recombination events within the XD chromosomes; these may have occurred before the establishment of the sterility mutation, or possibly may represent a low level of fertility of XD/XD females in the wild (although such females must be very rare, unless XD used to be at a higher frequency). Nevertheless, compared with the XST chromosomes the XD has severely reduced recombination. In the long-term, the costs of foregoing recombination can be substantial because nonrecombining chromosomes are likely to suffer low polymorphism, reduced potential for adaptation to changing conditions, and more frequent fixation of deleterious mutations due to Hill–Robertson interference (15, 27). The lack of recombination, in combination with its low equilibrium frequency, may make the XD chromosome particularly susceptible to accumulation of deleterious mutations, akin to the process seen in Y and neo-Y chromosomes (28). In these systems, however, it is possible for dosage compensation to evolve by coopting of the molecular machinery used to up-regulate the XST chromosome in males (29), but this is not possible for the XD in females. We therefore hypothesize that a positive feedback between declining chromosome frequency and increasing vulnerability to the spread of deleterious mutations will lead to a downward spiral in the fitness and frequency of the XD, eventually resulting in its loss from the species.
Preliminary evidence suggests that the XD of D. recens is already fixed for at least one deleterious mutation. XDs collected from throughout the range all carry the same recessive female sterility factor, resulting in sterility of XD/XD females. Because XD/XD females are sterile and recombination is suppressed in XD/XST females, neither this nor any other deleterious mutation can be eliminated from the XD via recombination. Regardless of whether this female sterile mutation was captured when an XD-associated inversion first arose, or arose within the XD lineage, this mutation will constrain the XD to low frequencies. If we assume that the dynamics of XD is governed solely by its transmission advantage in males and the sterility of XD/XD females, its expected equilibrium frequency is 25%. However, the current and evolutionarily recent frequency of XD is 2.8%, i.e., 9-fold less than expected, indicating that there must be additional deleterious fitness effects of the XD chromosome, such as lower fertilization success of XD males or reduced fitness of XD/XST females.
Although much of the XD chromosome evidently does not experience recombination, gene conversion between the XD and XST chromosomes may reduce the rate of XD degeneration. Gene conversion may play a substantial role in the maintenance of polymorphism, particularly in inversions and other regions of low recombination (21, 30, 31). Using the method of Betrán et al. (32), we analyzed each gene individually and identified seven unique gene conversion events, all of which are from XD to XST, and which range from 2 to 443 bp, with an average of 172 bp (SI Table 3). However, because all of the gene conversion events we identified are from XD to XST, these tracts may represent ancestral polymorphism and in any case do not represent rescue of XD by XST sequences.
There has been renewed interest in the effect of inversions on patterns of molecular evolution, particularly with respect to detecting the signal of coadapted gene complexes (33, 36). If the genes tied up within inversions do not interact epistatically, then differentiation among inversion types is expected to persist near the breakpoints but will break down throughout the inverted regions because of double crossovers and gene conversion events in heterozygotes (34, 35). Although allozymes were found in LD between inversion karyotypes (26), early studies of DNA polymorphism within and around inversions found LD only near the breakpoints (30). However, some more recent studies have revealed some LD at the DNA level among loci within inversions (36–38). Our results indicate that the D. recens XD haplotype is maintained in perfect LD across a chromosomal region with several inversions and that, on the XST chromosome, spans at least 130 cM. This LD is much more extensive than any other known drive or inversion system and may be the largest known segregating haplotype in any sexually outcrossing system. However, by preventing recombination over such a large genomic region, presumably to maintain a coadapted set of genes, the XD may ensure its own eventual demise as a result of an increasing load of deleterious mutations.
Materials and Methods
Markers and Map Order.
The X chromosome in D. recens is telocentric and equivalent to Muller's element A (39). The six loci were randomly chosen with respect to gene order and chromosomal location (because there was no genetic map for this species) and include portions of chorion protein 36 (cp36), elav (elav), period (per), runt, vermilion (v), and yellow (y). Primer sequences and fragment details are listed in SI Table 4. DNA was extracted from single males, and all PCR, sequencing, and data editing were performed by using standard protocols.
We determined the gene order and recombination distances among the molecular and available visible markers on an XST background. We crossed a single wild-type male to homozygous yellow (y) brown (b) females from a stock that was isogenic for a single y b XST chromosome, which produced y b/++ females that all carried the same two X chromosomes. We designed restriction digests based on single nucleotide differences between the male's allele and the corresponding allele on the y b chromosome. The genotypes of 230 of the sons of the y b/++ females were used to determine the distances among the markers. All map distances were calculated by using the Kosambi mapping function (25).
XD Characterization and Samples for Polymorphism Analyses.
From populations spanning most of the known range of D. recens (40) we characterized naturally occurring chromosomes as XD and XST. Wild-caught males were mated to laboratory virgin females, and the offspring sex ratio scored: males were characterized as carrying an XD if they produced ≥10 offspring, of which ≥90% were female; otherwise they were characterized as XST as long as they sired ≥10 offspring. All fly cultures were maintained at 22°C on Instant Drosophila Medium (Carolina Biological Supply, Burlington, NC) supplemented with commercial mushroom (Agaricus bisporus).
The 14 XD chromosomes used in this study come from populations across the range of the species, including Chebeague Island, ME (n = 2), Rochester, NY (n = 4), Mattawa, ON, Canada (n = 5), Smoky Mountains, TN (n = 1), Bemidji, MN (n = 1), and Edmonton, AB, Canada (n = 1). The 29 XST chromosomes were sampled randomly from the central populations in the range of D. recens, which do not show any signature of population structure (data not shown). These are from Bemidji, MN (n = 6), Bethlehem, NH (n = 7), Munising, MI (n = 7), and Rochester, NY (n = 9). One individual of the closely related speciesD. subquinaria was used for an outgroup.
Molecular Population Genetic Analyses.
From each of the 44 chromosomes we surveyed the pattern of DNA polymorphism at the six mapped molecular loci. DNA sequences were aligned manually, taking coding frame into account, and molecular evolutionary analyses were conducted by using DnaSP version 4.0 (41) unless otherwise noted. Sites with alignment gaps were excluded from the analyses. In addition, one XST sample was excluded from the v analyses because it contained an indel that spanned the entire 3′ half of the fragment, and a polymorphic glutamine repeat region was excluded from the elav sequence alignment. Polymorphism was measured at silent sites, including pairwise nucleotide diversity, π (42), and nucleotide site variability based on the number of segregating sites, θW (43). The frequency spectrum of polymorphisms was tested for departure from neutrality by using Tajima's D (44) using silent sites only. An overall D for the set of loci was determined by using the program HKA (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA) with 104 coalescent simulations without recombination within loci.
Phylogenetic relationships were constructed for each locus and for all six loci concatenated by chromosome (herein referred to as the concatenated data set) using parsimony with 10,000 bootstrap replicates in PAUP* v4.0b10 (45). We determined the degree of genetic differentiation between XD and XST samples using Snn (46) and KST* (47), with significance determined by using 10,000 permutations.
The population recombination parameter, ρ, was inferred for each locus by using the composite-likelihood method of Hudson (48), as implemented in the program LDhat (49). ρ is equal to 4Ne r, where r is the per-generation recombination rate, taking into account the lack of crossing over in males. Ne is the effective population size; for the X chromosome, this equals 0.75 of the autosomal value if there is no sexual selection (50). Fisher's exact test (51) was used to test whether pairs of variable sites within and between loci showed significant associations. We used the concatenated sequences, excluded singleton polymorphisms and sites with more than two alleles segregating, and applied a sequential Bonferroni correction (52) to account for multiple comparisons.
To infer the relative Ne of the XD versus the XST chromosome, we used simulations of the coalescent process using methods similar to those of Bachtrog and Charlesworth (53). This enabled us to not only account for the difference in recombination environments between the two types of chromosomes, but also to estimate confidence intervals for this difference. We simulated a single tree with six completely linked loci for the XD and six independent trees for the XST. We used a sample size of 14 chromosomes, using all of the XD data and a random sample of the XST data. For each locus the total number of segregating sites was assigned to the XD and XST trees in proportion to their length. After each run the number of segregating sites on the XD and XST was determined and the quantity ΔSsim = (SST − SD)/SST was computed. To estimate the reduction in Ne, the tree length was multiplied by a scaling factor k before mutations were laid down on the tree. The proportion of runs Mδ for which |ΔSobs− ΔSsim| ≤ 0.002 was tallied, and the MLE of k was determined by L(k|ΔS) = (Mδ/n), where n = 100,000 replicates.
Suppression of Recombination in XD/XST Females.
We used X-linked visible markers to test directly for suppressed recombination in heterozygous XST/XD females. Recombination between y and b on an XST background was determined by using offspring of heterozygous y b/++ females, and recombination on the heterozygous XST/XD background was determined by using the offspring of heterozygous y b/XD females. The rate of recombination and distance in cM between y and b were determined by scoring the offspring of these heterozygous females that had been mated to y b males. To estimate the recombination distances between y, b, and drive, sons of y b/XD females were crossed individually to virgin females, the offspring sex ratio was scored, and the male was genotyped for XD as described above.
Identification of XD Inversions.
To identify chromosomal inversions we used standard cytological techniques to prepare orcein-stained polytene chromosome squashes of third-instar larvae. The X chromosome was identified from male y b larvae because it consistently did not show regions of nonpairing, which were common among the autosomes, and it was paler in appearance than the autosomes. We crossed XD males to virgin XST y b females, which produced the XST/XD females that we then used to identify inversions associated with the XD.
Allelism of the XD Female Sterility Factor.
All of the XDs from this study that we have maintained in the laboratory harbor a recessive female sterility factor [n = 7 XDs from this study and 1 from Jaenike (20)]. We conducted complementation tests to determine whether this sterility factor was allelic in the various XD chromosomes. Because females that carry two copies of the same XD are sterile, to test whether two different XD chromosomes, e.g., XDi and XDj, are allelic, we crossed XDi/y b females to XDj males. Because the resulting F1 daughters were either XDi/XDj or XDi/y b, both of which appear to be wild type, we determined the genotype of each daughter by mating them to XST males and scoring their offspring for the presence of y b males. F1 daughters that produced y b sons were inferred to be XDj/y b, and those that produced only wild-type offspring were inferred to be XDi/XDj, but with sterility factors that complemented each other. If the sterility factors on the two different XD chromosomes were allelic and did not complement, then the female produced no offspring.
Supplementary Material
Acknowledgments
We thank M. Minhas and A. Melvin for laboratory assistance; C. Cornish, M. Minhas, and J. Savage for collecting flies; D. Bachtrog for sharing computer code; and P. Andolfatto, A. Betancourt, D. Charlesworth, J. Fry, C. Haag, J. Masly, A. Orr, and two anonymous reviewers for useful discussion and/or comments on the manuscript. This work was supported by a Royal Society USA fellowship (to K.A.D.) and National Science Foundation Grant DEB-0315521 (to J.J.). B.C. is supported by the Royal Society.
Abbreviations
- XD
driving X chromosome
- XST
standard wild-type chromosome
- LD
linkage disequilibrium
- Ne
effective population size
- MLE
maximum likelihood estimate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. EF188848–EF189104).
This article contains supporting information online at www.pnas.org/cgi/content/full/0605578104/DC1.
References
- 1.Crow JF. Sci Amer. 1979;240:134–143. doi: 10.1038/scientificamerican0279-134. [DOI] [PubMed] [Google Scholar]
- 2.Lyttle TW. Trends Genet. 1993;9:205–210. doi: 10.1016/0168-9525(93)90120-7. [DOI] [PubMed] [Google Scholar]
- 3.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Oxford Univ Press; 1930. [Google Scholar]
- 4.Hatcher MJ, Taneyhill DE, Dunn AM, Tofts C. Theor Popul Biol. 1999;56:11–18. doi: 10.1006/tpbi.1998.1410. [DOI] [PubMed] [Google Scholar]
- 5.Prout T, Bundgaard J, Bryant S. Theor Popul Biol. 1973;4:446–465. doi: 10.1016/0040-5809(73)90020-8. [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth B, Hartl D. Genetics. 1978;89:171–192. doi: 10.1093/genetics/89.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derome N, Métayer K, Montchamp-Moreau C, Veuille M. Genetics. 2004;166:1357–1366. doi: 10.1534/genetics.166.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaenike J. Annu Rev Ecol Syst. 2001;32:25–49. [Google Scholar]
- 9.Kusano A, Staber C, Chan HY, Ganetzky B. BioEssays. 2003;25:108–115. doi: 10.1002/bies.10222. [DOI] [PubMed] [Google Scholar]
- 10.Lyon MF. Annu Rev Gen. 2003;37:393–408. doi: 10.1146/annurev.genet.37.110801.143030. [DOI] [PubMed] [Google Scholar]
- 11.Palopoli MF, Wu C-I. Genetics. 1996;143:1675–1688. doi: 10.1093/genetics/143.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturtevant AH, Dobzhansky T. Genetics. 1936;21:473–490. doi: 10.1093/genetics/21.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash S. Genetics. 1974;77:795–804. doi: 10.1093/genetics/77.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacevic M, Schaeffer SW. Genetics. 2000;156:155–172. doi: 10.1093/genetics/156.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordo I, Charlesworth B. Curr Biol. 2001;11:R684–R686. doi: 10.1016/s0960-9822(01)00408-0. [DOI] [PubMed] [Google Scholar]
- 16.Schafer DJ, Fredline DK, Knibb WR, Green MM, Barker JSF. J Hered. 1993;84:188–194. doi: 10.1093/oxfordjournals.jhered.a111315. [DOI] [PubMed] [Google Scholar]
- 17.Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model. New York: Oxford Univ Press; 1997. [Google Scholar]
- 18.Staten R, Schully SD, Noor MAF. BMC Genet. 2004;5:12. doi: 10.1186/1471-2156-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubenko JS, Evgenev MB. Genetica. 1984;65:127–139. [Google Scholar]
- 20.Jaenike J. Amer Nat. 1996;148:237–254. [Google Scholar]
- 21.Andolfatto P, Wall JD. Genetics. 2003;165:1289–1305. doi: 10.1093/genetics/165.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andolfatto P, Przeworski M. Genetics. 2000;156:257–268. doi: 10.1093/genetics/156.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McVean GAT, Vieira J. Genetics. 2001;157:245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andolfatto P, Wall JD, Kreitman M. Genetics. 1999;153:1297–1311. doi: 10.1093/genetics/153.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosambi DD. Ann Eugen. 1944;12:172–175. [Google Scholar]
- 26.Krimbas CB, Powell JR. Drosophila Inversion Polymorphism (CRC, Boca Raton, FL). 1992 [Google Scholar]
- 27.Orr HA, Kim Y. Genetics. 1998;150:1693–1698. doi: 10.1093/genetics/150.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlesworth B, Charlesworth D. Philos Trans R Soc London Ser B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marin I, Siegal ML, Baker BS. BioEssays. 2000;22:1106–1114. doi: 10.1002/1521-1878(200012)22:12<1106::AID-BIES8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Andolfatto P, Depaulis F, Navarro A. Genet Res. 2001;77:1–8. doi: 10.1017/s0016672301004955. [DOI] [PubMed] [Google Scholar]
- 31.Schaeffer SW, Anderson WW. Genetics. 2005;171:1729–1739. doi: 10.1534/genetics.105.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betrán E. Rozas J, Navarro A, Barbadilla A. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobzhansky T. (Hereditas Lund, Swed) Suppl. 1949. pp. 210–224. [Google Scholar]
- 34.Ishii K, Charlesworth B. Genet Res. 1977;30:93–106. [Google Scholar]
- 35.Navarro A, Bardadilla A, Ruiz A. Genetics. 2000;155:685–698. doi: 10.1093/genetics/155.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaeffer SW, Goetting-Minesky MP, Kovacevic M, Peoples JR, Graybill JL, Miller JM, Kim K, Nelson JG, Anderson WW. Proc Natl Acad Sci USA. 2003;100:8319–8324. doi: 10.1073/pnas.1432900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munte A, Rozas J, Aguade M, Segarra C. Genetics. 2005;169:1573–1581. doi: 10.1534/genetics.104.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennington WJ, Partridge L, Hoffmann AA. Genetics. 2006;172:1655–1663. doi: 10.1534/genetics.105.053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson J, Stone W. Evolution in the Genus Drosophila. New York: Macmillan; 1952. [Google Scholar]
- 40.Jaenike J, Dyer KA, Cornish C, Minhas M. PLoS Biol. 2006;4:1852–1862. doi: 10.1371/journal.pbio.0040325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 42.Tajima F. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watterson GA. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 44.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swofford DL. Sunderland, MA: Sinauer; 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. [Google Scholar]
- 46.Hudson RR. Genetics. 2000;155:2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson RR, Boos DD, Kaplan NL. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 48.Hudson RR. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McVean G, Awadalla P, Fernhead P. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laporte V, Charlesworth B. Genetics. 2002;162:501–519. doi: 10.1093/genetics/162.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokal RR, Rohlf FJ. Biometry. 2nd Ed. San Francisco: Freeman; 1981. [Google Scholar]
- 52.Rice WR. Evolution. Vol. 43. Lawrence: Kans; 1989. pp. 223–225. [DOI] [PubMed] [Google Scholar]
- 53.Bachtrog D, Charlesworth B. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.