Abstract
Mg-chelatase catalyzes the insertion of Mg2+ into protoporphyrin IX at the first committed step of the chlorophyll biosynthetic pathway. It consists of three subunits: I, D, and H. The I subunit belongs to the AAA protein superfamily (ATPases associated with various cellular activities) that is known to form hexameric ring structures in an ATP-dependant fashion. Dominant mutations in the I subunit revealed that it functions in a cooperative manner. We demonstrated that the D subunit forms ATP-independent oligomeric structures and should also be classified as an AAA protein. Furthermore, we addressed the question of cooperativity of the D subunit with barley (Hordeum vulgare) mutant analyses. The recessive behavior in vivo was explained by the absence of mutant proteins in the barley cell. Analogous mutations in Rhodobacter capsulatus and the resulting D proteins were studied in vitro. Mixtures of wild-type and mutant R. capsulatus D subunits showed a lower activity compared with wild-type subunits alone. Thus, the mutant D subunits displayed dominant behavior in vitro, revealing cooperativity between the D subunits in the oligomeric state. We propose a model where the D oligomer forms a platform for the stepwise assembly of the I subunits. The cooperative behavior suggests that the D oligomer takes an active part in the conformational dynamics between the subunits of the enzyme.
INTRODUCTION
Mg-chelatase catalyzes the first committed step in chlorophyll biosynthesis through the ATP-dependent insertion of Mg2+ into protoporphyrin IX. The enzyme is strategically positioned at the branch point between chlorophyll and heme biosynthesis and appears to be of significant regulatory importance. The regulatory aspects may play an essential role in chloroplast maturation, which is known to be intimately related to chlorophyll biosynthesis (von Wettstein et al., 1995; Tanaka and Tanaka, 2006). Chloroplasts of higher plants develop from proplastids. In light-exposed tissues, the proplastids grow in size and differentiate into membrane structures that harbor the photosynthetic apparatus. If seedlings are germinated in the dark, the plastids develop into etioplasts containing perforated primary lamellar layers and crystalline prolamellar bodies. Mutants unable to synthesize Mg-protoporphyrin contain only unfenestrated thylakoids and lack prolamellar bodies (von Wettstein et al., 1995). Interestingly, Mg-protoporphyrin suppresses nuclear expression of Lhab1 and other genes encoding chloroplast-localized proteins (Kropat et al., 2000; Strand et al., 2003; Alawady and Grimm, 2005; Gadjieva et al., 2005), which suggests that Mg-protoporphyrin plays the essential role of signal molecule in the regulation of these processes. In addition, Mg-protoporphyrin and heme are known to regulate the first biosynthetic steps of their common pathway (Vasileuskaya et al., 2005). Recently, the large protein component of Mg-chelatase was reported to additionally function as an abscisic acid receptor (Shen et al., 2006). Together, these observations suggest that the understanding of the mechanism of Mg-chelatase is important not only for understanding chlorophyll biosynthesis but also for understanding the more general processes of chloroplast and plant development.
Much of the knowledge on Mg-chelatase originates from genetic and biochemical analysis of mutants. In the 1940s, a Chlorella vulgaris mutant, W5 brown, revealed protoporphyrin IX as a biosynthetic intermediate to chlorophyll (Granick, 1948). In the 1950s, barley (Hordeum vulgare) mutants accumulating protoporphyrin IX were grouped into three loci: Xantha-f, -g, and -h (Henningsen et al., 1993; von Wettstein et al., 1995). The observation suggested that Mg-chelatase was composed of three structural genes. The genes were eventually identified at the DNA level through studies of transposon-induced mutants of Rhodobacter capsulatus and Rhodobacter sphaeroides (Coomber et al., 1990; Gorchein et al., 1993; Bollivar et al., 1994). The vast collection of mutants provides a historical explanation to the rich variety of names given to the genes that encode Mg-chelatase. The three Mg-chelatase subunits are ∼140, 70, and 40 kD and are generally referred to as the H, D, and I subunits, respectively. In barley, the corresponding subunits are encoded by Xantha-f, -g, and -h. In R. capsulatus, the genes are referred to as bchH, bchD, and bchI (Bollivar et al., 1994). The 140-kD H subunit binds the protoporphyrin IX substrate and most likely also the Mg2+ substrate and is thus implicated as the catalytic component of the enzyme (Willows et al., 1996; Jensen et al., 1998; Willows and Beale, 1998; Karger et al., 2001). The 40-kD I subunit belongs to the functionally diverse superfamily of AAA proteins (ATPases associated with various cellular activities) (Fodje et al., 2001; Reid et al., 2003; Willows et al., 2004). AAA proteins are known to form ring-shaped oligomers and are represented in all kingdoms of life (Neuwald et al., 1999; Iyer et al., 2004). They are involved in processes like folding, assembly, and disassembly of protein complexes and usually undergo large conformational changes during their functional cycle (Vale, 2000; Hanson and Whiteheart, 2005).
Analysis of the amino acid sequence of the Mg-chelatase D subunit reveals three consecutive regions: an N-terminal domain homologous to the AAA module of the I subunit, a central acidic and Pro-rich region, and an integrin I domain with unknown function at the C terminus. The homology between the N terminus and the I subunit suggests that the D subunit also includes an AAA module and at least structurally belongs to the AAA protein family. This has recently been demonstrated by analysis of complexes of the I and D subunits employing the method of single-particle reconstruction from electron microscopy images (J. Lundqvist, E. Axelsson, A. Hansson, D. Birch, M. Hansson, S. Al-Karadaghi, and R.D. Willows, unpublished results). However, it should be noted that the D protein contains a divergent AAA domain and that it has no reported ATPase activity (Hansson and Kannangara, 1997; Jensen et al., 1999; Petersen et al., 1999a).
The Pro-rich region and the integrin I domain have been suggested to contribute to the formation of the ID complex (Fodje et al., 2001). Pro-rich regions are generally known to be involved in the interaction between proteins (MacArthur and Thornton, 1991) and in the stabilization of oligomeric complexes (Bergdoll et al., 1997), while integrin I domains function within αβ-heterodimeric transmembrane receptors involved in cell–cell and cell–matrix interactions (Hynes, 2002). A characteristic feature of integrin I domains is the presence of a metal ion–dependent adhesion site (MIDAS) (Lee et al., 1995). The MIDAS motif is characterized by the presence of two conserved sequence motifs (i.e., DXSXS and TD) separated from each other by ∼90 amino acid residues. The five conserved residues coordinate a Mg2+ or a Mn2+ ion. An interacting protein contributes the sixth ligand, a D or an E, which is often found within two integrin I domain recognition sequences (i.e., RGE/D and LDV). Interestingly, the 40-kD I subunit has a conserved RGE/D sequence, whereas LDV is found in the polypeptide of certain 140-kD H subunits. Interactions between the I and D subunits have been indirectly demonstrated in several studies, which showed that preincubation of these subunits in the presence of ATP and Mg2+ speeds up the onset of the Mg-chelatase reaction in vitro (Willows et al., 1996, 1999; Guo et al., 1998; Jensen et al., 1998). The interaction was also studied by employing a yeast two-hybrid system (Papenbrock et al., 1997).
Previous analyses of semidominant and recessive barley and maize (Zea mays) Mg-chelatase mutants deficient in the I subunit have also provided knowledge on the interactions between the subunits (Hansson et al., 1999, 2002; Sawers et al., 2006). The D subunit could not be detected in any of the studied barley I mutants, whereas wild-type levels of the D subunit were found in barley mutants that lacked the H subunit (Lake et al., 2004). This suggested that a functional I subunit is required for maintaining the D subunit in the cells. The semidominant and recessive mutations were of different character. The semidominant mutations were all point mutations resulting in changes of single amino acid residues, whereas the recessive mutations were deficient in transcription of the Xantha-h gene. Thus, the recessive xantha-h mutants contained no I protein, whereas wild-type levels were found in the semidominant mutants. For in vitro studies, the semidominant mutations found in barley and maize were constructed in existing expression systems for R. capsulatus bchI and Synechocystis chlI, respectively (Hansson et al., 2002; Sawers et al., 2006). The resulting I proteins showed no or reduced ATPase activity and could not contribute to the Mg-chelatase activity. However, the modified R. capsulatus proteins showed an ATP/ADP-dependant ability to form oligomers alone or with wild-type I subunits. Most interestingly, in vitro Mg-chelatase assays performed with mixtures of mutant and wild-type I subunits resulted in a significantly lower Mg-chelatase activity than expected based on the relative proportion of wild-type I subunit in the mixture. This inhibitory or poisoning effect of the mutant I subunits on the wild-type I subunits was >50% at a 1:1 ratio in the R. capsulatus system, whereas a threefold to ninefold excess of mutant subunits over wild-type protein was required to obtain a 50% inhibition in Synechocystis. These experiments demonstrated the dominant effect of the proteins in vitro, and it was concluded that the I subunits function in a concerted manner within the hexameric complex (Hansson et al., 2002).
Semidominant mutations may also be expected in the gene encoding the D subunit because the D subunit also includes an AAA module. However, only recessive mutations have been identified within this locus. In this study, we have characterized available barley xantha-g mutations at the DNA level and revealed a dominant phenotype in vitro. This suggests that the D subunits also function within a cooperative complex. Furthermore, we propose a model for the assembly of the active complex of Mg-chelatase with focus on the D subunit. In this model, the D subunit is suggested to form a platform for the stepwise assembly of the I subunits in the catalytic cycle.
RESULTS
Molecular Analysis of xantha-g Mutants
The barley mutants xantha-g28, -g37, -g44, -g45, and -g65 are recessive and lethal, and they accumulate protoporphyrin IX upon feeding with the chlorophyll biosynthetic precursor 5-aminolevulinic acid. Their ability to form chlorophyll has been analyzed (Henningsen et al., 1993). At low intensities of light, the leaky xantha-g45 mutant is capable of producing 20% of the chlorophyll found in wild-type leaves (Figure 1). The xantha-g44 mutant is slightly leaky and forms trace amounts of chlorophyll (2% of the wild type). By contrast, xantha-g28, -g37, and -g65 are nonleaky and do not synthesize chlorophyll.
Figure 1.
Phenotypes of Barley Wild-Type, xantha-g44, and xantha-g45 Seedling Leaves.
The plants were grown for 7 d in continuous light. The xantha-g44 mutant is slightly leaky and contains trace amounts of chlorophyll. The yellow color of carotenoids dominates the phenotype of this mutant. The xantha-g45 mutant is leakier and can form up to 20% of the chlorophyll present in the wild type. For this reason, xantha-g45 displays a pale green color.
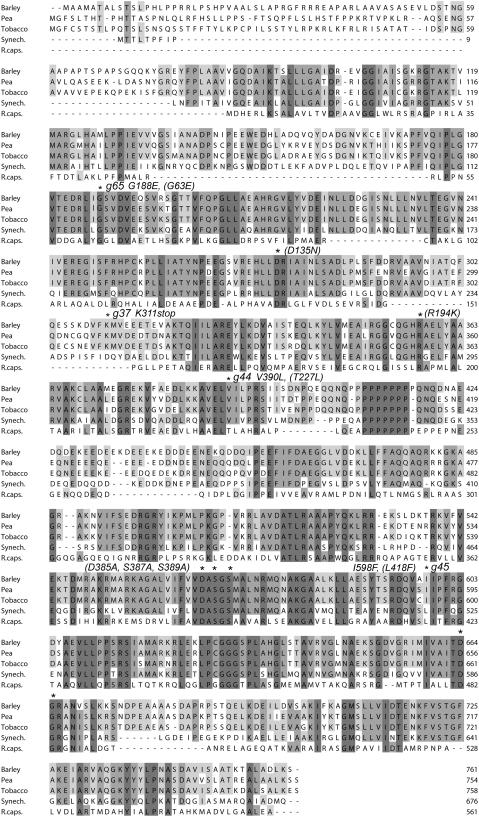
To perform genetic and biochemical analyses on the barley Mg-chelatase xantha-g mutants, it was essential to clone and sequence the chromosomal wild-type Xantha-g gene. This was done from a genomic barley DNA library in bacteriophage-λ. Oligonucleotides were constructed to amplify, clone, and sequence chromosomal DNA fragments from the five barley xantha-g mutants. A consensus sequence of the five mutants was deposited in the GenBank/EMBL data libraries (accession number AAZ32779). The sequenced region extends from 827 bp upstream of the putative ATG start codon of the 6940-bp Xantha-g gene to 226 bp downstream from the TAG stop codon (see Supplemental Figure 1 online). No obvious consensus sequence of a TATA box was identified. The restriction pattern of a DNA gel blotting experiment was in agreement with the presence of a single Xantha-g gene in barley located on the long arm of chromosome 5H (see Supplemental Figure 2 online). Fifteen Xantha-g exons were identified in the Xantha-g DNA sequence (see Supplemental Figure 3 online). The deduced D polypeptide sequence had 761 amino acid residues and included a putative chloroplast signal transit polypeptide of 43 residues (Emanuelsson et al., 1999). The barley D protein sequence displayed 80, 79, 53, and 23% identity to the orthologous proteins of tobacco (Nicotiana tabacum; Papenbrock et al., 1997), pea (Pisum sativum; Luo and Weinstein, 1997), Synechocystis sp PCC6803 (Jensen et al., 1996), and R. capsulatus (P26175), respectively (Figure 2).
Figure 2.
Alignment of the Amino Acid Sequences of Five Mg-Chelatase D Subunits.
Shadowed boxes highlight invariant and conserved residues. The amino acid residues that were affected by the barley xantha-g37, -g44, -g45, and -g65 mutations are indicated with asterisks as well as other residues that where modified in the study (R. capsulatus residues with numbers are given within parentheses). The MIDAS motif is formed by the invariant amino acid residues Asp-385, Ser-387, Ser-389, Thr-481, and Asp-482 (R. capsulatus numbering). The polypeptides were from barley, pea (Luo and Weinstein, 1997), tobacco (Papenbrock et al., 1997), Synechocystis sp PCC6803 (Jensen et al., 1996), and R. capsulatus (P26175).
Point mutations were identified in xantha-g37, -g44, -g45, and -g65, whereas no mutation was found in xantha-g28 within the analyzed DNA sequence. In the mutant xantha-g37, an aag codon encoding Lys-311 had been changed to a tag stop codon; in the mutant xantha-g44, a gtt codon had been changed to ctt (Val-390 to Leu); in the mutant xantha-g45, an att codon had been changed to ttt (Ile-598 to Phe); and in the mutant xantha-g65, a gga codon had been changed to gaa (Gly-188 to Glu). The residues Gly-188, Lys-311, and Val-390 were located within the N-terminal AAA domain, and Ile-598 was located in the C-terminal integrin I domain (Figure 2).
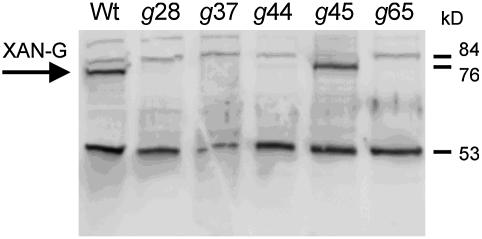
To determine whether the mutations affected the presence of the D protein in the homozygous xantha-g28, -g37, -g44, -g45, and -g65 mutants, a protein gel blot was performed using antibodies specific to the C-terminal part of the barley D subunit (Figure 3). In the two leaky xantha-g mutants, the presence of a full-length D protein was expected. This was clearly confirmed in xantha-g45, whereas only trace amounts of D protein were detected in xantha-g44 and none at all in the nonleaky mutants xantha-g28, -g37, and -g65.
Figure 3.
Protein Gel Blot Analysis of the Mg-Chelatase D Subunit.
Total cell extracts of barley wild type and the mutants xantha-g28, -g37, -g44, -g45, and -g65 were analyzed. Wild-type level of the D subunit was only found in xantha-g45, while trace amounts were detected in xantha-g44. The D subunit is indicated by an arrow. The cross-reacting protein at 53 kD demonstrates that similar amounts of protein extract (5 μg) were loaded in each lane.
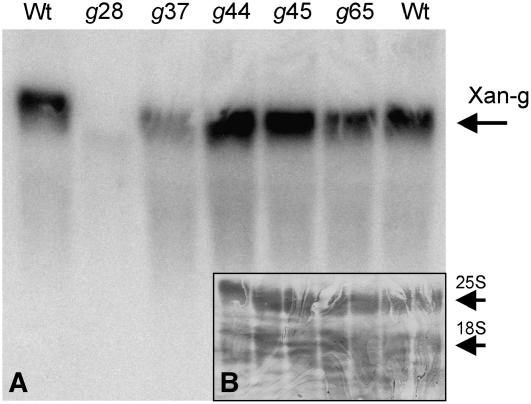
No mutation was identified within the analyzed DNA sequence of the xantha-g28 mutant, and at the same time, no 70-kD protein could be detected. To determine if the lack of protein was due to impaired transcription, it was necessary to analyze the presence of Xantha-g mRNA in the mutant xantha-g28. Total RNA was extracted from mutant leaves and analyzed by RNA gel blotting. No Xantha-g mRNA could be detected in xantha-g28 (Figure 4). This explained the lack of D protein and suggested that the xantha-g28 mutation was located in a cis-acting regulatory element upstream of the sequenced DNA region. A slightly reduced level of Xantha-g mRNA in xantha-g37 could possibly be explained by the phenomenon of non-sense-mediated mRNA decay (Isshiki et al., 2001; Gadjieva et al., 2004).
Figure 4.
RNA Gel Blot Analysis of Xantha-g mRNA in Barley Wild Type and the xantha-g28, -g37, -g44, -g45, and -g65 Mutants.
(A) No Xantha-g mRNA could be detected in xantha-g28. Most likely this strain had a mutation in the regulatory region of the Xantha-g gene upstream of the sequenced DNA region. Mutant xantha-g37 had a slightly reduced level of Xantha-g mRNA, which would be caused by non-sense-mediated mRNA decay.
(B) To make sure that an equal amount of total RNA had been used from each barley strain (21 μg), the blot was stained with methylene blue before being hybridized with the Xantha-g DNA probe. The nuclear-encoded 25S and 18S rRNA dominated the total RNA.
In Vitro Analysis
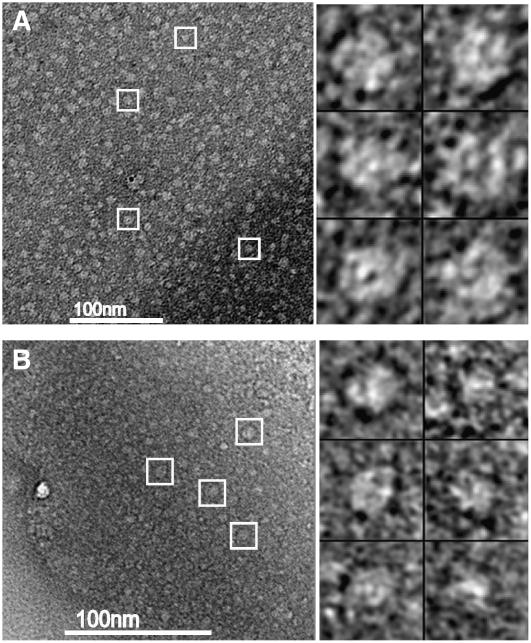
Previous successful studies by negative staining electron microscopy on the I subunit and on mixtures of I and D subunits encouraged us to investigate whether oligomeric structures could also be formed by the D subunit alone. The analysis was performed with R. capsulatus D subunit, as we have an expression system for the R. capsulatus Mg-chelatase subunits but not the barley subunits. R. capsulatus D protein was produced in Escherichia coli as His-tagged proteins and was purified from inclusion bodies by employing Ni-affinity chromatography and urea containing buffers. Active D protein was obtained by dialysis and adjusted to 0.1 μg/μL. The protein was added to a carbon-coated 400-mesh copper grid and was negatively stained with uranyl acetate. Images were collected at a magnification of 55,000 (Figure 5). The particles were similar in appearance to those of the I subunit, which after image processing revealed a closed hexameric ring structure (Willows et al., 2004). However, in contrast with the particles formed by I subunits, the D particles appeared to be more homogenous. That is, the D subunits seemed to form more uniform oligomers, whereas the I subunits often form incomplete ring structures. In addition, the D protein could form oligomeric structures even in the absence of ATP, contrasting it with the I subunit where ATP is essential for ring structure formation (Willows et al., 2004).
Figure 5.
Negative Staining Electron Micrographs of R. capsulatus D Oligomeric Complexes.
Oligomeric complexes formed in the presence (A) and absence (B) of ATP. Examples of particles are marked with white boxes. Six selected D particles from each micrograph are shown at the right.
Previously, an in vitro dissection of the semidominant barley mutations in the I subunit transferred to R. capsulatus bchI revealed that the I hexameric ring works cooperatively in Mg chelation (Hansson et al., 2002). Furthermore, two of the three semidominant mutations in the I subunit affect amino acid residues that are conserved in an alignment between the Mg-chelatase I and D polypeptides (D207N and R289K in the R. capsulatus I polypeptide). Together, these findings prompted us to investigate whether the D hexamer was also a cooperative AAA protein complex despite the fact that only recessive mutations have been described from the D locus.
The barley missense mutations xantha-g44, -g45, and -g65 were introduced in the R. capsulatus bchD gene by site-directed mutagenesis. The mutations corresponded to the modifications T227L, L418F, and G63E, respectively, in the R. capsulatus D subunit (Figure 2). We also performed mutations leading to the changes D135N and R194K, which corresponded to the semidominant mutations D207N and R289K, respectively, in the I subunit (Hansson et al., 2002). Furthermore, we constructed D385A, S387A, and S389A mutants, which affected residues of the proposed MIDAS motif in the C-terminal integrin I domain of the D subunit (Figure 2). Activity measurements were performed after refolding of recombinant wild-type and modified D subunits by rapid dilution in the presence of I protein (Willows and Beale, 1998).
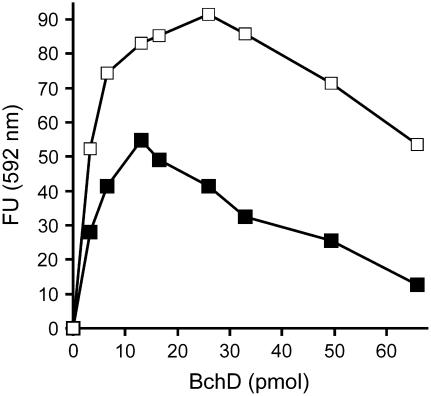
Addition of increasing amounts of D subunit to assays containing fixed amounts of I and H protein revealed an optimal amount of D subunit for the assay. When increasing the concentration of D subunit, the activity increased and reached a maximum at a point where the molar concentration of the D subunit was equal to the concentration of the I subunit. Thereafter, a further increase of the D subunit significantly inhibited the activity (Figure 6). Due to the negative effect of excessive D subunits, we always used a limiting amount of D in our activity analysis of the mutant proteins. In the 200-μL assays, 12.4 pmol H subunit, 5.0 pmol I subunit, and between 1.1 and 4.4 pmol D subunit were used. The addition of increasing amounts of mutant D subunits was always analyzed in parallel with a similar addition of extra wild-type D subunit.
Figure 6.
Measurements of Mg-Chelatase Activity at Various Concentrations of D Subunit.
The activity shows a maximum where the concentration of D subunit equals the concentration of I subunit. Filled squares: Assays contained 13 pmol of BchI and 26 pmol of BchH. Open squares: Assays contained 26 pmol of BchI and 26 pmol of BchH. The assays were stopped after 30 min by addition of alkali-acetone. The amount of formed Mg-protoporphyrin IX was recorded fluorometrically at 592 nm. FU, fluorescence units.
Mutations affecting the MIDAS motif were the only mutant D proteins that were unable to contribute to Mg-chelatase activity (Table 1). The proteins with the modifications T227L, L418F, and G63E, corresponding to the barley mutations xantha-g44, -g45, and -g65, respectively, had 40, 13, and 42% of the wild-type activity. The changes D134N and R193K led to 41 and 25% activity. This is in contrast with the situation with the I subunit where these mutations (D207N and R289K) totally abolished Mg-chelatase activity (Hansson et al., 2002).
Table 1.
Effect of Various Mutant D Subunits on Mg-Chelatase Activity upon Mixing before Refolding
| D Protein Analyzed | Relative Activity (% ± sd) | Amount of D Subunits in Assay (pmol)
|
|
|---|---|---|---|
| Wild-Type D | Mutant D | ||
| Wild type | 165 ± 9 | 4.4 | – |
| Wild type | 100 ± 2 | 2.2 | – |
| Wild type | 57 ± 1 | 1.1 | – |
| G63E | 42 ± 1 | – | 2.2 |
| D135N | 41 ± 1 | – | 2.2 |
| R194K | 25 ± 1 | – | 2.2 |
| T227L | 40 ± 5 | – | 2.2 |
| D385A | 2.6 ± 0.4 | – | 2.2 |
| S387A | 0 | – | 2.2 |
| S389A | 1.8 ± 0.3 | – | 2.2 |
| L418F | 13 ± 0.4 | – | 2.2 |
| G63E + wild type | 92 ± 3 | 2.2 | 2.2 |
| D135N + wild type | 85 ± 3 | 2.2 | 2.2 |
| R194K + wild type | 47 ± 2 | 2.2 | 2.2 |
| T227L + wild type | 92 ± 3 | 2.2 | 2.2 |
| D385A + wild type | 28 ± 1 | 2.2 | 2.2 |
| S387A + wild type | 56 ± 3 | 2.2 | 2.2 |
| S389A + wild type | 33 ± 1 | 2.2 | 2.2 |
| L418F + wild type | 48 ± 2 | 2.2 | 2.2 |
100% activity corresponds to 0.54 pmol Mg-protoporphyrin IX/(min × pmol D subunit) being formed. The amounts of I and H subunits in the assays were 5.0 and 12 pmol, respectively.
Modified D proteins, with testable activity, were generally very stable after refolding, as they could be stored at 4°C for a number of days without any significant change in activity. The one exception was the G63E protein, which gradually lost Mg-chelatase activity from 92% of wild-type activity when assayed immediately after refolding, to 67% when assayed ∼30 min after refolding, to 42% when assayed ∼1 h after refolding, and to 21% when assayed after 3 d at 4°C. The results presented in Table 1 are based on analyses performed 1 h after refolding.
To investigate whether the mutations had any dominant influences on the Mg-chelatase activity, assays were performed where 2.2 pmol wild-type D subunit was mixed with 2.2 pmol of mutant D subunit before refolding. The addition of all mutants significantly lowered the Mg-chelatase activity (Table 1). In other words, the mixtures showed a lower activity than if 2.2 pmol wild-type D was assayed alone. At the same time, the assay containing 4.4 pmol wild-type D subunit displayed a significantly higher activity, showing that the amount of D subunit was still rate-limiting in the assay.
The dominant behavior of the mutant D subunits in vitro demonstrated a concerted mechanism of the D oligomer. It should be noted that when the mutant and wild-type BchD proteins were refolded separately and then mixed, there was an additive effect on the Mg-chelatase activity (Table 2). This indicates that there existed no or a very slow exchange of subunits within the oligomeric complex once it had formed and that the dominant effect can only occur during the formation of the complex.
Table 2.
Effect of Various Mutant D Subunits on Mg-Chelatase Activity upon Mixing after Refolding
| Relative Activity (%) | Amount of D Subunits in Assay (pmol)
|
||
|---|---|---|---|
| D Protein Analyzed | Wild-Type D | Mutant D | |
| Wild type | 362 | 13 | – |
| Wild type | 208 | 6.5 | – |
| Wild type | 100 | 3.2 | – |
| G63E + wild type | 173 | 3.2 | 3.2 |
| D135N + wild type | 168 | 3.2 | 3.2 |
| R194K + wild type | 119 | 3.2 | 3.2 |
| T227L + wild type | 140 | 3.2 | 3.2 |
| D385A + wild type | 119 | 3.2 | 3.2 |
| S387A + wild type | 121 | 3.2 | 3.2 |
| S389A + wild type | 142 | 3.2 | 3.2 |
| L418F + wild type | 115 | 3.2 | 3.2 |
100% activity corresponds to 0.96 pmol Mg-protoporphyrin IX/(min × pmol D subunit) being formed. The amounts of I and H subunits in the assays were both 13 pmol.
DISCUSSION
On the Border between Dominance and Recessiveness
Dominant mutations can be expected in genes that encode oligomeric complex-forming proteins, which, in turn, function in a concerted manner. Hence, this relatively rare class of mutations can provide insight into the dynamics and function of cooperative oligomeric protein complexes. Oligomeric ring-shaped AAA protein complexes have often been found to undergo concerted conformational changes. One example is the N-ethylmaleimide sensitive factor (NSF) that has served as a reference point in studies on AAA proteins and is essential for membrane trafficking (Vale, 2000). Incorporation of inactive subunits in NSF has displayed a dominant influence on the complex, inhibiting the enzyme activity (Whiteheart et al., 1994). We have previously studied dominant mutations in the I subunit of Mg-chelatase (Hansson et al., 1999, 2002). This AAA protein family member is also sensitive to the addition of inactive building blocks, in vivo as well as in vitro (Hansson et al., 2002). Due to the high levels of sequence similarity between the I and D subunits of the Mg-chelatase, it was likely that the D subunit was also an AAA family member.
In this study, we demonstrated by negative staining electron microscopy that the D subunit could form oligomeric structures. Therefore, dominant mutations could be expected for the barley Xantha-g locus that encoded the D subunit. However, to the best of our knowledge, only recessive mutations in various species have been described for this gene.
This analysis reveals that the recessive behavior of the barley mutations xantha-g28, -g37, -g44, and -g65 in vivo is due to the absence of the resulting D proteins or to its significantly lowered amount in the case of xantha-g44. In other words, the hexameric D subunit complexes in heterozygote plants of these mutants never or very infrequently get inactive mutant subunits incorporated as part of the complex as a result of the mutant D subunits not being present in the cells. The absence of mutant D subunits was not surprising. The D protein appears to be very unstable in the plastids and is dependent on the I subunit (Lake et al., 2004). A slight change of the D subunit polypeptide due to a mutation might abolish the important interaction with the I protein and trigger an active degradation of the D protein. It has previously been shown that the D subunit could not be detected in any of the seven available barley xantha-h mutants that are deficient in the I subunit (Lake et al., 2004). This revealed that the I protein most probably protects the D subunit from proteolytic degradation, presumably by the formation of an ID complex. It could also reveal an unforeseen regulatory mechanism of chlorophyll biosynthesis in which the D subunit is rapidly degraded when not in a complex with the I subunit.
The only xantha-g mutant with a physiological level of the 70-kD protein was xantha-g45. Obviously, the xantha-g45 mutated protein is protected by the I subunit, which prevents its degradation. Most likely the D subunits resulting from the xantha-g45 allele can form complexes with wild-type D subunits in heterozygote mutants. These mixed complexes are likely to have a lower activity compared with pure wild-type complexes. However, due to the leaky nature of xantha-g45, the Mg-chelatase reaction never becomes rate-limiting for chlorophyll biosynthesis, resulting in a wild-type phenotype of the heterozygote and thus a segregation pattern of a recessive mutation. Alternatively, the hexameric complex formed by D subunits does not function in a concerted manner.
To determine if cooperativity occurs within a D hexamer, we made the barley xantha-g44, -g45, and -g65 missense mutations in the bchD gene of R. capsulatus (Figure 2). Similarly, we also made two of the three semidominant Xantha-h mutations (Hansson et al., 1999, 2002) in R. capsulatus bchD. This was possible as the affected residues were conserved in alignments between polypeptides of the I and D subunits. In yet another construct, we exchanged amino acid residues to abolish the suggested MIDAS motif of the D subunit. With the exception of the G63E-modified R. capsulatus D protein mimicking the barley mutant xantha-g65, all proteins were stable. Thus, the in vitro system made it possible to study the catalytic consequences of the mutations, which did not seem possible in vivo due to the active degradation of the D subunits not tied up in complexes with I subunits. With the exception of mutations affecting the MIDAS motif, all mutant proteins could contribute to the Mg-chelatase activity. When mutant and wild-type D subunits were mixed in a 1:1 ratio before refolding in Mg-chelatase activity measurements, an inhibition was observed. This demonstrated that the mutant subunits had a dominant influence on the complex. Most importantly, the inhibition suggests that the D hexamers do function in a concerted manner, which was unexpected from the recessive segregation pattern of the barley xantha-g mutants. This is a novel and important finding that must be considered in a mechanistic model of the Mg-chelatase.
Function of the D Subunit
A two-tiered hexameric ring structure is found in many type-2 AAA proteins, such as NSF, p97, and Clp (Vale, 2000). The type-2 AAA proteins are characterized by the presence of two consecutive AAA modules within the same polypeptide. The two modules form structural units within the different hexameric rings. Furthermore, the two modules are not identical. One AAA module has a high enzymatic activity and is involved in acting on the target molecule. The other often has low or no ATPase activity and has been suggested to serve a structural role in hexamer stability (Vale, 2000). The Mg-chelatase I and D subunits are both type-1 AAA proteins, as the polypeptides harbor only one AAA module. However, the similarity between the type-2 complexes and the ID complex suggests a similar function of the rings in a two-tiered arrangement. We know that the I hexamer is only formed in the presence of ATP/ADP and that it has a high ATPase activity, which suggests that the I subunit is the active domain within the ID complex.
In this work, we demonstrated that the D subunit was capable of forming oligomeric structures in the absence of ATP. No ATPase activity has been recorded for this subunit (Hansson and Kannangara, 1997; Jensen et al., 1999; Petersen et al., 1999a). These results fit with the D subunit functioning as a structurally important unit in the ID complex. We therefore suggest that the D subunits maintain their hexameric structure during the catalytic cycle, which functions as a platform for the assembly of the active I subunits. The unwillingness to exchange subunits between the catalytic cycles is supported by the additive activity observed when wild-type and mutant D subunits are refolded separately before being mixed and used in activity measurements (Table 2).
The D oligomer functioning as a platform for the assembly of I subunits is corroborated by the observation that an excess of D subunit relative to I subunit lowers the activity. This is due to fewer complete two-tiered hexameric ID complexes being formed upon increasing amounts of D (Figure 6). The same phenomenon has also been observed with the Synechocystis and the R. sphaeroides Mg-chelatase D subunits and is in contrast with the I and H subunits, which both show hyperbolic curves demonstrating saturation of these subunits (Jensen et al., 1998; Gibson et al., 1999). However, the dominant influence demonstrated in this work suggests that the D subunit is not simply a passive member of the ID complex.
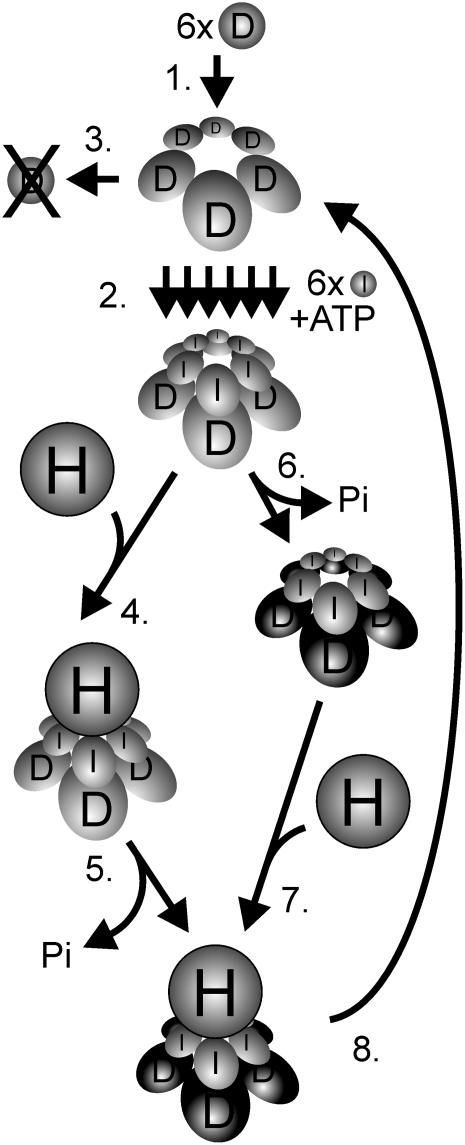
A combination of the results presented here and previously published biochemical data suggests the following consecutive steps in the interaction of the Mg-chelatase I and D subunits: The D subunits form a hexameric complex, where the exchange of subunits rarely occurs (Figure 7, step 1). The complex is formed in the absence of ATP and functions as a platform for the ATP and Mg2+-dependent stepwise assembly of the I monomers into a two-tiered ID hexameric double ring (Figure 7, step 2). Without the formation of an ID complex the D protein is degraded (Figure 7, step 3). The timing of the ATP hydrolysis by the I subunits is still not established. From structural studies it was proposed that ATP hydrolysis was blocked in an ID complex and triggered by interaction with the H subunit followed by catalysis (Fodje et al., 2001) (Figure 7, steps 4 and 5). However, from studies of I mutants with no ATPase activity, it seems that the binding of I to D is not enough to stabilize the D subunit (Lake et al., 2004). No D protein can be detected in such mutants; therefore, an ATPase-dependent process of a cooperative I hexamer seems to convert the D hexamer into a configuration that protects D from degradation (Figure 7, step 6). This suggests a model where the ATP-hydrolytic conformational change and stabilization of the D hexamer by the I hexamer occur prior to the binding of the H subunit (Figure 7, step 7). It is clear that binding of the H subunit is not a requirement for stabilization to occur, since the D subunit is present at wild-type levels in barley mutants without any H subunit (Olsson et al., 2004). Thus, in contrast with the I subunit, the H subunit has no influence on the survival of the D subunit.
Figure 7.
Model of the Catalytic Cycle of Mg-Chelatase with Focus on the D Subunit.
1, Six D subunits form a hexameric ring structure in an ATP-independent process. 2, In a Mg2+- and ATP-dependant process, six I subunits are stepwise assembled into a hexameric ring using the D hexameric structure as a platform. The two-tiered hexameric ring also forms in the presence of ADP, but further steps are abolished. 3, In the absence of functional I subunits, the D subunits are degraded. 4, Timing of ATP hydrolysis in relation to the addition of the H subunit is not established. From structural data, it is suggested that the ATP hydrolysis is stalled in the ID complex and, 5, triggered upon binding to the H subunit (Fodje et al., 2001). The H subunit holds the protoporphyrin IX and most likely also the Mg2+ substrate and has therefore been considered as the catalytic subunit. 6, From studies with ATPase-deficient I mutants, it seems that the formation of an ID complex is not enough to rescue the D subunit from degradation (Lake et al., 2004). Instead, a concerted ATPase-dependent action of the I proteins is likely to perform a structural change of the D proteins. After the conformational change, the D proteins are safe from degradation and, 7, bind to the H subunit. 8, After the formation of Mg-protoporphyrin, the complex disassembles except for the hexameric D ring, which is ready to function as a platform in a new catalytic cycle. The concerted behavior of the D hexamer could be expressed in step 6 if the cooperative activity of the I subunits required a concerted structural change of its D hexameric substrate. Alternatively, the concerted activity of the D hexamer is required in the catalytic interaction with the H subunit in steps 5 or 7. At present, it is unknown whether the H subunit docks to the I or D side of the complex, although docking to the I subunits is shown in the figure.
We are currently developing methods to involve the H subunit in our experiments to understand the mechanism of Mg-chelatase. Many basic questions remain to be answered. For instance, it is still an open question as to whether the H subunit binds to the D or the I side of the two-tiered complex. The similar structural arrangement of the two-tiered hexamers of type-2 AAA proteins and the two-tiered ID complex suggests that the H subunit docks to the ATPase-active I subunits. However, the inhibitory effect of mutant D subunits in mixed wild-type and mutant D hexameric rings indicates that the D hexamer also functions in a concerted manner. In other words, the D hexamer is not only a passive scaffold for the assembly of the I hexamer, but also an active component during catalysis that could include direct interaction with the H subunit. Alternatively, the cooperative behavior of the D subunits might be a consequence of their functioning as hexameric substrates of the I subunits, which, in turn, function as a concerted hexameric ring complex. This would imply that the structural change of the D subunits within the hexamer could only occur simultaneously and that the incorporation of mutant D subunits in the hexameric ring would convert the D complex into an inhibitor of the I hexamer. After the catalysis, the IDH complex falls apart, with the exception of the D hexamer, which is ready for another catalytic cycle (Figure 7, step 8).
METHODS
Plant Materials
Wild-type barley (Hordeum vulgare cv Svalöf's Bonus) and homozygous barley mutants xantha-g28, -g37, -g44, -g45, and -g65 were grown in moist vermiculite at 20°C for 7 d with 12-h cycles of light/dark (150 μE m−2 s−1). Wheat (Triticum aestivum) strains containing either a long or short arm of barley chromosomes (Islam et al., 1981) were used to determine the chromosomal location of the Xantha-g gene. The preparation of these disomic additional lines has been described elsewhere (Hansson et al., 1998).
Cloning and Sequencing
The Xantha-g gene was cloned from a barley genomic DNA library in bacteriophage λ (Lambda FIX II; Stratagene) using a 1723-bp SacI DNA fragment from plasmid pXg28a as a probe. Plasmid pXg28a is a pGEM-T derivative (Promega) containing a 1778-bp Xantha-g DNA fragment that was obtained using the published partial Xantha-g sequence (Petersen et al., 1999b). SacI cuts in the cloning cassette of the vector as well as in the end of the Xantha-g DNA insert. The 1723-bp SacI fragment contained 1683 bp of Xantha-g–specific DNA between bases 6291 and 7974 in the sequence published in this work. The DNA probe was labeled using the Random Primed DNA Labeling kit (Roche) and [α32P]dCTP 1.1 × 1017 Bq mol−1 (GE Healthcare). Escherichia coli MRA(P2) was infected with phage λ and plated on 18-cm Petri dishes to form 50,000 plaques in soft agar. Plaques from 10 plates were transferred to Hybond-N filters (GE Healthcare) and hybridized with radiolabeled DNA probe as described previously (Hansson et al., 1998). Positive clones were visualized by autoradiography. Regions containing positive clones were cut out from the agar plate and transferred to 1 mL SM buffer (50 mM Tris-HCl, pH 7.5, 20 mM MgSO4, 100 mM NaCl, and 0.01% [w/v] gelatin). Three additional rounds of screening with a decreasing number of plaques on each plate were performed until two single positive clones could be isolated. A restriction map was established for the insert of clone λXG7. Thereafter, inverse PCR (Ochman et al., 1988) could be performed on λXG7 to amplify various parts of the Xantha-g gene. Amplified DNA fragments were either sequenced directly or ligated into the pGEM-T vector (Promega) before sequencing with a Prism 3100 genetic analyzer and the BigDye Terminator v3.1 Cyclase Sequencing kit (Applied Biosystems). The sequence information from various parts of the Xantha-g gene in λXG7 was used to design oligonucleotides for amplification of chromosomal Xantha-g DNA of the xantha-g28, -g37, -g44, -g45, and -g65 mutants. Two independent PCR reactions of each chromosomal DNA fragment, ranging in size from 476 to 1847 bp, were performed to exclude false mutations introduced by the amplification procedure. Both DNA strands of the amplified fragments were sequenced.
Protein, DNA, and RNA Gel Blot Analyses
Total cell protein from barley leaves was extracted (Hansson et al., 1997) and separated by SDS-PAGE for protein gel blot analysis as described elsewhere (Lake et al., 2004). Antibodies were raised against the C-terminal half of the barley Xantha-g–encoded protein (Lake et al., 2004). Chromosomal DNA was extracted with cetyltrimethylammonium bromide as described by Murray and Thompson (1980). Twenty micrograms of chromosomal DNA was digested overnight by restriction endonucleases PstI and SphI and separated on a 0.8% (w/v) agarose gel before being transferred by capillary blotting to Hybond-N filters (GE Healthcare) using 10× SSC (Sambrook and Russell, 2001). Total RNA was isolated with Trizol reagent (Life Technology) following the manufacturer's instructions. Twenty-one micrograms of RNA were run on a 1% (w/v) agarose gel containing 6% (v/v) formamide and transferred by capillary blotting to Hybond-N filters using 10× SSPE (Sambrook and Russell, 2001). DNA and RNA gel blots were visualized by staining for 30 s in 0.1% (w/v) methylene blue and 0.3 M Na-acetate, pH 5, followed by washing in water. The probe used in the DNA gel blot and RNA gel blot analyses was identical to the probe used for cloning of the Xantha-g gene. The experimental procedure has been described in further detail elsewhere (Hansson et al., 1997).
Mg-Chelatase Activity Assay
Mg-chelatase assays were performed in triplicate in white, nontransparent, 96-well microtitre plates for fluorescence reading (BMG LabTechnologies). The total reaction volume was 200 μL for each sample. Samples were read at 1-min intervals during incubation at 30°C in a FLUOstar Galaxy fluorescence plate reader (BMG LabTechnologies) using a 420- to 10-nm excitation filter, a 600- to 10-nm emission filter, and a gain of 31. Standard assays contained 5.0 pmol of BchI, 2.2 pmol His-tagged BchD, and 12.4 pmol of His-tagged BchH in an assay buffer consisting of 0.1 M Tricine-NaOH, pH 8.0, 2 mM DTT, 15 mM MgCl2, 4 mM ATP, and 1.8 μM protoporphyrin IX. The assay was started by adding 180 μL of assay buffer containing protoporphyrin IX and HisBchH, which had been preincubated at 30°C for 15 min, to 20 μL of BchI-HisBchD premixes. The BchI-HisBchD premixes were prepared by rapidly diluting 3 μL of 19 μM HisBchD solubilized in 6 M urea with 250 μL of ice-cold 0.25 μM BchI in assay buffer without protoporphyrin IX (Willows and Beale, 1998). The quantities of HisBchD in the assay were varied by further dilution of this BchI-HisBchD premixture with the 0.25 μM BchI in assay buffer without protoporphyrin IX. The urea concentration was adjusted to 7.1 mM in all assays. The assays were linear for between 5 and 25 min, and the rate of Mg protoporphyrin formation was estimated from a standard curve. To check the validity of the fluorescence measurements with the FLUOstar Galaxy fluorescence plate reader, assays were routinely stopped after 30 min by transferring 100 μL of assay mixtures to 900 μL of 0.1 M NH3OH in acetone, centrifuging, and collecting emission spectra from 550 to 700 nm with the excitation set at 404 and 418 nm. The maxima at 595 and 630 nm correlated with the expected quantities of Mg protoporphyrin IX and protoporphyrin IX, respectively, based on the fluorescence measurements with the FLUOstar Galaxy fluorescence plate reader.
Site-Directed Mutagenesis, Expression, and Purification of Proteins
Site-directed mutagenesis was performed on plasmid pET15bRcBchD (Willows and Beale, 1998) using the QuikChange method (Stratagene). Plasmid pET15bRcBchD contains the Rhodobacter capsulatus bchD gene cloned in the pET15b vector (Novagen). To change the R. capsulatus bchD gene, new oligonucleotides were synthesized (see Supplemental Table 1 online). The constructs were confirmed by DNA sequencing of the entire gene. The wild-type and modified BchD were produced in E. coli BL21(DE3) (Studier and Moffatt, 1986) as recombinant His-tagged proteins and purified as previously described (Willows and Beale, 1998) with the exception that the purified His-tagged BchD proteins were immediately desalted into 50 mM MES-NaOH, 6 M urea, 15 mM MgCl2, 4 mM DTT, and 6% glycerol, pH 6.7. The BchH and BchI proteins were expressed and purified as described elsewhere (Willows and Beale, 1998).
Transmission Electron Microscopy
Carbon-coated 400-mesh copper grids were negatively charged using a Balzers SCD004 sputter coater instrument. A drop of 50 mM Tricine-NaOH, pH 8.0, 15 mM MgCl2, and 2 mM DTT was applied to the grid for 1 min, and the excess liquid was removed with a filter paper. Grids were also prepared with the same buffer supplemented with 2 mM ATP. Purified R. capsulatus BchD in urea was dialyzed overnight at 4°C against 1 liter of 50 mM Tricine-NaOH, pH 8.0, 15 mM MgCl2, 2 mM DTT, and ±2 mM ATP using a Spectra/Por membrane disc with 3500 D molecular cutoff (Spectrum Laboratories). The protein samples were centrifuged at 16,000g for 5 min prior to their addition to the grid and incubated for 1 min before the excess liquid was removed with a filter paper. Thereafter, the grid was negatively stained for 30 s using filtered 2% (w/v) uranyl acetate, and the excess liquid was removed using filter paper. The specimens were analyzed at a magnification of 55,000 using a 120-kV Philips CM-10 electron microscope.
Accession Numbers
The DNA sequence data of the barley Xantha-g gene reported in this article have been deposited in the GenBank/EMBL data libraries under accession number AAZ32779.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primers Used in Site-Directed Mutagenesis of R. capsulatus bchD.
Supplemental Figure 1. Nucleotide Sequence of the Barley Xantha-g Gene and the Derived Amino Acid Sequence.
Supplemental Figure 2. The Chromosomal Location of the Barley Xantha-g Gene as Analyzed by DNA Gel Blots.
Supplemental Figure 3. The Exon Arrangement of the Barley Xantha-g Gene, Encoding the 70-kD Subunit of Mg-Chelatase, and the Location of the Four Identified xantha-g Mutations.
Supplementary Material
Acknowledgments
This work was made possible thanks to generous support from the Anna and Edvin Berger Foundation to E.A., from the Swedish Research Council, the Crafoord Foundation, and the Nilsson-Ehle Foundation at the Royal Physiographic Society in Lund to M.H., and from the Australian Research Council Grant A09905713, a Macquarie University Research Development grant, and International Research Exchange Grant X00001636 to R.D.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mats Hansson (mats.hansson@biochemistry.lu.se).
Online version contains Web-only data.
References
- Alawady, A.E., and Grimm, B. (2005). Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 41 282–290. [DOI] [PubMed] [Google Scholar]
- Bergdoll, M., Remy, M.H., Cagnon, C., Masson, J.M., and Dumas, P. (1997). Proline-dependent oligomerization with arm exchange. Structure 5 391–401. [DOI] [PubMed] [Google Scholar]
- Bollivar, D.W., Suzuki, J.Y., Beatty, J.T., Dobrowolski, J.M., and Bauer, C.E. (1994). Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J. Mol. Biol. 237 622–640. [DOI] [PubMed] [Google Scholar]
- Coomber, S.A., Chaudhri, M., Connor, A., Britton, G., and Hunter, C.N. (1990). Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol. Microbiol. 4 977–989. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodje, M.N., Hansson, A., Hansson, M., Olsen, J.G., Gough, S., Willows, R.D., and Al-Karadaghi, S. (2001). Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J. Mol. Biol. 311 111–122. [DOI] [PubMed] [Google Scholar]
- Gadjieva, R., Axelsson, E., Olsson, U., and Hansson, M. (2005). Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester mutants. Plant Physiol. Biochem. 43 901–908. [DOI] [PubMed] [Google Scholar]
- Gadjieva, R., Axelsson, E., Olsson, U., Vallon-Christersson, J., and Hansson, M. (2004). Nonsense-mediated mRNA decay in barley mutants allows the cloning of mutated gene by a microarray approach. Plant Physiol. Biochem. 42 681–685. [DOI] [PubMed] [Google Scholar]
- Gibson, L.C.D., Jensen, P.E., and Hunter, C.N. (1999). Magnesium chelatase from Rhodobacter sphaeroides: Initial characterization of the enzyme using purified subunits and evidence for a BchI-BchD complex. Biochem. J. 337 243–251. [PMC free article] [PubMed] [Google Scholar]
- Gorchein, A., Gibson, L.C., and Hunter, C.N. (1993). Gene expression and control of enzymes for synthesis of magnesium protoporphyrin monomethyl ester in Rhodobacter sphaeroides. Biochem. Soc. Trans. 21 201S. [DOI] [PubMed] [Google Scholar]
- Granick, S. (1948). Protoporphyrin IX as a precursor of chlorophyll. J. Biol. Chem. 172 717–727. [PubMed] [Google Scholar]
- Guo, R., Luo, M., and Weinstein, J.D. (1998). Magnesium chelatase from developing pea leaves. Plant Physiol. 116 605–615. [Google Scholar]
- Hanson, P.I., and Whiteheart, S.W. (2005). AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 6 519–529. [DOI] [PubMed] [Google Scholar]
- Hansson, A., Kannangara, C.G., von Wettstein, D., and Hansson, M. (1999). Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc. Natl. Acad. Sci. USA 96 1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson, A., Willows, R.D., Roberts, T.H., and Hansson, M. (2002). Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA+ hexamers. Proc. Natl. Acad. Sci. USA 99 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson, M., Gough, S.P., Kannangara, C.G., and von Wettstein, D. (1997). Analysis of RNA and enzymes of potential importance for regulation of 5-aminolevulinic acid synthesis in the protochlorophyllide accumulating barley mutant tigrina-d12. Plant Physiol. Biochem. 35 827–836. [Google Scholar]
- Hansson, M., Gough, S.P., Kannangara, C.G., and von Wettstein, D. (1998). Chromosomal locations of six barley genes encoding enzymes of chlorophyll and heme biosynthesis and the sequence of the ferrochelatase gene identify two regulatory genes. Plant Physiol. Biochem. 36 545–554. [Google Scholar]
- Hansson, M., and Kannangara, C.G. (1997). ATPases and phosphate exchange activities in magnesium chelatase subunits of Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. USA 94 13351–13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen, K.W., Boynton, J.E., and von Wettstein, D. (1993). Mutants at xantha and albina Loci in Relation to Chloroplast Biogenesis in Barley (Hordeum vulgare L.). (Copenhagen, Denmark: The Royal Danish Academy of Sciences and Letters).
- Hynes, R.O. (2002). Integrins: Bidirectional, allosteric signaling machines. Cell 110 673–687. [DOI] [PubMed] [Google Scholar]
- Islam, A.K.M.R., Shepherd, K.W., and Sparrow, D.H.B. (1981). Isolation and characterization of euplasmic wheat-barley chromosome addition lines. Heredity 46 161–174. [Google Scholar]
- Isshiki, M., Yamamoto, Y., Satoh, H., and Shimamoto, K. (2001). Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol. 125 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, L.M., Leipe, D.D., Koonin, E.V., and Aravind, L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146 11–31. [DOI] [PubMed] [Google Scholar]
- Jensen, P.E., Gibson, L.C., Henningsen, K.W., and Hunter, C.N. (1996). Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J. Biol. Chem. 271 16662–16667. [DOI] [PubMed] [Google Scholar]
- Jensen, P.E., Gibson, L.C., and Hunter, C.N. (1999). ATPase activity associated with the magnesium-protoporphyrin IX chelatase enzyme of Synechocystis PCC6803: Evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP-dependent interaction of the ChlI and ChlD subunits. Biochem. J. 339 127–134. [PMC free article] [PubMed] [Google Scholar]
- Jensen, P.E., Gibson, L.C.D., and Hunter, C.N. (1998). Determinants of catalytic activity with the use of purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from Synechocystis PCC6803. Biochem. J. 334 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger, G.A., Reid, J.D., and Hunter, C.N. (2001). Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry 40 9291–9299. [DOI] [PubMed] [Google Scholar]
- Kropat, J., Oster, U., Rüdiger, W., and Beck, C.F. (2000). Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24 523–531. [DOI] [PubMed] [Google Scholar]
- Lake, V., Olsson, U., Willows, R.D., and Hansson, M. (2004). ATPase activity of magnesium chelatase subunit I is required to maintain subunit D in vivo. Eur. J. Biochem. 271 2182–2188. [DOI] [PubMed] [Google Scholar]
- Lee, J.O., Rieu, P., Arnaout, M.A., and Liddington, R. (1995). Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18). Cell 80 631–638. [DOI] [PubMed] [Google Scholar]
- Luo, M., and Weinstein, J.D. (1997). Cloning and sequencing of a cDNA encoding the putative Mg-chelatase subunit D (accession no. AF014399) from pea (Pisum sativum L. cv. Spring). Plant Physiol. 115 315. [Google Scholar]
- MacArthur, M.W., and Thornton, J.M. (1991). Influence of proline residues on protein conformation. J. Mol. Biol. 218 397–412. [DOI] [PubMed] [Google Scholar]
- Murray, H.G., and Thompson, W.F. (1980). Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 9 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald, A.F., Aravind, L., Spouge, J.L., and Koonin, E.V. (1999). AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9 27–43. [PubMed] [Google Scholar]
- Ochman, H., Gerber, A.S., and Hartl, D.L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, U., Sirijovski, N., and Hansson, M. (2004). Characterization of eight barley xantha-f mutants deficient in magnesium chelatase. Plant Physiol. Biochem. 42 557–564. [DOI] [PubMed] [Google Scholar]
- Papenbrock, J., Gräfe, S., Kruse, E., Hanel, F., and Grimm, B. (1997). Mg-chelatase of tobacco: Identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 12 981–990. [DOI] [PubMed] [Google Scholar]
- Petersen, B.L., Kannangara, C.G., and Henningsen, K.W. (1999. a). Distribution of ATPase and ATP-to-ADP phosphate exchange activities in magnesium chelatase subunits of Chlorobium vibrioforme and Synechocystis PCC6803. Arch. Microbiol. 171 146–150. [DOI] [PubMed] [Google Scholar]
- Petersen, B.L., Møller, M.G., Jensen, P.E., and Henningsen, K.W. (1999. b). Identification of the Xan-g gene and expression of the Mg-chelatase encoding genes Xan-f, -g and -h in mutant and wild type barley (Hordeum vulgare L.). Hereditas 131 165–170. [Google Scholar]
- Reid, J.D., Siebert, C.A., Bullough, P.A., and Hunter, C.N. (2003). The ATPase activity of the ChlI subunit of magnesium chelatase and formation of a heptameric AAA+ ring. Biochemistry 42 6912–6920. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sawers, R.J., Viney, J., Farmer, P.R., Bussey, R.R., Olsefski, G., Anufrikova, K., Hunter, C.N., and Brutnell, T.P. (2006). The maize oil yellow1 (oy1) gene encodes the I subunit of magnesium chelatase. Plant Mol. Biol. 60 95–106. [DOI] [PubMed] [Google Scholar]
- Shen, Y.-Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443 823–826. [DOI] [PubMed] [Google Scholar]
- Strand, Å., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83. [DOI] [PubMed] [Google Scholar]
- Studier, F.W., and Moffatt, B.A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189 113–130. [DOI] [PubMed] [Google Scholar]
- Tanaka, A., and Tanaka, R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9 248–255. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. (2000). AAA proteins: Lords of the ring. J. Cell Biol. 150 F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileuskaya, Z., Oster, U., and Beck, C.F. (2005). Mg-protoporphyrin IX and heme control HEMA, the gene encoding the first specific step of tetrapyrrole biosynthesis, in Chlamydomonas reinhardtii. Eukaryot. Cell 4 1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart, S.W., Rossnagel, K., Buhrow, S.A., Brunner, M., Jaenicke, R., and Rothman, J.E. (1994). N-ethylmaleimide-sensitive fusion protein: A trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willows, R.D., and Beale, S.I. (1998). Heterologous expression of the Rhodobacter capsulatus BchI, -D, and -H genes that encode magnesium chelatase subunits and characterization of the reconstituted enzyme. J. Biol. Chem. 273 34206–34213. [DOI] [PubMed] [Google Scholar]
- Willows, R.D., Gibson, L.C.D., Kanangara, C.G., Hunter, C.N., and von Wettstein, D. (1996). Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur. J. Biochem. 235 438–443. [DOI] [PubMed] [Google Scholar]
- Willows, R.D., Hansson, M., Beale, S.I., Laurberg, M., and Al-Karadaghi, S. (1999). Crystallization and preliminary X-ray analysis of the Rhodobacter capsulatus magnesium chelatase BchI subunit. Acta Crystallogr. Sect. D Biol. Crystallogr. 55 689–690. [DOI] [PubMed] [Google Scholar]
- Willows, R.D., Hansson, A., Birch, D., Al-Karadaghi, S., and Hansson, M. (2004). EM single particle analysis of the ATP-dependent BchI complex of magnesium chelatase: An AAA+ hexamer. J. Struct. Biol. 146 227–233. [DOI] [PubMed] [Google Scholar]
- von Wettstein, D., Gough, S., and Kannangara, C.G. (1995). Chlorophyll biosynthesis. Plant Cell 7 1039–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.