Abstract
BEL1-like transcription factors interact with Knotted1 types to regulate numerous developmental processes. In potato (Solanum tuberosum), the BEL1 transcription factor St BEL5 and its protein partner POTH1 regulate tuber formation by mediating hormone levels in the stolon tip. The accumulation of St BEL5 RNA increases in response to short-day photoperiods, inductive for tuber formation. RNA detection methods and heterografting experiments demonstrate that BEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction. This movement of RNA to stolon tips is correlated with enhanced tuber production. Overexpression of BEL5 transcripts that include the untranslated sequences of the BEL5 transcript endows transgenic lines with the capacity to overcome the inhibitory effects of long days on tuber formation. Addition of the untranslated regions leads to preferential accumulation of the BEL5 RNA in stolon tips under short-day conditions. Using a leaf-specific promoter, the movement of BEL5 RNA to stolon tips was facilitated by a short-day photoperiod, and this movement was correlated with enhanced tuber production. These results implicate the transcripts of St BEL5 in a long-distance signaling pathway that are delivered to the target organ via the phloem stream.
INTRODUCTION
As part of an elaborate long-distance communication system, plants have evolved a unique signaling pathway that takes advantage of connections in the vascular tissue, predominately the phloem. This information pathway has been implicated in regulating development, responding to biotic stress, delivering nutrients, and as a vehicle commandeered by viruses for spreading infections (Lough and Lucas, 2006). There are several experimental examples demonstrating that cosuppression of expression mediated by systemic acquired gene silencing involves RNA transport within the phloem (Sonoda and Nishiguchi, 2000; Crete et al., 2001).
Unlike cell-autonomous animal systems, plants move RNA and proteins from the cell of origin through the phloem to target sites to activate environmentally regulated and developmental processes. A recent study on flower induction presented evidence consistent with the transport of the mRNA of the FLOWERING LOCUS T (FT) gene to the shoot apex, where downstream floral genes were activated (Huang et al., 2005). These results suggest that the FT mRNA is an important component of the elusive flowering signal that is activated by photoperiod and moves from the leaf to the shoot apex.
Techniques involving grafting showed that specific RNAs can move long distances through the phloem. Scions (the upper portion of the graft) of cucumber (Cucumis sativus) grafted onto pumpkin (Cucurbita pepo) stocks provided direct evidence that specific pumpkin mRNAs were translocated through the heterograft (Ruiz-Medrano et al., 1999; Xoconostle-Cazares et al., 1999). RNAs present in phloem sap moved selectively into apical tissues of heterografted scions. The discovery of the RNA binding protein Cm PP16 provided additional support for the long-distance transport of RNA in pumpkin (Xoconostle-Cazares et al., 1999). A unique set of proteins is selectively transported through phloem cells to mediate destination-specific processes (Aoki et al., 2005). In this system, whereas shootward translocation appeared to be passively carried by bulk flow, rootward movement of two phloem RNA binding proteins, Cm PP16-1 and Cm PP16-2, was selectively controlled. The data indicated that this selective movement is regulated by protein–protein interaction in the phloem sap. It was proposed that lateral transfer of Cm PP16-1 and Cm PP16-2 might be controlled selectively at the node or by selective exit from the sieve tube system.
An important family of transcription factors that regulate the developmental events in apical meristems is the Knox (for knotted-like homeobox) gene family (Reiser et al., 2000). Long-distance movement of RNA was reported for a KNOTTED1-like homeobox gene of tomato (Solanum lycopersicum), LeT6 (Kim et al., 2001). The transport of this Knox RNA occurred in an acropetal direction and induced developmental changes in the wild-type scion consistent with the Mouse-ear phenotype. Knotted1 RNA binds to its protein to facilitate cell-to-cell movement (Kim et al., 2005). There are several examples of other transcription factors, functional in meristems, whose mRNA can be transported from cell to cell or over long distances (Kim et al., 2001; Lucas et al., 1995; Ruiz-Medrano et al., 1999; reviewed in Haywood et al., 2002). RNA for GAI, a DELLA-type protein involved in GA signaling, moves through the phloem to the shoot apex in Arabidopsis thaliana, tomato, and pumpkin (Haywood et al., 2005). Delivery of gai RNA mediated highly reproducible changes in leaf phenotype. Studies like these have demonstrated that RNA entry into functional sieve elements occurs via a selective mechanism and that the movement to the target organ is a regulated process.
Knox genes belong to the TALE (for three amino acid loop extension) superclass of transcription factors (Bürglin, 1997). TALE transcription factors are distinguished by a very high level of sequence conservation in the DNA binding region, designated the homeodomain, consisting of three α-helices (Kerstetter et al., 1994). The two main groups of transcription factors of the TALE superclass in plants are the KNOX and BEL1 types (Bürglin, 1997), and protein members of these two groups have been identified that physically interact. Interactions between KNOX and BEL proteins have been documented in barley (Hordeum vulgare), Arabidopsis, maize (Zea mays), and potato (Solanum tuberosum) (Müller et al., 2001; Bellaoui et al., 2001; Smith et al., 2002; Chen et al., 2003). The KNOX protein of potato, designated POTH1, regulates plant growth by controlling gibberellic acid (GA) synthesis and enhancing cytokinin levels (Rosin et al., 2003a). Transgenic overexpression of POTH1 produced plants that exhibited reduced expression of a key enzyme in GA biosynthesis, aberrant leaf morphology, and enhanced tuber production (Rosin et al., 2003a). Using POTH1 as bait, seven distinct BEL1-like proteins of potato were identified (Chen et al., 2003). Similar to overexpression mutants of POTH1, transgenic lines that overexpressed one of the potato BEL1 partners, St BEL5, exhibited enhanced tuber formation and increased cytokinin levels (Chen et al., 2003). Transcription assays with the BEL and KNOX proteins of potato indicate that, in tandem, they bind specific DNA sequences of the promoter of ga20 oxidase1 (ga20ox1) to repress its activity (Chen et al., 2004). These results indicate that the tandem interaction of St BEL5 and POTH1 is essential for regulation of the expression of their target gene, ga20ox1, and that they cannot act alone.
Tuber formation in potatoes is a complex developmental process that requires the interaction of environmental, biochemical, and genetic factors (reviewed in Ewing and Struik, 1992). In potato genotypes such as S. tuberosum ssp andigena, short-day (SD) photoperiods are strictly required for tuber formation, whereas long-day (LD) conditions inhibit tuberization. Under inductive conditions, a transmissible signal originating from the leaf is activated that initiates cell division via cytokinins and a change in the orientation of cell growth in the subapical region of the stolon tip via a reduction in GA levels (Xu et al., 1998a).
In this signal transduction pathway, perception of the appropriate environmental cues is mediated by phytochrome and gibberellins (Jackson et al., 1996; Jackson and Prat, 1996). High levels of gibberellins are correlated with the inhibition of tuberization, whereas low levels are associated with induction at both the site of perception (the leaf) and in the target organ, the stolon apex, site of the newly formed tuber (Jackson and Prat, 1996; Xu et al., 1998b). The results of Martinez-Garcia et al. (2002) suggested that CONSTANS was involved in regulating the production of the long-distance signal that activates tuber formation.
The identity of the tuberization signal is unknown, but recent reports on the role of FT in flowering have shown that long-distance movement of specific RNAs may act as a signal that mediates growth responses regulated by photoperiod (Huang et al., 2005). This report documents the presence of BEL1-like mRNAs in phloem cells of potato, photoperiod-mediated movement of this RNA to its target organ, and the capacity of transgenic lines that overexpress the full-length St BEL5 transcript to overcome the negative effects of LD conditions on tuberization.
RESULTS
Accumulation and Location of St BEL5 RNA in Wild-Type Plants
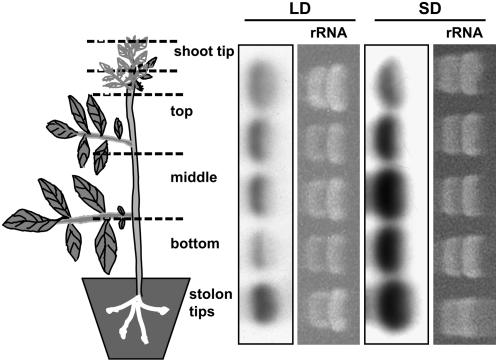
Previous work has demonstrated that BEL5 RNA is ubiquitous in potato plants and that its accumulation is enhanced in leaves and stolons by SD conditions (Chen et al., 2003). To examine this observation in more detail, an analysis of St BEL5 mRNA accumulation in stems and stolons from wild-type plants of S. tuberosum ssp andigena grown under both LD and SD conditions was undertaken. As expected, levels of St BEL5 RNA increased under SD conditions (Figure 1). St BEL5 RNA levels accumulated in an increasing concentration gradient from the shoot tip (low levels) through the stem to the stolon tip in response to SD conditions (Figure 1). Under LD conditions, the highest levels of RNA were observed in shoot tips and stolons with the lowest levels in the lower portion of the stem (Figure 1).
Figure 1.
Accumulation of St BEL5 RNA in Stems and Stolons of Wild-Type Plants under LD and SD Photoperiod Conditions.
Plants of the photoperiodic responsive line S. tuberosum ssp andigena were grown in the greenhouse under LDs until the 12-leaf stage and then moved to a growth chamber under LD (16 h light/8 h dark) or SD (8 h light/16 h dark) conditions. After 12 d, tissue was harvested from shoot tips, stem sections, and stolon tips, and RNA was extracted and subjected to electrophoresis on a denaturing gel. Hybridization was performed on RNA gel blots with a 32P-labeled DNA probe specific for St BEL5. Ten micrograms of total RNA was loaded per lane. Equal loading was verified by visualizing rRNA loading under UV light.
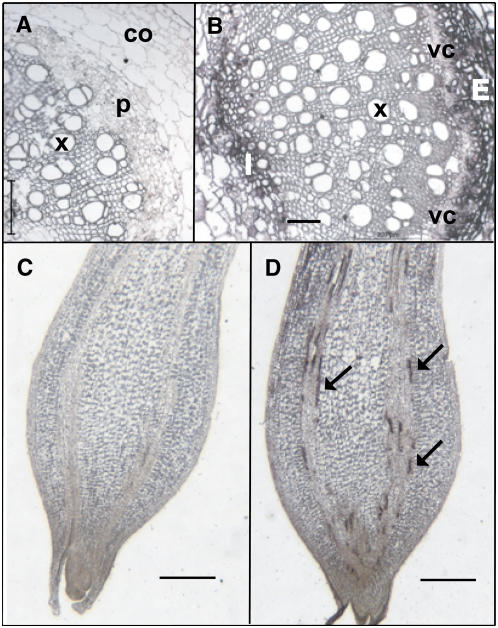
To determine the precise location of transcripts of St BEL5 in stems and stolons, in situ hybridization was performed on both stem and stolon sections of wild-type plants. In transverse sections of stems, BEL5 RNA was detected in the internal (Figure 2B) and external phloem (Figure 2B), characteristic of the potato vascular system. No signal was detected in pith, cortex, or xylem cells or when using a sense probe for hybridization (Figure 2A). In stolon tips that are in the early stages of tuber formation (marked by swelling in the stolon tip; Figures 2C and 2D), St BEL5 RNA was detected in the discontinuous sieve elements of both internal and external phloem (Figure 2D, arrows). Most of the signal was detected in the region of most active cell growth during tuberization (Xu et al., 1998a), the subapical region. Very little, if any, transcript was detected in the apical meristem of the stolon (Figure 2D). These results clearly demonstrate the presence of St BEL5 mRNA in phloem cells of potato stem and stolons.
Figure 2.
In Situ Hybridizations of Sections of the Potato Stem and Stolon Tip of Wild-Type Plants during Early Tuber Formation.
Sections were hybridized with a digoxygenin-labeled 0.2-kb RNA copy of sequence from the 3′ untranslated region of St BEL5 ([A] and [C], sense riboprobe; [B] and [D], antisense riboprobe). The presence of St BEL5 mRNA is indicated by the dark stain under bright-field microscopy. Bars = 100 μm in (B) and 0.5 mm in (C) and (D). The stolon sections ([C] and [D]) are from an SD-grown plant in the early stages of tuber formation. The stem sections ([A] and [B]) are from LD-grown plants. Similar results were obtained in stem sections from SD-grown plants.
(A) Transverse section of an LD stem; a negative control. p, phloem cells; co, cortex; x, xylem.
(B) Transverse section of an LD stem with antisense probe. E, external phloem cells; I, internal phloem cells; x, xylem; vc, vascular cambium.
(C) and (D) Longitudinal sections of newly tuberizing stolon tips. Sieve elements in (D) were identified by a comparison to longitudinal sections of stolon tips in Cutter (1978). The arrows in (D) indicate positive St BEL5 signal.
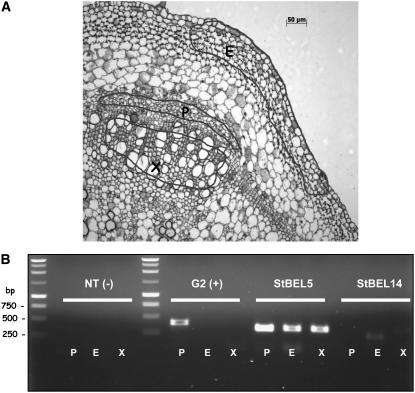
To increase the sensitivity of detection for specific RNAs in a defined number of homogeneous cells derived from stems, we made use of laser microdissection coupled with laser pressure catapulting (LPC) (Niyaz et al., 2005) from transverse paraffin sections of wild-type potato stems on the PALM Microlaser system (Plant Sciences Institute, Iowa State University). For microdissection, focused laser light was used to excise selected cells from regions of the xylem (Figure 3A), phloem (Figure 3A), or epidermis (Figure 3A). RT-PCR of RNA extracted from specific stem cells of both LD- and SD-grown plants showed that St BEL5 RNA was present in xylem, phloem, and epidermal cells (Figure 3B). All seven of the RNAs of the BEL1 family from potato (Chen et al., 2003) were tested, and only RNA of St BEL14 was not detected in phloem cells. St BEL14 transcripts were detected only in epidermal cells (Figure 3B). Previously, the RNA from St BEL14 was detected in flowers, leaves, and roots (Chen et al., 2003). RNA for the protein partner of St BEL5, POTH1, was also detected in phloem cells (data not shown). RNA from a nitrate transporter (NT; specific to roots; Nazoa et al., 2003) was used as a negative control, and a G2-like transcription factor, an RNA specific to phloem cells (Zhao et al., 2005), was used a positive control (Figure 3B). The results of the laser capture microdissection/RT-PCR analysis verify the presence of St BEL5 RNA in phloem cells of potato stems.
Figure 3.
Laser Microdissection and LPC to Identify Specific RNAs in Phloem Cells of Potato Stems.
Microdissection was performed on paraffin-imbedded transverse sections of potato stems of wild-type plants (S. tuberosum ssp andigena) using the PALM Microlaser system (Plant Sciences Institute, Iowa State University). For microdissection, focused laser light was used to excise selected cells from regions of the xylem, phloem, or epidermis (X, P, and E in [A], respectively). After microdissection, the sample cells were directly catapulted into an appropriate collection device. RNA was extracted from these cells and used as a template for RT-PCR using gene-specific primers for each RNA sample (B). NT is nitrate transporter (specific for root cells). G2 is the G2-like transcription factor (specific for phloem cells). Identity of PCR products was confirmed by sequencing isolated bands. Data shown are from SD-grown plants. Identical results were obtained when using material from LD-grown plants.
Movement of a Transcript for the St BEL5 Coding Sequence across a Graft Union
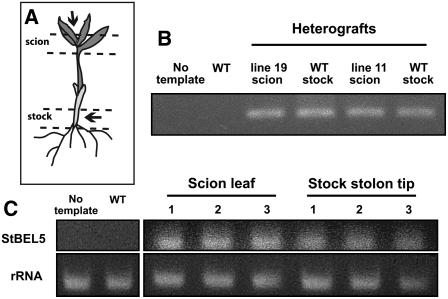
Because of the RNA gradient in stems and the location of St BEL5 RNA, we considered that St BEL5 RNA could be moving through the phloem toward the base of the plant. Previous work has shown that transgenic lines overexpressing St BEL5 coding sequence (cds) produced more tubers at a faster rate than controls (Chen et al., 2003). Other than the effect on tuber formation, these St BEL5 lines exhibited phenotypes similar to wild-type plants. Here, we made use of these transgenic lines to examine RNA mobility across a graft union. Scions of overexpression lines for St BEL5 were grafted onto wild-type stocks in vitro (Figure 4A). The grafted plants were cultured under SDs for 8 d and harvested. RNA was extracted from scion and stock tissue as indicated by the dashed lines in Figure 4A. RNA was then used as template in RT-PCR with gene-specific nested primers. Nonplant nopaline synthase sequence attached to all transgenic RNAs produced by our transgenic lines was used as a transcript-specific primer to discriminate from the native St BEL5 RNA. All PCR products detected in scion (positive control) and stock (test for movement) RNA samples represent tagged, transgenic RNA.
Figure 4.
Movement of St BEL5-cds Transcripts across a Graft Union in Vitro and in Soil-Grown Plants.
Using heterografts with tissue culture plants (A) and RT-PCR with gene-specific primers, RNA for St BEL5 moves across a graft union toward the base of the plant ([B], WT stock lanes). All PCR products detected in scion (positive control) and stock (test for movement) RNA samples represent tagged transgenic RNA. Arrows in (A) indicate where samples for RNA extraction were taken. Lines 19 and 11 are transgenic potato lines that overexpresses St BEL5 mRNA coding sequence only. RNA from line 19 and line 11 scion samples ([A], top arrow) was used as a positive control. Wild-type RNA from stock material ([A], bottom arrow) was sampled for both heterografts. Heterografts were cultured in vitro for 10 d under SD conditions (8 h light/16 h dark). PCR was performed twice off cDNA template made from RNA and reverse transcriptase. Two different gene-specific primers were used with a nonplant DNA tag specific for the transgenic RNA (designated NT-2 in Methods). RNA from wild-type/wild-type autografts was used as a negative control (WT lane). Ethidium bromide–stained PCR product of expected size (360 bp) is shown in (B). The identity of these bands was verified with a DNA gel blot using a St BEL5–specific probe. RT-PCR results from whole plant heterografts of St BEL5-cds/wild type are shown in (C). Scion leaf samples were from overexpression lines of St BEL5-cds from three separate heterografts, whereas wild-type stock samples were harvested from 5.0 mm of the stolon tip after 14 d of SD conditions. Transcript-specific primers were used for St BEL5 and for the rRNA reactions. No PCR product was detected from reactions without template or from RNA extracted from wild-type/wild-type autografts.
Using two different transgenic lines, St BEL5 RNA moved through the phloem stream in the stem toward the base of the plant (Figure 4B). Movement was also tested in soil-grown plants, and these heterografts confirmed the movement of St BEL5 RNA through the stem to wild-type stolon tips (Figure 4C). This movement was correlated with a twofold increase in tuber yields of St BEL-cds/wild-type heterografts compared with wild-type/wild-type autografts.
Analysis of Full-Length St BEL5 RNA
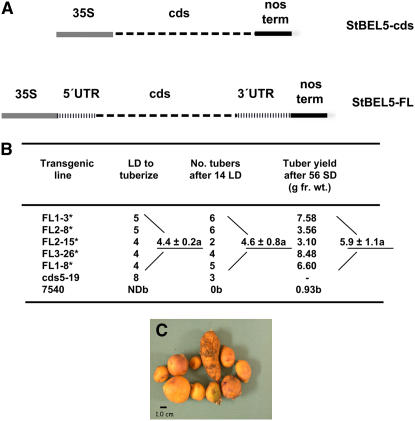
Initial analyses of yield and RNA mobility were performed with a truncated form of the St BEL5 RNA, ∼2000 nucleotides long and containing only the complete coding sequence of St BEL5 (Figure 5A). Because there is such wide variation among the potato BEL RNAs in the length of their untranslated regions (UTRs) (e.g., 505 nucleotides for the 3′ UTR of St BEL5 versus 60 and 90 nucleotides for St BEL30 and -14, respectively), we speculated that the 3′ UTR of St BEL5 could control its expression and/or movement. Therefore, a full-length construct of St BEL5 was cloned into the binary vector pCB201 and driven by the cauliflower mosaic virus (CaMV) 35S promoter to assess its effect on development (Figure 5A). This construct included 146 nucleotides of 5′ UTR and the complete 505 nucleotides of 3′ UTR. To assess photoperiod effects, transformation was implemented on the photoperiod-responsive potato cultivar S. tuberosum ssp andigena (Banerjee et al., 2006). In SD-adapted genotypes like ssp andigena, SD photoperiods (<12 h of light) are required for tuber formation, whereas under LD conditions, no tubers are produced.
Figure 5.
Rate of Tuberization of Full-Length St BEL5 Transgenic Lines.
Previous work on the analysis of transgenic lines that overexpressed St BEL5 was done with the coding sequence only (A) (Chen et al., 2003). In this experiment, transgenic lines overexpressing the full-length St BEL5 transcript (A), including ∼650 nucleotides of the UTRs (accession number AF406697) were tested. Line 7540 is nontransformed S. tuberosum ssp andigena. Evaluations were performed on 10 plants per line cultured on Murashige and Skoog (MS) medium with 6% sucrose (B). Plants were scored for time to tuber formation under LDs (earliness), number of tubers after 14 LDs, and total tuber yield after 56 d under SD conditions in vitro. The greenhouse LD experiment was with soil-grown plants. The top five lines (asterisks) tuberized as soil-grown plants in 10-cm pots after 8 weeks under LD conditions in the greenhouse. Most of these tubers ranged from 1.5 to 2.5 cm in diameter (C). Overexpression lines of the coding sequence only (Chen et al., 2003) did not form tubers under LDs even after 6 months in the greenhouse. The means of the St BEL5-FL lines for earliness, tuber number, and yield were significantly greater than the 7540 control plants at P = 0.05 (Fisher's test), and this difference is designated by the letters a or b. ND, not detected.
More than 50 putative full-length St BEL5 overexpression lines (St BEL5-FL) were screened, and 10 lines were identified to characterize in more detail (Banerjee et al., 2006). Several of these lines were selected on the basis of enhanced tuber formation under LDs in vitro. Tuber formation under in vitro conditions, however, is not an uncommon observation, so we used a number of parameters for assessing a plant's capacity for tuber induction, including both in vitro and soil-grown culture. The least to most inductive conditions proceed from LD soil-grown to LD in vitro to SD soil-grown to SD in vitro. Under the noninductive conditions of in vitro LDs, full-length St BEL5 transgenic lines tuberized sooner (earliness) and to a greater degree (tuber number) than control plants and the transgenic St BEL5-cds line 5-19 (Figure 5B). Under SD in vitro conditions, tuber yields for these lines were greater than controls (up to ninefold more in some cases). Seven St BEL5-FL lines (out of 12 tested) were identified that formed tubers on soil-grown plants in 10-cm pots after 8 weeks under LD conditions in the greenhouse. Tubers produced from these plants ranged from 1.5 to 2.5 cm in diameter and were comparable in appearance and size to tubers formed from wild-type plants under SD conditions (Figure 5C). Wild-type andigena lines do not make tubers under LD conditions. Overexpression coding sequence lines of St BEL5 were not able to form tubers after 6 months under LDs in the greenhouse (Chen et al., 2003). Addition of both the 3′ and 5′ UTRs in overexpression lines of St BEL5 clearly affected the timing and extent of tuber formation under both LD and SD conditions. The St BEL5-FL transgenic lines exhibited increased earliness, tuber numbers, and overall yields. Only the addition of the St BEL5 UTRs to transgenic constructs produced soil-grown plants that were able to overcome the negative effects of LD conditions on tuber formation.
Movement of the Full-Length Transcript of St BEL5 across a Graft Union to Stolon Tips
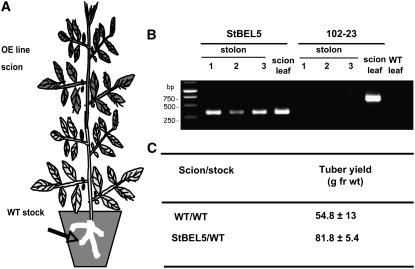
Based on the patterns of RNA accumulation and localization in the phloem, we considered that St BEL5 RNA was moving through the phloem toward the base of the plant to the target organ, the stolon tip. The data of Figure 4 were generated using a truncated form of the St BEL5 transcript. To examine the movement of full-length St BEL5 RNA through the phloem and to assess the effect of this movement on tuber production, heterografts of scions from overexpression lines and wild-type stock were analyzed (Figure 6A). A transgenic potato line overexpressing antisense sequence of a MADS box RNA, POTM1 (Rosin et al., 2003b), was used as a negative control. Grafted plants were allowed to heal and acclimate in the greenhouse before being transferred to SD conditions in a growth chamber. After 12 d under SD conditions, stolon tips 5.0 mm in length were harvested (Figure 6A, arrow) and the RNA extracted. Again, a nonplant sequence tag fused to all transgenic RNAs was used as an RNA-specific primer to discriminate from native BEL RNAs. RT-PCR with transcript-specific nested primers revealed that in three out of four different grafted plants, the full-length St BEL5 transcript moved to the stolon tip (Figure 6B). No POTM1 RNA was detected in stolon tips from four separate heterografts of the negative control. Wild-type scions were grafted onto wild-type stocks to assess the effect on tuber yields. After only 28 d, the St BEL5 full-length heterografts exhibited a 50% increase in tuber production compared with the wild-type control (Figure 6C). These results demonstrate movement of the full-length St BEL5 transcript through phloem to the target organ, the stolon tip, and correlation of this movement to enhanced tuber production.
Figure 6.
Movement of Full-Length Transcripts of St BEL5 across a Graft Union and Its Effect on Tuber Yield.
Soil-grown wild-type stock plants were grafted with scions from an St BEL5 overexpression line (5FL1-3) in the greenhouse. Both stock and scion material were grown initially under LDs. Grafts were sealed with plastic soda straw sections. Plants were placed in plastic bags, and graft unions were allowed to form. After 4 weeks of LDs in the greenhouse, grafted plants were transferred to a growth chamber and acclimated under LD conditions for 1 week before transfer to SD conditions. Leaf and stolon tip samples (arrow in [A]) were harvested after 12 d of SD conditions and the RNA extracted. RT-PCR with gene-specific primers was performed for both a negative control (a potato MADS box gene, 102-23) and test samples (St BEL5-FL). Grafts made from an overexpression line for an antisense sequence of a potato MADS box gene (line 102-23) were used as a nonmobile control. RNA from scion leaf samples was used as a positive control (scion leaf). Wild-type RNA from stolon tips, 0.5 cm in length, was sampled for both heterografts and used in the RT-PCR reactions. PCR was performed twice off template made from RNA and reverse transcriptase. Two different gene-specific primers were used with a nonplant DNA tag specific for the transgenic RNA to discriminate from the native RNA. Three plants were assayed for both heterografts and are designated 1, 2, and 3. RNA from leaves of a wild-type/wild-type autograft was used as a negative PCR control (WT leaf lane). Similar negative results were obtained with RNA from autograft stolons. For tuber yields, plants were harvested after 28 d, and the mean of three plants was calculated for wild-type and St BEL5 grafted plants. Wild-type scions grafted onto wild-type stocks were used as the yield controls. Identity of PCR products was confirmed by blot hybridization with a gene-specific probe.
RNA Accumulation in Transgenic Lines Expressing St BEL5 Transcripts with and without the UTRs
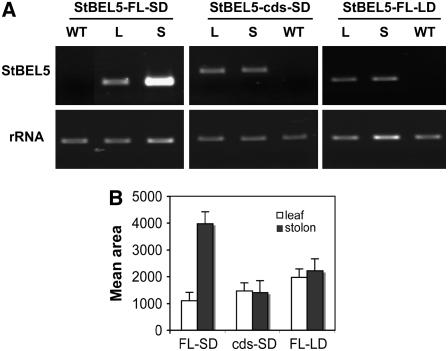
To better understand the functional role of the St BEL5 UTRs in enhancing tuber formation, we compared the relative accumulation of St BEL5 RNAs in whole plants of transgenic lines that expressed only the coding sequence (St BEL5-cds) or the coding sequence plus both the 5′ UTR and the 3′ UTR (St BEL5-FL). As with previous transgenic lines used here, both constructs were driven by the CaMV 35S promoter in the binary vector pCB201. Transgenic plants with each of the two constructs were grown under SDs for 14 d, and the RNA was extracted from new leaves and 5.0-mm stolon tips from three separate plants for both constructs (Figures 7A and 7B, lanes L and S for St BEL5-FL-SD and St BEL5-cds-SD). One-step RT-PCR was performed using a nonplant sequence tag fused to all transgenic RNAs (designated NT-2) and a gene-specific primer from either the open reading frame (St BEL5-cds-SD) or the 3′ UTR (St BEL5-FL) of St BEL5. Use of the NT-2 primer again makes it possible to discriminate transgenic RNA from native BEL5 RNAs. PCR products were scanned, quantified, and normalized with 18S rRNA product. In the samples from SD plants, more PCR product for the St BEL5-FL transcript was detected in stolons (Figures 7A and 7B, lane S for St BEL5-FL-SD) compared with leaves (Figures 7A and 7B, lane L for St BEL5-FL-SD), whereas the yield of product for the St BEL5-cds RNA from leaf and stolon exhibited, as expected, a ratio of ∼1:1 (Figures 7A and 7B, St BEL5-cds-SD). When transgenic lines expressing the St BEL5-FL transcript were grown under 14 LDs, preferential accumulation of the transgenic RNA in stolons did not occur and the ratio was ∼1:1 (Figures 7A and 7B, St BEL5-FL-LD). No PCR product was detected from native St BEL5 RNA extracted from leaves of wild-type plants (Figure 7A, WT lanes). All PCR reactions were optimized to yield product in the linear range (31 cycles for both BEL5 RNAs and 17 cycles for the rRNA). These results demonstrate that addition of the UTRs leads to preferential accumulation of RNA in stolon tips and that photoperiod appears to regulate this process. These results are consistent with the accumulation pattern of St BEL5 RNA in wild-type plants presented in Figure 1.
Figure 7.
RNA Accumulation in Transgenic Lines Expressing St BEL5 Transcripts with (St BEL5-FL) and without (St BEL5-cds) the UTRs of the RNA and the Effect of Photoperiod.
Transgenic plants were grown under SDs for 14 d, and the RNA was extracted from 0.5-cm stolon tips (S) and new leaves (L) from three separate plants for each construct (A). For the photoperiod experiment, transgenic plants with the St BEL5-FL construct were grown under LDs for 14 d ([B], St BEL5-FL-LD), and the RNA was extracted from 0.5-cm stolon tips and new leaves from three separate plants. The significant accumulation of St BEL5 RNA in stolon tips shows that signal in the stolon is dependent on photoperiod. One-step RT-PCR was performed using 20 ng of total RNA, a nonplant sequence tag fused to all transgenic RNAs (designated NT-2), and a gene-specific primer from either the open reading frame of St BEL5 (for St BEL-cds) or from the 3′ UTR (for St BEL5-FL). Use of the NT-2 primer makes it possible to discriminate transgenic RNA from native BEL5 RNAs. No PCR product was detected from native St BEL5 RNA extracted from leaves of wild-type plants ([A], WT lanes). The PCR reactions were normalized using rRNA primers. All PCR reactions were standardized and optimized to yield product in the linear range (31 cycles for both BEL5 RNAs and 17 cycles for the rRNA). Expected size of the full-length product was 375 and 550 nucleotides for the cds product. Both constructs were driven by the CaMV 35S promoter in the binary vector pCB201. The 5′ UTR of St BEL5 is 146 nucleotides, and the 3′ UTR is 505 nucleotides. Homogenous PCR products were quantified (B) using ImageJ software (Abramoff et al., 2004) and normalized using the rRNA values. The results of (A) are a representative sample of one of the biological replicates for each treatment. Standard errors of the means of the three biological replicates are shown in (B). Open bars, leaf; closed bars, stolon.
Movement of St BEL5 RNA in Transgenic Lines Using a Leaf-Specific Promoter
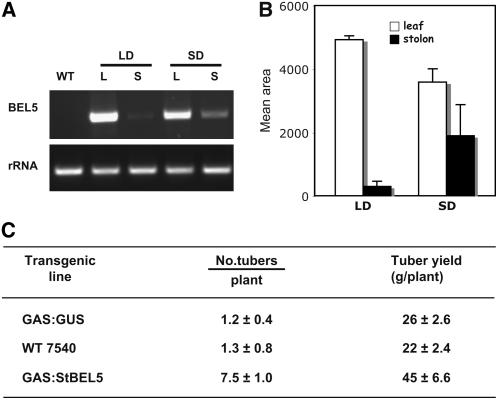
To confirm that the preferential accumulation of full-length St BEL5 RNA in Figure 7 was due to photoperiod-mediated movement, the leaf-specific galactinol synthase (GAS) promoter from Cucumis melo was used to drive St BEL5 expression in the companion cells of leaf minor veins (Haritatos et al., 2000; Ayre et al., 2003). The GAS:St BEL5 (full-length) construct was cloned into pBI101.2 and transformed into S. tuberosum ssp andigena, line 7540. Plants were grown under either SD or LD conditions, leaves and stolon tips were harvested after 10 d, and plants were scored for earliness after 10 SDs (number of tubers formed) and yield of tubers (g plant−1) after 28 SDs. One-step quantitative RT-PCR was performed on three replicates using a nonplant sequence tag fused to all transgenic RNAs (designated NT-2) and a gene-specific primer from the 3′ UTR (GSP1) of St BEL5 (Figure 8A). PCR products were scanned, quantified, and normalized with 18S rRNA product. In the samples from plants grown under SD, the ratio of quantified St BEL5 RNA that moved to stolons in relation to the source leaf was sevenfold greater in SD plants compared with LD plants (Figure 8B), indicating enhanced mobility of the St BEL5 RNA under SD conditions. Enhanced levels of mobile RNA were observed in samples of larger sized tubers harvested from GAS:St BEL5 plants. No PCR product was detected from native St BEL5 RNA extracted from leaves of wild-type plants (Figure 8A, WT). All PCR reactions were optimized to yield product in the linear range (39 cycles for the BEL5 RNA and 17 cycles for the 18S rRNA). This increased RNA mobility was correlated with earliness (more tubers after 10 d of SD conditions) and tuber yields in these SD-grown GAS:St BEL5 lines when compared with controls (Figure 8C). These results demonstrate the photoperiod-mediated movement of St BEL5 RNA and are consistent with the results of the preferential accumulation presented in Figure 7 using a CaMV 35S promoter and the concentration gradient of wild-type St BEL5 RNA shown in Figure 1.
Figure 8.
The Effect of Photoperiod on RNA Accumulation in Stolon Tips of Transgenic Lines Expressing Full-Length St BEL5 Transcripts Driven by the Leaf-Specific Cm GAS Promoter (Ayre et al., 2003).
Transgenic plants were grown under greenhouse conditions until the 12- to 14-leaf stage and then grown under SDs or LDs for 10 d before harvest. The RNA was extracted from 0.5-cm stolon tips (S) and new leaves (L) from three separate plants for each construct. Harvested plants were scored for tuber numbers after 10 d and tuber yields after 28 d under SD conditions (C). One-step RT-PCR (A) was performed using 500 ng of total RNA, a nonplant sequence tag fused to all transgenic RNAs (designated NT-2), and a gene-specific primer from the 3′ UTR of St BEL5 (GSP1). Use of the NT-2 primer makes it possible to discriminate transgenic RNA from native BEL5 RNAs. No PCR product was detected from native St BEL5 RNA extracted from leaves of wild-type plants ([A], WT lane). All PCR reactions were standardized and optimized to yield product in the linear range (39 cycles for the BEL5 RNA and 17 cycles for the rRNA). Expected size of the BEL5 product was 550 nucleotides. The full-length BEL5 construct was driven by the Cm GAS promoter in the binary vector pBI101.2. Homogenous PCR products were quantified (B) using ImageJ software (Abramoff et al., 2004) and normalized using the rRNA values. The results of (A) are a representative sample of one of the biological replicates for each treatment. Standard errors of the means of the three biological replicates are shown ([B] and [C]). Open bars, leaf; closed bars, stolon. GUS expression was detected in leaves but not stolons of GAS:GUS transgenic lines of S. tuberosum ssp andigena (data not shown).
Promoter Activity of St BEL5
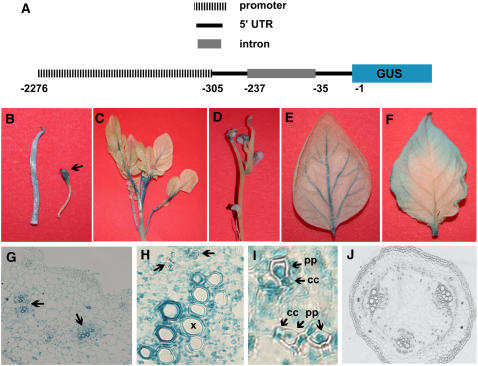
To determine the source of St BEL5 RNA synthesis, the transcriptional activity of St BEL5 was assessed. Upstream regulatory sequence of the St BEL5 gene was isolated and sequenced using Universal GenomeWalker technology (BD Biosciences). A 202-nucleotide intron was identified within 103 bp of the 5′ UTR. This 5′ UTR sequence plus the intron was included along with the 1.97-kb upstream sequence to drive β-glucuronidase (GUS) expression in the binary vector pBI101 (Figure 9A). This construct was transformed into S. tuberosum ssp andigena line 7540 via Agrobacterium tumefaciens–mediated transformation (Banerjee et al., 2006). Approximately 20 lines were screened, and four were selected for further characterization. Promoter activity was assayed using histochemical staining for GUS expression.
Figure 9.
GUS Activity of the Upstream Region of St BEL5.
The promoter sequence used in this construct to drive GUS expression contained a 202-nucleotide intron located within 103 nucleotides of the 5′ UTR and 1971 nucleotides of genomic sequence upstream from the 5′ UTR (A). GUS activity was detected in stolons and newly formed tubers ([B], arrow) and in leaf veins and petioles ([C] to [E]). The plant in (D) was cultured in vitro under LD conditions. All other material was from soil-grown plants. The leaf in (F) is 30 d older than the leaf in (E). Transverse section of a petiole exhibiting staining throughout the tissue, with the greatest level of activity around vascular cells is shown in (G) (arrows). Under higher magnification, blue staining was detected in companion cells (cc), in parenchyma-associated phloem cells (pp) ([H] and [I]), and in xylem cell walls (x) (H). No staining was observed in transverse sections of internodal regions of the stem (J). Bars = 100 μm in (G) and (J), 20 μm in (H), and 5.0 μm in (I).
GUS activity was observed consistently in stolons from both tuberizing and nontuberizing plants (Figure 9B). New tubers exhibited intense staining (Figure 9B, arrow). GUS accumulation driven by the St BEL5 promoter was highest in the veins of leaves, petioles (Figures 9C to 9E), and roots (see Supplemental Figure 2 online). No staining was observed along the internodes of stems (Figures 9D and 9J). Photoperiod had very little effect on activity, but we did observe slightly more intense staining in leaves from LD plants compared with SD plants. As leaves matured, levels of activity were reduced (Figure 9F). Transverse sections of petioles and stems provided more detailed information on the localization of promoter activity. In petioles, staining was observed throughout the tissue, with the greatest concentration in the vascular bundles (Figure 9G, arrows). A higher magnification revealed the presence of blue staining in companion cells and parenchyma-associated phloem cells (Figures 9H and 9I; Cutter, 1978) and in the walls of xylem cells (Figure 9H). No activity was observed in transverse sections of stem internodes under any conditions (Figure 9J).
DISCUSSION
Are the Patterns of RNA Accumulation and Transcriptional Activity of St BEL5 Consistent with Its Putative Role as a Mobile RNA?
The results of this report suggest that the RNA of St BEL5 functions as a long-distance signal that regulates tuber formation. Let us consider in more detail the source of RNA and its overall pattern of accumulation to determine if these results are consistent with such a role. The abundant accumulation of St BEL5 RNA in the stem and its location in phloem cells is consistent with a role as a mobile RNA. In fact, all of the St BEL RNAs except one were detected in phloem cells, presenting the intriguing possibility that St BEL mRNAs serve as mobile signals in a variety of developmental and signaling processes. BEL1-like genes are known to function in floral development (Bhatt et al., 2004; Smith et al., 2004; Kanrar et al., 2006). Could BEL RNAs be moving into floral meristems to regulate inflorescence architecture or to affect the competence to respond? Preliminary results in potato do not rule out this possibility. St BEL5 promoter activity in the flower was restricted to the stigma tip and anthers, whereas mRNA was detected in sepals and petals. In preliminary grafting experiments, St BEL5 RNA from transgenic stock was detected in wild-type scions. POTH1 RNA was also detected in stems and in phloem cells. Is its RNA mobile? A previous report verified movement of a tomato Knox-like fusion RNA across a graft union and into the shoot apical meristem (Kim et al., 2001). Using in vitro heterografts, POTH1 mRNA movement was confirmed as well, but its physiological role is not clear.
That St BEL5 RNA was detected in the microdissected xylem and epidermal cells of the stem was unexpected. It is plausible that BEL5 RNA in the phloem could move cell-to-cell to adjacent regions like the xylem and epidermis. Both xylem and phloem cells arise from the vascular cambium (Figure 2B). The external phloem of potato is situated between these areas (Figure 3A), and it has been established that xylem and phloem cells are interconnected. Unlike mature xylem cells, newly formed immature xylem cells and xylem-associated parenchyma are very likely to exhibit the capacity for intercellular communication critical for differentiation (Sieburth and Deyholos, 2006). Vascular strands arising from leaf traces and juvenile stem bundles of Zinnia elegans contain xylem parenchyma cells with wall in-growths, characteristic of transfer cells (Dahiya et al., 2005). Transfer cells form at sites of intensive trafficking and provide efficient and rapid transport of solutes. Transfer cell arrangement of the nodal complex appears to facilitate the exchange of solutes between phloem and xylem so that solutes can reach developing sinks (Gunning et al., 1970).
Based on expression profiles, numerous regulatory genes, including Leu-rich repeat transmembrane protein kinases, protein phosphatases, ubiquitin E3 ligases, and transcription factors belonging to the MYB, MADS, bZIP, WRKY, Dof, NAC, and bHLH families, are represented in both xylem and phloem cells and likely perform roles in vascular tissue development (Zhao et al., 2005). These genes may be involved in vascular tissue-specific functions or in secondary metabolic pathways that contribute to the structural and functional features unique to vascular tissues. St BEL5, in combination with Knox partners, could be involved in a similar function. Three Knox genes of poplar (Populus tremula), Ptt KNOX1, Ptt KNOX2, and Ptt KNOX6, showed high expression in procambial cells, suggesting a role for Knox genes in cambial meristems (Schrader et al., 2004).
Through their duplex interactions, the numerous BEL1-like (seven have been identified) and Knox (six identified) proteins present in potato provide for a diverse array of functions. For example, Brevipedicellus, a knotted1-like transcription factor of Arabidopsis, regulates several genes involved in lignin biosynthesis, an important biochemical pathway of secondary cell wall growth (Mele et al., 2003). As the BEL1/Knox interaction is ubiquitous in the plant kingdom, these results are consistent with the activity of the St BEL5 promoter in xylem cell walls of the petiole (Figure 9H) and RNA accumulation in xylem stem cells (Figure 3). The salient point is that if St BEL5 RNA is moving through the plant from the leaf to the stolon tip, it must at least be present in the phloem. This point was confirmed by the results of both Figures 2 and 3.
What does the pattern of transcription tell us about the role of St BEL5? Light appears to activate restricted transcription in the veins of leaves and the main vascular conduit from leaves to stems, the petiole. Transcription was detected in at least some phloem-associated cells (Figures 9H and 9I). Again, the location of transcription in leaf veins and in phloem cells is consistent with a mechanism of regulated mobility.
Signaling molecules, like RNAs, are ferried through numerous gateways scattered throughout the body of the plant (Ding et al., 2003). Plasmodesmata play a key role in controlling, both temporally and spatially, the transport of signaling molecules from cell-to-cell in different regions of the plant. Within phloem tissue, the sieve element/companion cell complex plays a central role in trafficking a wide range of RNAs and proteins throughout the plant (Oparka and Turgeon, 1999). Screening for these mobile molecules occurs at checkpoints or surveillance fields located at meristems and junctions between different organs, such as a petiole and a stem or a stem and a flower (Lucas et al., 2001; Ding et al., 2003). A surveillance mechanism controls the selective trafficking of RNAs into the shoot apical meristem, for example, as shown by the fact that viruses are prevented from invading cells of the apical meristem (Foster et al., 2002). Trafficking can even be controlled within an organ. Potato spindle tuber viroid RNA can move into sepals but not the other floral organs (Zhu et al., 2002). The build-up of St BEL5 transcript through synthesis in leaf veins and petioles establishes a pool of RNA that may be selectively delivered into the phloem of the stem for long-distance transport.
Transcription of St BEL5 in the underground organs, stolons and roots, appears to follow a different mechanism for activation. Clearly, light is not involved. It is conceivable that some unidentified developmental factor, possibly even the St BEL5/Knox complex itself, regulates St BEL5 transcription in stolons and roots. Activity of the complex in target cells could be further regulated by the required presence of both partners and the appropriate subcellular localization (Bhatt et al., 2004; Chen et al., 2004).
The Function of St BEL5 RNA Movement
Whereas there are numerous reports available on the long-distance movement of RNAs in plants (Lucas et al., 1995; Ruiz-Medrano et al., 1999; reviewed in Haywood et al., 2002), including those that code for transcription factors (Ruiz-Medrano et al., 1999; Kim et al., 2001; Nakajima et al., 2001; Wu et al., 2003; Haywood et al., 2005), the rationale for movement of some of these RNAs is not clear. In this case, however, it is logical that the movement of St BEL5 RNA could act as a signal that mediates tuber development. Overexpression lines of St BEL5 exhibited enhanced tuber production (Chen et al., 2003), but other than this phenotype, these transgenic lines were normal. Overexpression lines of POTH1 and other Knox genes produced plants with abnormal leaf architecture (Parnis et al., 1997; Nishimura et al., 2000; Rosin et al., 2003a). No such foliar aberrations have ever been observed in St BEL5 overexpression lines. The St BEL5 protein works with one of its Knox partners, POTH1, to target specific genes. Using the ga20ox1 promoter as a model target, no effect on transcription was observed without the involvement of both partners (Chen et al., 2004). RNA levels for POTH1 are constitutive in stolons, whereas RNA accumulation of St BEL5 increases in response to SDs, the environmental signal for tuber formation. In this report, the rationale for the movement of the St BEL5 RNA as a signal for tuber formation, in conjunction with its Knox partner, is compelling.
The Photoperiod Effect of the UTRs of St BEL5 RNA
It is worth noting that transgenic constructs with the addition of the 5′ and 3′ UTRs of the mRNA of St BEL5 exhibited preferential accumulation of transgenic RNA in stolons and that this process was facilitated by SD conditions (Figure 7). This photoperiod effect on RNA accumulation was consistent with the accumulation pattern of St BEL5 transcripts in wild-type plants (Figure 1) and the preferential movement of St BEL5 RNA to stolons under SDs in the GAS:St BEL5 plants (Figure 8). Such preferential accumulation of RNA occurs in wild-type plants despite the fact that the promoter of St BEL5 is most active in leaf veins and petioles (Figure 9). The regulated accumulation of St BEL5-FL RNA in the transgenic lines most likely explains the enhanced capacity for tuber formation in these plants (Figures 5B and 5C).
There are at least two plausible explanations for the effect of the full-length transcript. The first is that the extra UTR sequence may improve the efficiency of translation, making more of the protein available to bind with putative target genes and enhance the tuberization response. There are numerous reports identifying RNA sequences that mediate enhanced protein synthesis (Gualerzi et al., 2003; Arroyo-Helguera et al., 2005; Barreau et al., 2006; Ortega et al., 2006). A second possible explanation that is supported by results from the movement experiments of Figures 7 and 8 is that the UTRs of St BEL5 RNA facilitate preferential movement of the transcript through phloem to the stolon tip under SDs. This targeted movement would increase the concentration of the transcript in stolon tips, leading to increased translation and subsequent enhanced activity by the BEL5 transcription factor in coordination with its partner. We have observed that the mRNA of the St BEL5 partner, POTH1, accumulated constitutively in stolons regardless of the photoperiod or light conditions (Rosin et al., 2003a).
While there are numerous reports on full-length mobile RNAs in plants, there is a scarcity of information on the RNA sequence that mediates this process (Haywood et al., 2005). Such sequences, designated zip code elements, have been identified in the RNAs of animals and function in recognizing RNA binding proteins (reviewed in Jansen, 2001). Whereas these zip codes may be located anywhere in the transcript, they are most predominant in the 3′ UTR (Macdonald and Kerr, 1997; Chartrand et al., 1999; Saunders and Cohen, 1999; Corral-Debrinski et al., 2000; Thio et al., 2000). Viroid RNA motifs have been identified that facilitate selective movement through plant cells (Qi et al., 2004). These short sequences most likely mimic endogenous plant RNA motifs that are recognized by cellular factors for transport. We propose that a zip code–like element exists in one or both of the UTRs of St BEL5 mRNA to mediate its long-distance movement.
Seven BEL RNAs of potato have been identified and exhibit a wide variety in RNA accumulation patterns and in the length of their 3′ UTR, ranging from 60 nucleotides in St BEL30 to 505 nucleotides in St BEL5. St BEL14, the BEL RNA that was not detected in phloem cells (Figure 3), has a 3′ UTR of only 90 nucleotides. Three sequence-related BELs, St BEL5, -11, and -29, are the only BELs with RNA levels that increase in response to SD conditions (Chen et al., 2003). Their 3′ UTRs are the longest of the group, 505, 290, and 385 nucleotides, respectively. Remarkably, these RNAs and St BEL5-like RNAs of tobacco (Nicotiana tabacum) and tomato exhibit conservation of sequence in this UTR. Within the potato group, two motifs, UAGGUUA and ACUUCU, are present and repeated in the 3′ UTRs of all three RNAs. Of course, it is often the secondary or tertiary structure of the RNA that is important for recognition by a protein partner. MFOLD modeling (Zuker, 2003) of the St BEL5 transcript revealed three major areas of structural conservation, one within the coding sequence and within both UTRs (see Supplemental Figure 1 online). Based on the proximity of the 5′ and 3′ UTR structures in 40 MFOLD configurations, 78% placed the stem/loop structures of the UTR sequences in very close proximity (see Supplemental Figure 1 online). This proximity of the 5′ and 3′ UTRs is not evident in the MFOLD configurations of the RNA sequence of St BEL14. Establishing the function of these complex stem/loop structures in any kind of recognition system with a protein chaperone will require further sustained, in-depth experimentation.
Our results indicate that both the coding sequence and full-length transcripts of St BEL5 can move through the plant across a graft union. Assuming that the UTRs play a role in facilitating long-distance movement, how can both transcript types move to stolons? This could be explained if both regions contained sequences involved in RNA movement. For example, there could be a protein binding motif in the coding sequence. Such motifs are present in the coding region of mobile RNAs of animals (Chartrand et al., 1999). In addition, using the CaMV 35S promoter in grafting experiments can be misleading because this promoter is very active in the phloem. Expressing test RNA in phloem (and any other cells not consistent with the wild-type transcriptional source) allows the RNA to bypass putative checkpoints (Ding et al., 2003) that are ordinarily effective in wild-type lines. Whatever motifs are present, the data from Figure 7 suggest that the UTRs play a major role in preferential accumulation of the St BEL5 transcript.
Our current model for this signaling pathway is based on the initial transcriptional activation of the St BEL5 gene in the veins of leaves and petioles under LD conditions, a process probably mediated by light quantity. If St BEL5 is involved in tuber formation, such promoter activation would coincide with the increased production of photoassimilate in leaves that will imminently be available for the newly formed tuber sink. The switch to SDs facilitates movement of the St BEL5 RNA through the petiole junction into the stem by mediating the activation or expression of appropriate RNA binding proteins. This leads to increased RNA levels in stems under SDs (Figure 1). Under these conditions, RNA may then be escorted to site-specific targets, like stolon tips, via protein chaperones. Enhanced translation then occurs on-site, leading to efficient binding to a Knox partner(s) and subsequent subcellular localization in the nucleus and the regulation of transcription for any number of BEL/Knox target genes.
METHODS
Plant Transformation
The full-length sequence of St BEL5 (accession number AF406697), including 5′ and 3′ UTR sequences, was cloned into the binary vector pCB201 (Xiang et al., 1999) and driven by the CaMV35S promoter with a nos terminator. The cassette was then transferred to Agrobacterium tumefaciens strain GV2260. Transformation of the potato species Solanum tuberosum ssp andigena (line 7540) was performed as described by Banerjee et al. (2006). Plants were maintained in growth chambers and in the greenhouse. For vector construction of GAS:BEL5, the Cm GAS promoter of 1.8 kb was PCR amplified using plasmid pSG3K101 (Ayre et al., 2003) as template and primer pair 5′XbaI 5′-GCTCTAGATGACTTGGATTAATTCTCTAAC-3′ and 3′SmaI 5′-AACCCGGGATTGACTTTGGTGCTTT-3′ and cloned into pBI101.2. Subsequently, the reporter GUS gene present in the vector was replaced by full-length St BEL5. St BEL5 was amplified using 5′SmaI 5′-CATCCCGGGTCAGTCTGACAAGAAGGCAA-3′ and 3′SacI 5′-CGAGAGCTCGCTAATCTAATAATGATAGCAC-3′. A 351-bp block located within this 1.8-kb fragment was established as the smallest element necessary and sufficient for minor-vein expression in the leaf (Ayre et al., 2003). Transformation and transgenic plantlet regeneration were performed as previously described (Banerjee et al., 2006). GUS expression was detected in the minor veins of leaves but not stolons of GAS:GUS transgenic lines of S. tuberosum ssp andigena (data not shown).
Micrografting
For in vitro micrografting, tissue culture plants of S. tuberosum ssp andigena (line 7540) and St BEL5 lines (5-11 and 5-19, cds only) served as rootstock and scion, respectively. Stock plants were grown for 3 to 4 weeks as described (Banerjee et al., 2006). In vitro micrografting was performed in Magenta boxes containing 80 mL of semisolid medium of MS salts and vitamins, 2% sucrose, and 0.2% gelrite. Wild-type stock plants were decapitated ∼3.0 cm above the root shoot joint and served as rootstock. A longitudinal slit of ∼1.0-cm deep was made in the center of the cut end of the rootstock, and these grafts were placed in Magenta boxes. Scion (2 to 3 cm in length) was prepared by cutting off the shoot tip of St BEL5 lines (5-11 and 5-19), and a V-shape cut was made at the cut end before they were inserted into slits of the rootstock. The grafted cultures were incubated in growth chambers under a SD photoperiod. Cultures were incubated at a light intensity of 35 μmol m−2 s−1 cool white fluorescent light at 27°C. Because of the high humidity and ample supply of nutrients, in vitro grafts exhibited a very high rate of success. After 10 d of incubation, shoot tip and rootstock tissue samples were harvested as shown in Figure 4A for total RNA extraction and RT-PCR analysis.
Grafting of Soil-Grown Plants
Grafting of soil-grown plants was performed in 15-cm plastic pots containing Sunshine LC-1 Mix (Sun-Gro Horticulture). In this experiment, St BEL5 line 5FL1-3 (full-length sequence), S. tuberosum ssp andigena (line 7540), and POTM1-1 line 102-23 (Rosin et al., 2003b) were multiplied from tubers and maintained in the greenhouse until grafts were made. Scions from 5FL1-3 and 102-23 plants were prepared by making a V-shape cut on the exposed end of the excised shoot apex (∼3 inches in length). Stock plants for all the grafts were prepared by decapitating the shoot apex of andigena line 7540 ∼10 cm above the soil, and ∼1.0- to 1.5-cm deep slits were made. Scions were inserted into the slits of the rootstock, and the pots were immediately enclosed in transparent polyethylene bags. In this way, 20 grafts were made (eight for each 5FL1-3/wild-type and 102-23/wild-type heterograft types and four for wild-type/wild-type autografts). Grafts were maintained and gradually hardened in the greenhouse for 2 weeks until the unions were strong and plants reached the 8- to 10-leaf stage. Thereafter, all the grafts were moved to a growth chamber set at a LD photoperiod and incubated for 10 d. After 10 d of incubation, the photoperiod condition was changed to SDs. Leaves (from the shoot apex) and stolon tips (4 to 5 mm) were harvested from heterografts after 12 d of SD conditions. Total RNA was extracted and subjected to RT-PCR analysis. Tuber yields of the remaining grafted plants were evaluated after 28 d of SD conditions.
RT-PCR Analysis of Tissue Samples Harvested from Both in Vitro and Soil-Grown Grafted Plants
Total RNA was isolated from harvested tissues (scion leaves and stock stolon tips or stem sections) using the RNeasy Plant Mini kit (Qiagen). Each RNA sample was treated with Ambion Turbo DNase to eliminate DNA contamination. One microgram of total RNA was used as the template for reverse transcription. The cDNAs were synthesized using a gene-specific primer (GSP1: 5′-ATCATAGGAGAAAAGAAGTGGAGA-3′) and a transgenic RNA-specific reverse primer, NT-2. Reactions were performed using the Superscript III one-step RT-PCR system (Invitrogen) with Platinum taq DNA polymerase in an MJ Research PTC-200 thermocycler. The RT reaction mixture was subjected to the following conditions: 50°C for 30 min, 94°C for 2 min, 94°C for 15 s, 56°C for 30 s, 68°C for 1 min, and 40 cycles followed by one cycle of incubation at 68°C for 5 min. One microliter of RT product was used as template for the second PCR with a nested gene-specific primer (GSP2: 5′-TTTCTTTTGGGTTGGCTTGGAGT-3′) and the reverse vector-specific primer (NT-2: 5′-GCGGGACTCTAATCATAAAAAC-3′). NT-2 is sequence from the nos terminator that is on the binary vector and is transcribed as a portion of the transgenic RNA of St BEL5. The PCR was performed at the following conditions: 94°C for 3 min, 94°C for 30 s, 56°C for 30 s, 68°C for 1 min, 35 cycles, and followed by one cycle at 68°C for 10 min. Primers for RT-PCR analyses were designed from the 3′ UTR of St BEL5 and were synthesized at the DNA Synthesis and Sequencing Facility (Iowa State University). Primers for analysis of 102-23 POTM1-1 lines were designed from the antisense sequence of POTM1-1 (U23757). Sequences of the forward primers were 5′-AATGGATTCATGCATCAATTG-3′ and 5′-TCCAAATCTTCTCCCACATA-3′, respectively. Again, NT-2 was used as the reverse primer.
Primers used in the RT-PCR for in vitro grafted samples were GSP1, GSP2, and NT-2. Conditions of the RT-PCR were the same as previously described except with 40 cycles followed by one cycle of incubation at 72°C for 10 min. Nested PCR was performed under the same conditions with 35 cycles. The PCR products were subjected to electrophoresis in a 1.2% agarose gel and photodocumented under UV light.
Evaluation of Tuber Formation
Evaluation of in vitro tuber formation was performed on 10 plants per independent line cultured on MS medium with 6% sucrose. Cultures were maintained in a growth chamber under either 16 h light/8 h dark (LD) or 8 h light/16 h dark (SD) and incubated at a light intensity of 35 μmol m−2 s−1 cool white fluorescent light at 27°C. Cultures were evaluated daily and scored for the number of tubers that formed.
In Situ Hybridization Analysis
Preparation of tissue samples and in situ hybridizations were performed as described (Cañas et al., 1994). Digoxygenin-UTP–labeled RNA probes, both sense and antisense, were transcribed with RNA polymerases according to instructions (Roche Biochemicals). Unincorporated nucleotides were removed over a Sephadex G-50 column. For immunological detection, the slides were incubated in buffer 1 (1% blocking solution, 100 mM Tris, pH 7.5, and 150 mM NaCl) for 1 h and then equilibrated with buffer 2 (100 mM Tris, pH 7.5, 150 mM NaCl, 0.5% BSA, and 0.3% Triton X-100). Tissue sections were then incubated with antidigoxygenin-alkaline-phosphatase conjugate diluted 1:1000 in buffer 2 in a humidified box for 2 h, then washed three times for 20 min in 100 mM Tris, pH 7.5, and 150 mM NaCl. The tissue sections were equilibrated in buffer 3 (100 mM Tris, pH 9.5, 100 mM NaCl, and 50 mM MgCl2) for 10 min and then incubated in 3.2 μg/mL of 5-bromo-4-chloro-3-indolyl-phosphate; 6.6 μg/mL nitro-blue tetrazolium salt in buffer 3 in a humidified box for 13 h (above-ground tissues) or 7 h (underground tissues). Accumulation of St BEL5 mRNA is visualized as a dark stain under bright-field illumination. Sections were viewed and photodocumented with a Leitz 35-mm camera on a Leitz Orthoplan light microscope.
Laser Capture Microdissection
S. tuberosom ssp andigena line 7540 plants were grown in soil in the greenhouse until plants reached the 10- to 15-leaf stage and then transferred to a growth chamber under SD conditions (8 h light/16 h dark, 21°C) or LD conditions (8 h dark/16 h light, 21°C) for 10 d. Stem samples were harvested and cross sections were trimmed to 5.0 mm in thickness and fixed 24 h at 4°C in 15 mL of freshly prepared 3:1 (v/v) ethanol-acetic acid (Farmer's fixative). This fixative was infiltrated into the sections under vacuum (400 mm of Hg) for 15 min on ice. Fixed tissue was then dehydrated at room temperature in a graded series of ethanol (1 h each at 75%, 85%, 100%, 100%, and 100%), followed by an ethanol/xylene series (1 h each at 75:25%, 50:50%, 25:75% 0:100%, 0:100%, and 0:100%). Flakes of Paraplast-XTra tissue embedding medium (Fisher Scientific) were added to the final step. Once the flakes melted at room temperature, liquefied Paraplast-XTra was added, and sample vials were placed in the 60°C oven. The medium was replaced at 1.5-h intervals until all xylene residue was absent. Samples were positioned in pure Paraplast-XTra, and sections were cut on a rotary microtome (AO Spencer 820 Microtome; American Optical), floated in water on Probe-on microscope slides at 42°C to stretch ribbons, air-dried, and stored. Slides were deparaffinized twice for 10 min each in 100% xylene and air-dried in the hood for 5 min. Sections were deparaffinized just prior to laser microdissection.
For microdissection, the PALM Laser Microbeam instrument was employed. This system consists of a low-heat UV (337 nm nitrogen) laser and an inverted microscope. A pulsed UV nitrogen laser beam is first focused through the objective lens to a <1.0-μm diameter beam spot that ablates the target without heating adjacent material. LPC, a high photonic pressure force, is then used to capture the target cells in the lid of an LPC microfuge tube. Cells were selected using the graphics tools of P.A.L.M. RoboSoftware. After selection, specific cells from the phloem, xylem, and epidermis were isolated separately by the laser microbeam and collected by LPC into the lid of a 0.5-mL reaction tube (Zeiss) filled with 40 μL of ethanol, placed in a holder located closely above the slide.
RNA from the ethanol/acetic acid–fixed cells is extracted and isolated using the PicoPure RNA isolation kit and protocol (Arcturus). Picopure isolated samples were treated with the RNase-Free DNase Set kit (Qiagen). For RT-PCR, RNA was isolated from phloem, xylem, and epidermal cells from potato stem sections. To evaluate the purity of cell types, the samples were analyzed by RT-PCR for the presence of the phloem-specific potato RNA (accession number TC118156) that is similar to the G2-like transcription factor of Arabidopsis thaliana (Zhao et al., 2005) and root-specific potato RNA (accession number CK267169), homolog to the NT gene of Arabidopsis (Nazoa et al., 2003) as a negative control. RNA was reverse-transcribed using the one-step reverse transcriptase kit (Invitrogen) and 0.25 μM gene-specific primers. The PCR conditions were adjusted based on the primers used. Gene-specific primers were as follows: G2 (accession number TC118156); G2-F, 5′-ACAACCGCACAAAGAATTTAATG-3′ and G2-R, 5′-TGTTCTCCACATATGTTCAAAT-3′ (400 bp); for NT (accession number CK267169), NT-F, 5′-TGGTGTTACTGGTAGAGAA-3′, and NT-R, 5′-TCTGTAAAGAAGCGAGGT-3′ (737 bp); for St BEL5, BEL3UTRa-F, 5′-ATACCAGAAAGTCTCG-3′, and BEL3UTRa-R, 5′-AATACTACTAGTTGTATCAAT-3′ (310 bp); and for St BEL14, BEL14-F1418, 5′-ACAACATGGTGGAAGTG-3′, and BEL14-R1610, 5′-CCAGCCAAATCATGAAG-3′ (193 bp).
Quantitative One-Step RT-PCR Analysis
Total RNA was isolated from leaves and stolons of overexpressing transgenic plants of full-length St BEL5 and St BEL5 cds grown under SDs for 14 d. RNA samples were treated with Ambion Turbo DNase to eliminate DNA contamination. Twenty nanograms of total RNA was used as the template for RT for all 18S rRNA and both St BEL5 RNA samples. The RT reaction mixture for one-step reaction for all three RNAs was subjected to the following conditions (50°C for 30 min, 94°C for 2 min, 94°C for 15 s, 56°C for 30 s, 68°C for 30 s, followed by one cycle of incubation at 68°C for 5 min). After determining the linear range for both PCR products, 17 PCR cycles were used for the 18S rRNA and 31 cycles for amplification of the BEL5 product. St BEL5 gene-specific primers (GSP1 and GSP2) and a nonplant vector-specific nos terminator primer sequence (NT-2) were used. Primers for 18S rRNA were provided in the manufacturer's kit (Ambion QuantumRNA Universal 18S, catalog number 1718). The product sizes for St BEL5-FL and St BEL-cds were ∼375 and 550 bp, respectively; whereas, the 18S rRNA product size was ∼315 bp. For the GAS:St BEL5 quantitative RT-PCR analyses, GSP1 and NT-2 were used as primers. PCR reactions were again standardized and optimized to yield product in the linear range (39 cycles for the St BEL5 RNA and 17 cycles for the rRNA). Expected size of the St BEL5 product in the GAS:StBEL5 transgenic lines was 550 bp.
Promoter Isolation and Construction of the pBI101-BEL5pr:GUS Construct
The promoter of St BEL5, including part of the 5′ UTR and the first intron, was isolated using the Universal GenomeWalker kit (BD Biosciences Clontech). DNA was isolated from young leaves of greenhouse-grown plants of S. tuberosum ssp andigena (line 7540) by the CTAB method. The manufacturer's protocol (Clontech) was followed to construct the libraries. The primary PCR amplification of St BEL5 promoter fragments from the different GenomeWalker libraries was performed using an St BEL5 gene-specific primer (GSP1: 5′-TCCCATGATTATGACGTTGTTGATG-3′) and adaptor primer 1 (AP1). The primary PCR reaction was performed with the following conditions: seven cycles (94°C for 2 s and 72°C for 3 min) followed by 32 cycles (94°C for 2 s and 67°C for 3 min) and final extension of 4 min at 67°C. A nested PCR was then performed using 1.0 μL of a 1:10 dilution of the primary PCR product as a template and using a second St BEL5–specific primer (GSP2: 5′-CCGAGGTTCCTTGATAGTACATATCTG-3′) and adaptor primer AP2 (provided in the kit). The second PCR reaction was performed with the following conditions: five cycles (94°C for 2 s and 72°C for 3 min) followed by 20 cycles (94°C for 2 s and 67°C for 3 min) and final extension of 4 min at 67°C. The reactions were performed in the GeneAmp 2400 PCR system (Perkin-Elmer). A DNA fragment of 2275 bp was amplified from the genomic library and cloned into pCR2.1 vector of the TA cloning kit (Invitrogen). Both strands of the insert were completely sequenced at the DNA Facility of Iowa State University. The sequence was analyzed, and a 202-bp intron was identified, located within the 5′ UTR of the St BEL5 cDNA (Figure 9A).
Analysis of Promoter Activity
The 2275-bp genomic fragment upstream of the translation start site was PCR amplified using primers 5′-CACTAGTTTCGGGTTTCTCTTTTATC-3′ and 5′-GACTAGTGCTGGTCCTACGAACCTTG-3′. The amplified promoter fragments were cloned into pBI101 vector (Jefferson et al., 1987) and mobilized to Agrobacterium strain GV2260 by chemical transformation (An et al., 1988). The construct was then transferred to S. tuberosum ssp andigena using an Agrobacterium-mediated transformation protocol (Banerjee et al., 2006). The kanamycin-resistant and GUS-positive lines were selected and used for promoter analysis. Histochemical staining of GUS was performed using 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrates (Jefferson et al., 1987). Samples were then cleared with 100% ethanol. For histological analysis, samples were stained with GUS assay buffer. After destaining, the samples were dehydrated through an ethanol series and embedded in paraffin. Ten-micron sections were prepared and viewed after deparaffinization under bright-field microscopy with a Zeiss Axioplan 2 microscope and photodocumented using a Nikon COOLPXX 995 digital camera.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF406697 (St BEL5) and AF406700 (St BEL14).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Models of St BEL5 RNA Structures Configured Using MFOLD.
Supplemental Figure 2. GUS Activity of the Upstream Region of St BEL5 in Roots.
Supplementary Material
Acknowledgments
We thank Brian A. Campbell for his assistance with photodocumentation and Fredy Romero for his help with statistical analyses. We thank Robert Turgeon for providing the Cm GAS promoter. This work was supported by National Science Foundation Award 0344850 in the Division of Integrative Organismal Biology. S.-G.S. was supported by funds from the Korea Research Foundation (KRF-2005-013-C00034).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David J. Hannapel (djh@iastate.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics International 11 36–42. [Google Scholar]
- An, G., Ebert, P.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–19.
- Aoki, K., Suzui, N., Fujimaki, S., Dohmae, N., Hayashi, H., Yamaya, T., and Sakakibaraa, H. (2005). Destination-selective long-distance phloem proteins. Plant Cell 17 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo-Helguera, O., Mejia-Viggiano, C., Varela-Echavarria, A., Cajero-Juarez, M., and Aceves, C. (2005). Regulatory role of the 3′ untranslated region (3′UTR) of rat 5′ deiodinase (D1). Effects on messenger RNA translation and stability. Endocrine 27 219–225. [DOI] [PubMed] [Google Scholar]
- Ayre, B.G., Blair, J.E., and Turgeon, R. (2003). Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiol. 133 1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A.K., Prat, S., and Hannapel, D.J. (2006). Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 170 732–738. [Google Scholar]
- Barreau, C., Paillard, L., and Osborne, H.B. (2006). AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 33 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Etchells, J.P., Canales, C., Lagodienko, A., and Dickinson, H. (2004). VAAMANA—A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111. [DOI] [PubMed] [Google Scholar]
- Bürglin, T.R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas, L.A., Busscher, M., Angenent, G.C., Beltran, J.P., and van Tunen, A.J. (1994). Nuclear localization of the petunia MADS box protein FBP1. Plant J. 6 597–604. [Google Scholar]
- Chartrand, P., Meng, X.-H., Singer, R.H., and Long, R.M. (1999). Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9 333–336. [DOI] [PubMed] [Google Scholar]
- Chen, H., Banerjee, A.K., and Hannapel, D.J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38 276–284. [DOI] [PubMed] [Google Scholar]
- Chen, H., Rosin, F.M., Prat, S., and Hannapel, D.J. (2003). Interacting transcription factors from the TALE superclass regulate tuber formation. Plant Physiol. 132 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski, M., Blugeon, C., and Jacq, C. (2000). In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20 7881–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crete, P., Leuenberger, S., Iglesias, V.A., Suarez, V., Schob, H., Holtorf, H., van Eeden, S., and Meins, F. (2001). Graft transmission of induced and spontaneous post-transcriptional silencing of chitinase genes. Plant J. 28 493–501. [DOI] [PubMed] [Google Scholar]
- Cutter, E.G. (1978). Structure and development of the potato plant. In The Potato Crop: The Scientific Basis for Improvement, P.M. Harris, ed (New York: John Wiley & Sons), pp. 70–152.
- Dahiya, P., Milioni, D., Wells, B., Stacey, N., Roberts, K., and McCann, M.C. (2005). A RING domain gene is expressed in different cell types of leaf trace, stem, and juvenile bundles in the stem vascular system of zinnia. Plant Physiol. 138 1383–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Qi, Y. (2003). Symplasmic protein and RNA traffic: Regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6 596–602. [DOI] [PubMed] [Google Scholar]
- Ewing, E.E., and Struik, P.C. (1992). Tuber formation in potato: Induction, initiation and growth. Hortic. Rev. (Am. Soc. Hortic. Sci.) 14 89–198. [Google Scholar]
- Foster, T.M., Lough, T.L., Emerson, S.J., Lee, R.H., Bowman, J.L., Forster, R.L.S., and Lucas, W.J. (2002). A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, C.O., Giuliodori, A.M., and Pon, C.L. (2003). Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331 527–539. [DOI] [PubMed] [Google Scholar]
- Gunning, B.E.S., Pate, J.S., and Green, L.W. (1970). Transfer cells in vascular system of stems: Taxonomy, association with nodes, and structure. Protoplasma 71 147–171. [Google Scholar]
- Haritatos, E., Ayre, B.G., and Turgeon, R. (2000). Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 123 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.), S303–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Huang, T., Bohlenius, H., Eriksson, S., Parcy, F., and Nilsson, O. (2005). The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309 1694–1696. [DOI] [PubMed] [Google Scholar]
- Jackson, S.D., Heyer, A., Dietze, J., and Prat, S. (1996). Phytochrome B mediates the photoperiodic control of tuber formation in potato. Plant J. 9 159–166. [Google Scholar]
- Jackson, S.D., and Prat, S. (1996). Control of tuberisation in potato by GAs and phytochrome B. Physiol. Plant 9 407–412. [Google Scholar]
- Jansen, R.P. (2001). mRNA localization: Message on the move. Nat. Rev. Mol. Cell Biol. 2 247–256. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar, S., Onguka, O., and Smith, H.M. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 1163–1173. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R., Vollbrecht, E., Lowe, B., Veit, B., Yamaguchi, J., and Hake, S. (1994). Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y., Rim, Y., Wang, J., and Jackson, D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289. [DOI] [PubMed] [Google Scholar]
- Lough, T.J., and Lucas, W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2 849–857. [DOI] [PubMed] [Google Scholar]
- Macdonald, P.M., and Kerr, K. (1997). Redundant RNA recognition events in bicoid mRNA localization. RNA 3 1413–1420. [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Virgos-Soler, A., and Prat, S. (2002). Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc. Natl. Acad. Sci. USA 99 15211–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., and Hake, H. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27 13–23. [DOI] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 307–311. [DOI] [PubMed] [Google Scholar]
- Nazoa, P., Vidmar, J.J., Tranbarger, T.J., Mouline, K., Damiani, I., Tillard, P., Zhuo, D., Glass, A.D., and Touraine, B. (2003). Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: Responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52 689–703. [DOI] [PubMed] [Google Scholar]
- Nishimura, A., Tamaoki, M., Sakamoto, T., and Matsuoka, M. (2000). Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol. 41 583–590. [DOI] [PubMed] [Google Scholar]
- Niyaz, Y., Stich, M., Sagmuller, B., Burgemeister, R., Friedemann, G., Sauer, U., Gangnus, R., and Schutze, K. (2005). Noncontact laser microdissection and pressure catapulting: Sample preparation for genomic, transcriptomic, and proteomic analysis. Methods Mol. Med. 114 1–24. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., and Turgeon, R. (1999). Sieve elements and companion cells—Traffic control centers of the phloem. Plant Cell 11 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, J.L., Moguel-Esponda, S., Potenza, C., Conklin, C.F., Quintana, A., and Sengupta-Gopalan, C. (2006). The 3′ untranslated region of a soybean cytosolic glutamine synthetase (GS) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J. 45 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnis, A., Cohen, O., Gutfinger, T., Hareven, D., Zamir, M.D., and Lifschitz, E. (1997). The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Pelissier, T., Itaya, A., Hunt, E., Wassenegger, M., and Ding, B. (2004). Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Sánchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42 151–166. [PubMed] [Google Scholar]
- Rosin, F.M., Hart, J.K., Horner, H.T., Davies, P.J., and Hannapel, D.J. (2003. a). Overexpression of a Knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol. 132 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin, F.M., Hart, J.K., Van Onckelen, H., and Hannapel, D.J. (2003. b). Suppression of a vegetative MADS Box gene of potato activates axillary meristem development. Plant Physiol. 131 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cázares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACPmRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Saunders, C., and Cohen, R.S. (1999). The role of oocyte transcription, the 5′ UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell 3 43–45. [DOI] [PubMed] [Google Scholar]
- Schrader, J., Nilsson, H., Mellerowicz, E., Berglund, A., Nilsson, P., Hertzberg, M., and Sandberg, G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth, L.E., and Deyholos, M.K. (2006). Vascular development: The long and winding road. Curr. Opin. Plant Biol. 9 48–54. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M., Campbell, B.C., and Hake, S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14 812–817. [DOI] [PubMed] [Google Scholar]
- Sonoda, S., and Nishiguchi, M. (2000). Graft transmission of post-transcriptional gene silencing: Target specificity for RNA degradation is transmissible between silenced and non-silenced plants, but not between silenced plants. Plant J. 21 1–8. [DOI] [PubMed] [Google Scholar]
- Thio, G.L., Ray, R.P., Barcelo, G., and Schupbach, T. (2000). Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 15 435–446. [DOI] [PubMed] [Google Scholar]
- Wu, X., Dinneny, J.R., Crawford, K.M., Rhee, Y., Citovsky, V., Zambryski, P.C., and Weigel, D. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130 3735–3745. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40 711–717. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Xu, X., van Lammeren, A.A.M., Vermeer, E., and Vreugdenhil, D. (1998. b). The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol. 117 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Vreugdenhil, D., and van Lammeren, A.A.M. (1998. a). Cell division and cell enlargement during potato tuber formation. J. Exp. Bot. 49 573–582. [Google Scholar]
- Zhao, C., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.