Abstract
The entry of Salmonella typhimurium into nonphagocytic cells requires a panel of bacterial effector proteins that are delivered to the host cell via a type III secretion system. These proteins modulate host–cell signal-transduction pathways and the actin cytoskeleton to induce membrane ruffling and bacterial internalization. One of these bacterial effectors, termed SipA, is an actin-binding protein that is required for efficient Salmonella entry into host cells. We report here that SipA forms a complex with T-plastin on bacterial infection. Formation of such a complex, which requires the presence of F-actin, results in a marked increase in the actin-bundling activity of T-plastin. We also report that T-plastin is recruited to S. typhimurium-induced membrane ruffles by a CDC42-dependent signaling process and is required for bacterial entry. We propose that modulation of the actin-bundling activity of T-plastin by SipA results in the stabilization of the actin filaments at the point of bacterial–host cell contact, which leads to more efficient Salmonella internalization.
Keywords: bacterial pathogenesis, actin cytoskeleton, type III secretion, signal transduction
Microbial pathogens have evolved sophisticated mechanisms to interact with their hosts. This is particularly true for pathogens such as Salmonella spp. that have had a long-standing association with their hosts (1). These bacteria are the cause of a variety of diseases ranging from localized gastroenteritis (e.g., food poisoning) to more life-threatening illnesses such as typhoid fever. Central to the pathogenicity of Salmonella spp. is their ability to gain access to nonphagocytic cells (2). This process requires a panel of bacterial proteins that are delivered to the host cell via a type III protein secretion system (2). These effector proteins trigger host-cell signaling pathways resulting in profuse actin cytoskeleton rearrangements and membrane ruffles at the point of bacterial–host cell contact that ultimately leads to bacterial uptake (3–5).
Recent studies have provided significant insights into the host-cell signaling events leading to bacterial uptake. Contact of Salmonella typhimurium with host cells results in rapid calcium fluxes and the production of arachidonic acid metabolites that are required for Salmonella-induced cytoskeletal reorganization (6). In addition, the GTP-binding proteins CDC42 and Rac-1 play an essential role in the signaling events leading to bacterial internalization (7).
Although several substrate proteins of the invasion-associated type III secretion system have been identified (8–15), it is not known how many of these proteins are directly involved in modulating host cellular functions. A number of secreted proteins have been implicated in playing a role in either protein secretion or protein translocation into the host cell (16, 17). In contrast, other secreted proteins play no role in these processes and are therefore likely candidates to have effector function inside the host cell. One such protein, SopE, has recently been shown to function as an activator of small GTPases of the Rho subfamily such as CDC42 and Rac-1 (18). Another secreted protein, SipA, (10), fits the criteria for a candidate effector protein in S. typhimurium. This protein shares some amino acid sequence similarity to IpaA, a Shigella spp. protein that has been shown to modulate bacterial entry into epithelial cells by binding to vinculin (19). We have recently reported that SipA also enhances bacterial entry. However, the Salmonella protein exerts its effect by binding directly to actin, resulting in a significant decrease in the critical concentration required for actin polymerization as well as a marked inhibition of actin depolymerization (20). We report here that an additional mechanism by which SipA modulates S. typhimurium entry into cultured epithelial cells involves the complexing of SipA with the actin-bundling protein T-plastin. Formation of such a complex results in a significant increase in the actin-bundling activity of T-plastin, thus leading to more efficient bacterial uptake.
MATERIALS AND METHODS
Bacterial Strains and Quantitation of Bacterial Entry.
The wild-type S. typhimurium strain SL1344 and its sipA mutant derivative SB848 have been described (20, 21). Salmonella internalization into HeLa cells was measured as described (7).
Construction of Plasmids and Transfection of Eukaryotic Cells.
A plasmid carrying the full-length cDNA encoding human T plastin was kindly provided by Paul Matsudaira (Massachusetts Institute of Technology, Cambridge, MA). A hemagglutinin (HA) epitope-tagged derivative of human T-plastin was constructed by fusing the entire T-plastin coding sequence to the HA tag by using the tagging vector pJ3H (22). A dominant-negative HA-tagged form of human T-plastin was constructed by cloning the coding sequence of its N-terminal half (residues 1–381) into pJ3H. His-tagged T-plastin was constructed by fusing the entire T-plastin-coding region into the His-tagging vector pQE30 (Qiagen, Chatsworth, CA). Plasmids expressing glutathione S-transferase (GST)-SopE (18), GST-SipA (20) or the dominant-negative mutant form of CDC42Hs (CDC42HsN17) (7) have been described. Transfection of COS-1 cells was carried out as described (23).
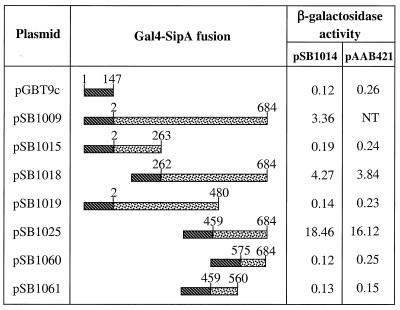
Yeast Two-Hybrid Screen. The Gal4-based yeast two-hybrid system was used following standard procedures (24). The bait plasmid (pSB1009) was constructed by fusing the entire coding sequence for SipA without the start codon to the yeast Gal4 binding domain in pGBT9c (24). A HeLa cell cDNA library, which was constructed by oligo(dT)-priming in pGADGH (CLONTECH), was kindly provided by Xosé Bustelo (State University of New York, Stony Brook). A total of 4 × 107 transformants were screened in the yeast indicator strain Y153 by using the cotransformation protocol as described (CLONTECH). Of 18 putative positive clones, 1 (pSB1014) had β-galactosidase activity that was significantly higher than the background and met all of the standard two-hybrid specificity tests. Sequencing analysis of the insert in pSB1014 revealed that it contains the C-terminal half of human T-plastin (residues 330 to 630) and a 1.6-kilobase untranslated DNA fragment at the 3′ end. The presence of a long untranslated region in the T-plastin message has been reported (25).
A series of SipA deletions was constructed by using either restriction enzyme digestion or PCR. Plasmid pAAB421, which encodes full-length actin fused to the Gal4 activation domain was kindly provided by Alison Adams (University of Arizona, Tucson, AZ). Levels of β-galactosidase activity were measured by using the Miller assay (57).
Purification of Recombinant Proteins from Escherichia coli.
GST, GST-SipA and GST-SopE proteins were purified as described (26) by using Glutathione Sepharose 4B (Amersham Pharmacia). To purify proteins free of the GST domains, fusion proteins were cleaved by adding thrombin (Sigma), which was subsequently removed by running the sample through a benzamidine Sepharose 6B affinity column (Amersham Pharmacia). His-tagged T-plastin protein was expressed in Escherichia coli strain BL21 and purified by using Ni-NTA agarose (Qiagen, Chatsworth, CA). All purified proteins were dialyzed extensively and resuspended in Dulbecco’s PBS with 1 mM DTT.
GST Pull-Down and Coimmunoprecipitation Assays.
GST pull-down and coimmunoprecipitation assays were carried out as described (22). Plastin, actin, α-actinin, and vinculin were detected by Western blotting by using rabbit anti-plastin (kindly provided by Paul Matsudaira, Massachusetts Institute of Technology, Cambridge, MA), rabbit anti α-actinin or mouse monoclonal anti-vinculin or anti-actin antibodies (Sigma). When indicated, latrunculin A (Molecular Probes) was added at a final concentration of 10 μg/ml.
Actin-Bundling Assay.
Actin-bundling activity of plastin was determined by using the low-speed differential centrifugation assay as described (27). All samples and solutions were precleared at 12,000 × g for 15 minutes before being added to the reaction. To investigate the effect of SipA on the bundling activity of T- plastin, we first determined the minimum amount of T-plastin capable of bundling F-actin under our assay conditions. We next determined the minimum amount of SipA necessary to promote actin-bundling activity of T-plastin (0.055 μM), which by itself was not capable of bundling actin. A concentration of SipA (1.5 μM) that gave the maximum bundling activity was chosen for all of the subsequent assays. SipA protein was incubated at room temperature for 15 minutes with varying amounts of T-plastin and precleared at 12,000 × g for 15 minutes before being added to the reaction. The bundling reaction contained 50 mM sodium phosphate buffer (pH 7.0), 100 mM KCl, 0.1 mM ATP, 0.5 mM EGTA, 1 mM MgCl2, and 4.4 μM rabbit skeletal muscle G-actin (Cytoskeleton, Denver) in a total volume of 50 μl. The final mixture was incubated at room temperature for 90 minutes to allow bundle formation. Samples were centrifuged at 12,000 × g for 15 minutes, and supernatants were separated from pellets. The proteins in the supernatants and the pellets were separated on SDS/10% PAGE gels and stained with brilliant blue R-250 (Sigma), and the relative amounts of actin in each fraction (pellet and supernatant) were estimated by scanning densitometry. To examine the effect of SipA on the bundling activity of T-plastin by electron microscopy, F-actin (2 μM) was incubated with different concentrations of His-tagged T-plastin in the presence or absence of SipA (2 μM) in actin polymerization buffer (20 mM Pipes, pH 7.0/75 mM KCl/2 mM MgCl2/0.1 mM EGTA/0.1 mM DTT/0.05 mM ATP). The mixture was incubated for 30 minutes to allow bundle formation at room temperature. Samples were negatively stained with 1% uranyl acetate and mounted onto glow discharge grids, which were nitrocellulose- and carbon-coated.
RESULTS
SipA Interacts with T-Plastin in a Yeast Two-Hybrid Screen of a HeLa Cell cDNA library.
To identify host cellular proteins that interact with SipA, we conducted a yeast two-hybrid screen (28) of a HeLa cell cDNA library by using a fusion of the DNA-binding domain of Gal4 and SipA (Gal4-SipA) as bait. A clone was identified encoding the last 300 aa of T-plastin (fimbrin), which specifically interacts with the Gal4-SipA chimeric protein. T-plastin is a member of the highly conserved plastin/fimbrin family of actin-binding proteins, which also includes the isoforms L- and I-plastin (25, 29–33). All isoforms of plastin have the ability to cross-link F-actin in very tight parallel bundles (27, 30, 34, 35). Although plastin (fimbrin, I-plastin) was first described as a component of the microvilli of intestinal epithelial cells, it is also prominently associated with actin microfilaments in membrane ruffles, filopodia, stereocilia, and cell adhesion sites of a variety of cell types (32, 33, 35–37). Deletion analysis determined that the last 226 aa of SipA were necessary and sufficient to mediate its interaction with T-plastin in the yeast two-hybrid assay (Fig. 1).
Figure 1.
Interaction of SipA with T-Plastin and actin in the yeast two-hybrid system. Plasmids expressing different truncated forms of SipA (stippled box) fused to the Gal4-binding domain (hatched box) as indicated were transformed into a yeast indicator strain expressing a fusion between the Gal4 activation domain and the C-terminal 300 aa of human T-plastin (pSB1014) or the entire yeast actin (pAAB421). The β-galactosidase activity is expressed in Miller units.
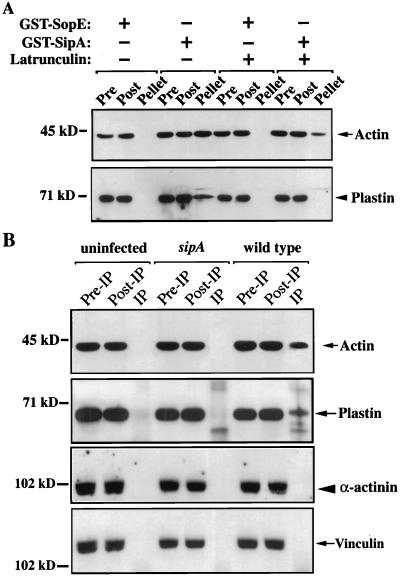
Interaction of SipA with T-plastin in Vivo and in Vitro. We analyzed the interaction between SipA and plastin by several in vitro and in vivo binding assays. HeLa cell lysates were incubated with either purified GST-SipA or GST-SopE (as negative control) immobilized on glutathione-agarose beads. Plastin readily bound to beads coated with GST-SipA but not to beads coated with GST-SopE (Fig. 2A). Consistent with previous results (38), GST-SipA, but not GST-SopE pulled down F-actin (Fig. 2A).
Figure 2.
Interaction of SipA with plastin in vivo and in vitro. (A) Interaction of SipA with plastin examined with a GST pull-down assay. The presence of actin and plastin bound to the GST-SipA beads (Pellet), unbound in HeLa cell extracts (Post), or before the pull-down (Pre) was detected by immunoblot with an anti-actin (Upper) or anti-plastin (Lower) antibody. (B) SipA-plastin complex formation in vivo. HeLa cells were infected with either wild-type S. typhimurium or the sipA mutant strain, and the interaction in cell lysates between SipA and actin, plastin, α-actinin, or vinculin was investigated by coimmunoprecipitation analysis. IP, immunoprecipitate.
We examined whether SipA interacts with plastin during S. typhimurium infection. HeLa cells were infected with wild-type S. typhimurium or an isogenic sipA mutant strain, and the interaction of SipA and plastin was investigated by coimmunoprecipitation analysis of cell lysates. Plastin was detected in SipA immunoprecipitates of cells infected with wild-type S. typhimurium but not in immunoprecipitates of uninfected cells or cells infected with the sipA mutant strain (Fig. 2B). In contrast, other actin-binding proteins such as α-actinin and vinculin were not detected in the immunoprecipitated complex, indicating that only a subset of actin-binding proteins are able to associate with the SipA-actin complex after Salmonella infection (Fig. 2B).
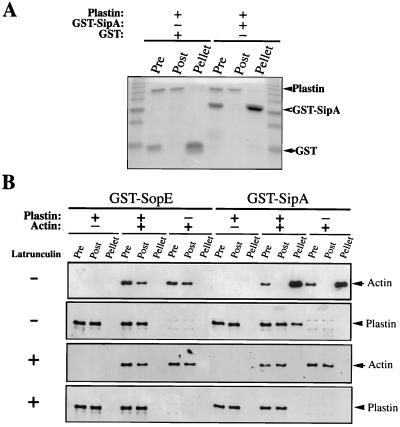
Interaction of SipA with T-plastin Is Mediated by F-Actin. To test whether SipA binds plastin directly, we used a GST pull-down assay by using proteins purified from E. coli. Purified His-tagged T-plastin was incubated with purified GST-SipA or GST alone. As shown in Fig. 3A, GST-SipA did not bind to T-plastin under these assay conditions. We also failed to detect direct binding of SipA to plastin by using a gel overlay assay of HeLa cell lysates (data not shown). Taken together, these results indicate that SipA most likely does not bind to plastin directly. Therefore, complex formation between SipA and T-plastin probably occurs through the interaction of SipA with another plastin-binding protein.
Figure 3.
(A) SipA does not bind to T-plastin directly. Purified T-plastin was incubated with purified GST-SipA or GST alone, and the presence of T-plastin bound to the GST-SipA beads was examined by Coomassie blue staining. (B) SipA and T-plastin complex formation is mediated by F-actin. GST-SipA pull down assay by using extracts of E. coli expressing human T-plastin in the presence or absence of F- actin. The presence of actin and plastin bound to the GST-SipA (or GST-SopE as a control) beads (Pellet) or unbound in the extracts (Post) or before adding the GST beads (Pre) was probed by immunoblot. Experiments were carried out in the presence (+) or in the absence (−) of latrunculin A.
A well characterized property of the plastin/fimbrin family of proteins is the ability to bind and bundle F-actin (33). We have previously shown that SipA is able to bind F-actin (20). We therefore investigated the possibility that SipA may form a complex with plastin in the presence of F-actin. We carried out a GST-SipA pull-down assay in the presence or absence of F-actin by using extracts of E. coli expressing human T-plastin. As previously shown, GST-SipA was able to pull down F-actin (Fig. 3B). In addition, SipA was able to pull down plastin from E. coli extracts in the presence but not in the absence of F-actin (Fig. 3B). In contrast, the control protein GST-SopE was not able to pull down T-plastin either in the presence or in the absence of F-actin (Fig. 3B). To further confirm that the formation of the SipA-plastin complex depends on F-actin, GST-SipA pull-down experiments were carried out in the presence of latrunculin A, a marine toxin that disrupts actin filaments (39). Addition of latrunculin A disrupted the interaction of GST-SipA with actin either in E. coli extracts (Fig. 3B) or in HeLa cell lysates (Fig. 2A). Furthermore, latrunctulin A prevented the interaction of GST-SipA with plastin both in E. coli (Fig. 3B) or HeLa cell extracts (Fig. 2A). These results indicate that the formation of the SipA-plastin complex is mediated by F-actin. This is further supported by the observation that the minimum domain of SipA required for its interaction with plastin in the yeast two-hybrid assay completely overlaps with the minimal domain required for its interaction with actin (Fig. 1).
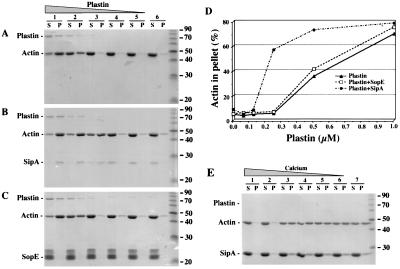
SipA Enhances the Actin-Bundling Activity of T-Plastin. The requirement of SipA for efficient actin cytoskeleton rearrangements and bacterial entry into cultured cells (20) coupled to its ability to interact with T-plastin suggested that SipA may exert its function not only by stabilizing actin filaments but also by modulating the actin-bundling activity of T-plastin. To test whether SipA affects the actin-bundling activity of T-plastin, we used a low-speed cosedimentation actin-bundling assay. At low speeds (12,000 × g), F-actin remains in the supernatant unless present in highly cross-linked bundles (27, 40–42). As shown in Fig. 4A and D, no significant amount of actin was detected in the low-speed centrifugation pellet when the concentration of plastin was <0.3 μM. However, when the concentration of plastin was increased to 0.5 μM, ≈50% of the actin present in the reaction was recovered in the low-speed centrifugation pellet (Fig. 4A and D). Addition of SipA to the reaction significantly increased the bundling-activity of plastin. In the presence of SipA, >50% of F- actin was recovered in the low-speed centrifugation pellet even at a concentration of T-plastin (0.25 μM) that was below the minimum required for detectable actin-bundling activity of T-plastin in the absence of SipA (Fig. 4B and D). Consistent with previous observations (20), no actin was recovered in the low-speed centrifugation pellet, indicating that SipA alone has no bundling-activity in the absence of T-plastin (Fig. 4B and D). Addition of SopE (18), an S. typhimurium-secreted protein that had been purified in an identical manner to SipA, failed to increase the bundling-activity of T-plastin when examined by the same low-speed centrifugation assay. (Fig. 4C and D). Furthermore, the increased bundling-activity of T-plastin by SipA was inhibited by elevated Ca2+ concentration (Fig. 4E). These results are consistent with previous observations that the actin-bundling activity of T-plastin (P. Matsudaira, personal communication) or its related isoform L-plastin (27) are greatly diminished when Ca2+ levels are >0.25 mM.
Figure 4.
Modulation of the actin-bundling activity of T-plastin by SipA. The bundling activity T-plastin at various concentrations alone (A) or in the presence of SipA (1.5 μM) (B) or SopE (1.5 μM) (as negative control) (C) was examined by using a low-speed cosedimentation assay. The concentration of T-plastin ranged from 1 μM (lane 1) to 0.0625 μM (lane 5) in a series of 1:2 dilutions. The presence of actin in supernatants (S) or pellets (P) of the cosedimentation assay was determined by Coomassie blue staining. The quantitation of the experiments in A–C is presented in D and is expressed as % of the total amount of actin in the reaction that is present in the pellet fraction. (E) Calcium dependence of the actin-bundling activity of T-plastin in the presence of SipA. The actin-bundling activity of T-plastin (0.25 μM) in the presence of SipA (3 μM) was examined by using a low-speed cosedimentation assay as described above. The concentration of calcium ranged from 1 mM (lane 1) to 0.03125 mM (lane 6) in a series of 1:2 dilutions. Samples in lanes 6 of A–C and lane 7 in E represent bundling reactions in the absence of T-plastin and calcium respectively. Equivalent results were obtained in three repetitions of this experiment.
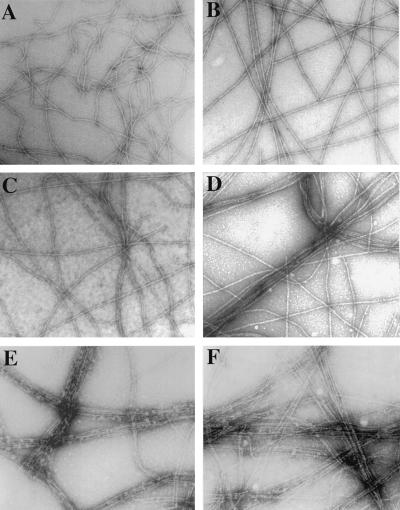
We examined the effect of low concentrations of plastin on the assembly of F-actin bundles in the presence or absence of SipA by electron microscopy. When F-actin was incubated with low concentrations of plastin (0.125 μM) under assembly-inducing conditions, only a meshwork of actin filaments with occasional aggregates were observed, with no filaments arranged in tight bundles (Fig. 5C). In contrast, incubation of F-actin with the same concentration of plastin in the presence of SipA resulted in abundant bundles of parallel actin filaments (Fig. 5D). This arrangement closely resembled the organization of actin filaments when actin was incubated with high (0.5 μM) concentrations of T-plastin alone (Fig. 5E). The addition of SipA to bundling reactions containing high concentrations of plastin did not further increase the bundling activity, although the bundles appear to be straighter (Fig. 5F) presumably because of the ability of SipA to straighten F-actin filaments (22). No bundling was observed on incubation of F-actin with SipA alone (Fig. 5B). Taken together, these results demonstrate that SipA significantly increases the actin-bundling activity of T-plastin.
Figure 5.
Effect of SipA on the actin-bundling activity of T-plastin visualized by electron microscopy. Electron micrographs of negatively stained F-actin (2 μM) incubated with no (A and B), low (0.125 μM; C and D) or high (0.5 μM; E and F) concentrations of T-plastin in the presence (B, D, and F) or absence (A, C, and E) of SipA (2 μM). Highly organized F-actin bundles are visible in the presence of SipA and with low concentrations of T-plastin (C) which, by itself, does not induce actin-bundling. Tighter bundles are found in the actin bundles formed in the presence of SipA and high concentration of T-plastin (D), although the same concentration of SipA alone (B) had no effect on bundling. (Bar = 0.1 μm.)
T-Plastin Is Required for S. typhimurium Entry into Cultured Cells.
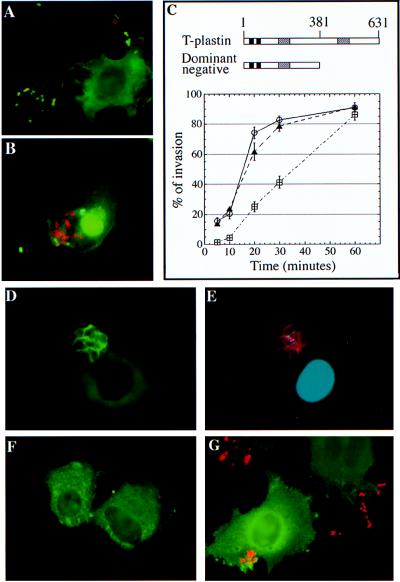
The requirement of SipA for efficient S. typhimurium internalization into nonphagocytic cells coupled to its ability to modulate the actin-bundling activity of T-plastin prompted us to investigate the role of T-plastin in Salmonella entry into cultured cells. COS-1 cells were transiently transfected with plasmids expressing a dominant negative form of T-plastin (43, 44), and the ability of S. typhimurium to enter into those cells over time was examined by using a fluorescence microscopy assay (7). Expression of a dominant-negative form of T-plastin significantly reduced the levels of S. typhimurium internalization (Fig. 6A) when compared with the vector control (Fig. 6B) or to the transient expression of wild-type plastin (Fig. 6C). The reduced level of invasion was more pronounced (≈10-fold) after short infection times but was almost completely overcome after 60 minutes of infection (Fig. 6C). These results indicate that T-plastin is necessary for efficient S. typhimurium entry into host cells. The delayed bacterial invasion observed in the presence of dominant-negative T-plastin is similar to the delayed internalization displayed by the sipA-null mutant (20).
Figure 6.
Role of T-plastin in S. typhimurium entry into cultured cells. COS-1 cells were transfected with plasmids encoding HA-epitope-tagged wild-type human T-plastin (C, ○), its dominant-negative form (A and C, ⊞), or the same vector encoding the green fluorescent protein (B and C, ▴). Transfected cells were infected with wild-type S. typhimurium with a multiplicity of infection of 50 (C). Data presented in C are averages of two independent experiments. In each experiment, a minimum of 300 COS-1 cells was counted in every category at each time point. Recruitment of plastin to the S. typhimurium-induced membrane ruffles. COS-1 cells were transfected with a plasmid encoding an HA-epitope-tagged human T-plastin either alone (D–F) or together with a plasmid expressing CDC42HsN17 (5-fold excess) (G). Transfected cells were infected with wild-type S. typhimurium for 20 minutes (D, E, and G) or left uninfected (F), and cells were fixed and stained with a monoclonal antibody directed to the HA epitope tag to visualize T-plastin (green, D, F, and G), rhodamine phalloidin to visualize F-actin (E), and 4′,6′-diamidino-2-phenylindole (DAPI) (E) or rhodamine-labeled anti S. typhimurium O antigen antibody to stain bacteria (G).
T-Plastin Is Recruited to S. typhimurium-Induced Membrane Ruffles. It has been previously shown that plastin is localized to ruffling membranes (32, 35, 37); therefore, we investigated whether T-plastin was recruited to the S. typhimurium-induced membrane ruffles. COS-1 cells were transfected with plasmids expressing HA epitope-tagged T-plastin and subsequently infected with different strains of S. typhimurium. T-plastin was evenly distributed in uninfected cells (Fig. 6F). However, infection of transfected cells with wild-type S. typhimurium but not a type III secretion-defective invA mutant strain (data not shown) efficiently induced recruitment of T-plastin (Fig. 6D) to membrane ruffles (Fig. 6E). The S. typhimurium sipA mutant strain was also capable of inducing the recruitment of T-plastin to membrane ruffles (data not shown). These results indicate that SipA is not directly responsible for the recruitment of T-plastin to membrane ruffles and also suggest that such a recruitment may be the result of signaling events triggered by other Salmonella effector proteins. Consistent with this hypothesis, T-plastin was recruited to membrane ruffles induced by the transient expression in COS cells of sopE, which encodes a Salmonella effector protein that functions as an exchange factor for Rho GTPases such as CDC42 (data not shown).
To further investigate the role of S. typhimurium-induced host-cell signaling in T-plastin recruitment, we examined the involvement of the small GTP-binding protein CDC42 in this process (7). CDC42 has been shown to be absolutely required for S. typhimurium-induced host-cell signaling and membrane ruffling (7). COS-1 cells were cotransfected with plasmids expressing HA epitope-tagged T-plastin and a dominant-negative mutant of CDC42Hs (CDC42HsN17). These cells were subsequently infected with wild-type S. typhimurium. Expression of CDC42HsN17 completely abrogated the bacterially induced redistribution of T-plastin (Fig. 6G), indicating that S. typhimurium-mediated signaling through CDC42 is responsible for the recruitment of T-plastin to membrane ruffles.
DISCUSSION
It is now apparent that microbial pathogens that have had long-standing associations with their animal or plant hosts have evolved sophisticated mechanisms to engage them in highly fine-tuned interactions (1). The interaction of S. typhimurium with host cells is a remarkable example of these sophisticated adaptations. Contact of S. typhimurium with host cells results in the activation of the type III protein secretion and translocation machinery that directs the delivery of bacterial effector proteins into the host cell (45, 46). These proteins, in turn, stimulate or interfere with a variety of host cellular functions. For example, one of these effector proteins, SopE, activates small GTP-binding proteins from the Rho subfamily by stimulating GTP/GDP nucleotide exchange (18). This activation results in actin cytoskeleton rearrangements, membrane ruffling, and the stimulation of nuclear responses. We have shown here that SipA, another target of the type III protein secretion system (10), is also capable of modulating the host cellular cytoskeleton to promote efficient bacterial internalization. Interestingly, SipA exerts its effect by influencing a completely distinct yet functionally related step in the cascade of events that leads to membrane ruffling and bacterial uptake.
Through a yeast two-hybrid screen of a HeLa cell cDNA library for proteins capable of interacting with SipA, we identified T-plastin, an actin-binding protein that bundles actin into very tight parallel bundles (25, 29–33). Further in vitro and in vivo biochemical assays confirmed that SipA is capable of forming complexes with T-plastin. However, the interaction between these two proteins appears to be mediated by F-actin. The observation that addition of latrunculin A, which disrupts F- actin, prevented the formation of the SipA-plastin complex supports this hypothesis. Most likely, actin is also responsible for bridging the interaction between SipA and T-plastin in the yeast two-hybrid system.
Differential sedimentation and electron microscopy assays demonstrated that the formation of the SipA-actin-plastin complex results in a significant increase in the actin-bundling activity of T-plastin. The activity of actin-bundling proteins, including plastin have been shown to be modulated by a variety of factors such as phosphorylation, cytosolic Ca2+ levels or the function of associated actin-binding proteins (40, 41, 47–53). It is not clear how SipA increases the bundling activity of T-plastin, but it is possible that by binding actin, SipA may induce a conformational change in F-actin that facilitates the actin-bundling activity of T-plastin. This is supported by the observation that in the presence of SipA, the actin filaments appeared straighter, and the helical twist and subunit structure along the filament length is much less obvious than in the absence of SipA (20). However, our data do not completely exclude the possibility that by binding to actin, SipA undergoes a conformational change that renders it able to bind T-plastin, thereby increasing its bundling activity.
At least three isoforms of plastin have been described, I-, L-, and T-plastin. T-plastin is found in all solid tissues, I-plastin is mostly found in the brush border of absorptive intestinal and kidney cells, and L-plastin is found in hematopoeitic cells. Differences in primary sequence, tissue distribution, subcellular localization, and phosphorylation patterns suggest the possibility of distinct roles for individual plastin isoforms in the organization of the cytoskeleton (27, 29, 30, 35, 54, 55). Nevertheless, our results do not address the possibility that SipA may differentially modulate the bundling activities of different plastin isoforms. In addition, our data do not address the possibility that SipA may modulate the activity of other actin-bundling proteins. However, another actin-bundling protein such as α-actinin was not found in immune complexes obtained with an anti-SipA antibody from cells infected with S. typhimurium. These results suggest that the SipA-actin complex may only associate with a discrete subset of actin-binding proteins.
Plastin has also been shown to localize to membrane ruffles and filopodia induced by a variety of stimuli including the activation of the small GTP-binding protein CDC42, suggesting that it serves an important role in the formation of these actin cytoskeleton structures (32, 35, 37). Consistent with this hypothesis, we have shown that T-plastin is required for efficient S. typhimurium-induced cytoskeletal rearrangements and bacterial internalization, processes that require the activity of CDC42 (7). Furthermore, we have shown that T-plastin is recruited to S. typhimurium-induced membrane ruffles in a CDC42-dependent but SipA-independent manner. These results indicate that SipA forms complexes with T-plastin after the cytoskeletal proteins are recruited to the membrane ruffles as a consequence of CDC42-dependent signaling events triggered by other S. typhimurium effector proteins such as the Rho GTPase exchange factor SopE (18). A role for T-plastin in the internalization of the bacterial pathogen Shigella spp. has also been proposed (56). These bacteria encode a protein, IpaA, that has some sequence similarity to SipA. However, the function of the IpaA protein appears to be different from that of SipA because the Shigella protein has been shown to bind vinculin (19). We were not able to detect any interaction between S. typhimurium SipA and vinculin.
Our results presented here further support a two-step mechanism for efficient S. typhimurium entry into host cells. On contact with host cells, S. typhimurium triggers signal-transduction pathways through the delivery of effector proteins such as SopE via its type III protein secretion and translocation apparatus. These signaling pathways, which are dependent on the function of CDC42, lead to the recruitment of a number of cellular proteins. The recruited proteins include actin and T-plastin, which mediate the actin cytoskeleton rearrangements that result in membrane ruffling. These cytoskeletal rearrangements are further modulated by the bacterial effector SipA, which reduces the critical concentration of actin, stabilizes actin filaments, and increases the actin-bundling activity of T-plastin. These SipA activities enhance the actin cytoskeleton reorganization and the formation of the membrane ruffles that eventually internalize the bacteria. Although it is not known how the modulation of the actin-bundling activity of T-plastin by SipA affects the bacterial internalization process, it is possible that the augmented bundling activity serves to increase the stability of the actin filaments that drive and support the growth of the membrane ruffles and filopodia induced by S. typhimurium. This stabilization may cause a more pronounced outward extension of these cellular processes, thereby facilitating bacterial uptake. Indeed, it has been shown that actin-bundling proteins can increase F-actin stability by inhibiting their depolymerization without affecting their assembly (49). Thus, by modulating the actin-bundling activity of T-plastin, SipA may help to increase the net accumulation of actin filaments at the point of bacterial–host cell contact or even to influence the position and polarity of these cross-linked filaments. Stabilizing the actin filaments at the bacterially induced membrane ruffles may also facilitate the persistence of these filaments even when the concentration of G- actin has fallen below the critical concentration for polymerization. Although more studies will be required to substantiate this hypothesis, this model is consistent with the observation that the actin cytoskeleton rearrangements induced by the S. typhimurium sipA mutant are more diffuse than those induced by the wild-type strain (20). The ability of S. typhimurium to influence different events in the complex process of actin-cytoskeleton-dependent membrane ruffling is a further demonstration of the exquisite adaptation of this pathogen to its host.
Acknowledgments
We thank Paul Matsudaira for kindly providing anti-plastin antibodies and a T-plastin cDNA clone, Xose Bustelo for the HeLa cell cDNA library, Alison Adams for the yeast actin clone, and members of the Galán laboratory for critical review of this manuscript. This work was supported by Public Health Service Grants AI30492 and GM52543 from the National Institutes of Health to J.E.G. and DK-25387 to M.S.M.
ABBREVIATIONS
- HA
hemagglutinin
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Galán J E, Bliska J B. Annu Rev Cell Dev Biol. 1996;12:219–253. doi: 10.1146/annurev.cellbio.12.1.221. [DOI] [PubMed] [Google Scholar]
- 2.Galán J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi A. Am J Pathol. 1967;50:109–136. [PMC free article] [PubMed] [Google Scholar]
- 4.Finlay B B, Ruschkowski S. J Cell Sc. 1991;99:283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 5.Ginocchio C, Pace J, Galán J E. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pace J, Hayman M J, Galán J E. Cell. 1993;72:505–514. doi: 10.1016/0092-8674(93)90070-7. [DOI] [PubMed] [Google Scholar]
- 7.Chen L M, Hobbie S, Galán J E. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 8.Collazo C M, Zierler M K, Galán J E. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaniga K, Uralil J, Bliska J B, Galán J E. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaniga K, Trollinger D, Galán J E. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaniga K, Tucker S C, Trollinger D, Galán J E. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pegues D A, Hantman M J, Behlau I, Miller S I. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 13.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 14.Hardt W-D, Galán J E. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardt W-D, Urlaub H, Galán J E. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collazo C, Galán J E. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collazo C, Galán J E. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 18.Hardt W-D, Chen L-M, Schuebel K E, Bustelo X R, Galán J E. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 19.Tran Van Nhieu G, Ben-Ze’ev A, Sansonetti P J. EMBO J. 1997;16:2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou D, Mooseker M, Galán J E. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 21.Hoiseth S K, Stocker B A. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 22.Sells M A, Chernoff J. Gene. 1995;152:187–189. doi: 10.1016/0378-1119(94)00685-l. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel P L, Fields S. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 25.Lin C S, Park T, Chen Z P, Leavitt J. J Biol Chem. 1993;268:2781–2792. [PubMed] [Google Scholar]
- 26.Guan K-L, Dixon J E. Ann Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 27.Namba Y, Ito M, Zu Y, Shigesada K, Maruyama K. J Biochem. 1992;112:503–507. doi: 10.1093/oxfordjournals.jbchem.a123929. [DOI] [PubMed] [Google Scholar]
- 28.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 29.Lin C S, Aebersold R H, Kent S B, Varma M, Leavitt J. Mol Cell Biol. 1988;8:4659–4668. doi: 10.1128/mcb.8.11.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C S, Shen W, Chen Z P, Tu Y H, Matsudaira P. Mol Cell Biol. 1994;14:2457–2467. doi: 10.1128/mcb.14.4.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Arruda M V, Watson S, Lin C S, Leavitt J, Matsudaira P. J Cell Biol. 1990;111:1069–1079. doi: 10.1083/jcb.111.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bretscher A, Weber K. J Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glenney J R, Kaulfus P, Matsudaira P T, Weber K. J Biol Chem. 1981;256:9283–9288. [PubMed] [Google Scholar]
- 34.Bretscher A. Proc Natl Acad Sci USA. 1981;78:6849–6853. doi: 10.1073/pnas.78.11.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arpin M, Friederich E, Algrain M, Vernel F, Louvard D. J Cell Biol. 1994;127:1995–2008. doi: 10.1083/jcb.127.6.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsudaira P T, Burgess D R. J Cell Biol. 1979;83:667–673. doi: 10.1083/jcb.83.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutartre H, Davoust J, Gorvel J-P, Chavrier P. J Cell Sci. 1996;109:367–377. doi: 10.1242/jcs.109.2.367. [DOI] [PubMed] [Google Scholar]
- 38.Zhou D, Hardt W-D, Galán J E. Infect Immun. 1999;67:1974–1981. doi: 10.1128/iai.67.4.1974-1981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spector I, Shochet N R, Kashman Y, Groweiss A. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 40.Matsumura F, Yamashiro-Matsumura S. J Biol Chem. 1986;261:4655–4659. [PubMed] [Google Scholar]
- 41.Sasaki Y, Hayashi K, Shirao T, Ishikawa R, Kohama K. J Neurochem. 1996;66:980–988. doi: 10.1046/j.1471-4159.1996.66030980.x. [DOI] [PubMed] [Google Scholar]
- 42.Yamakita Y, Ono S, Matsumura F, Yamashiro S. J Biol Chem. 1996;271:12632–12638. doi: 10.1074/jbc.271.21.12632. [DOI] [PubMed] [Google Scholar]
- 43.Goldsmith S C, Pokala N, Matsudaira P, Almo S C. Proteins. 1997;28:452–453. doi: 10.1002/(sici)1097-0134(199707)28:3<452::aid-prot13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 44.Goldsmith S C, Pokala M, Shen W Y, Fedorov A A, Matsudaira P, Almo S C. Nat Struct Biol. 1997;4:708–712. doi: 10.1038/nsb0997-708. [DOI] [PubMed] [Google Scholar]
- 45.Ginocchio C, Olmsted S B, Wells C L, Galán J E. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 46.Zierler M K, Galan J E. Infect Immun. 1995;63:4024–4028. doi: 10.1128/iai.63.10.4024-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacaud M, Derancourt J. Biochemistry. 1993;32:3448–3455. doi: 10.1021/bi00064a031. [DOI] [PubMed] [Google Scholar]
- 48.Glenney J R, Bretscher A, Weber K. Proc Natl Acad Sci USA. 1980;77:6458–6462. doi: 10.1073/pnas.77.11.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zigmond S H, Furukawa R, Fechheimer M. J Cell Biol. 1992;119:559–567. doi: 10.1083/jcb.119.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grazi E, Trombetta G, Guidoboni M. Biochem Biophysic Res Comm. 1990;167:1109–1114. doi: 10.1016/0006-291x(90)90637-3. [DOI] [PubMed] [Google Scholar]
- 51.Messier J M, Shaw L M, Chafel M, Matsudaira P, Mercurio A M. Cell Motil Cytoskeleton. 1993;25:223–233. doi: 10.1002/cm.970250303. [DOI] [PubMed] [Google Scholar]
- 52.Maciver S K, Wachsstock D H, Schwarz W H, Pollard T D. J Cell Biol. 1991;115:1621–1628. doi: 10.1083/jcb.115.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mische S M, Mooseker M S, Morrow J S. J Cell Biol. 1987;105:2837–2845. doi: 10.1083/jcb.105.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hisano T, Ono M, Nakayama M, Naito S, Kuwano M, Wada M. FEBS Lett. 1996;397:101–107. doi: 10.1016/s0014-5793(96)01150-7. [DOI] [PubMed] [Google Scholar]
- 55.Park T, Chen Z P, Leavitt J. Cancer Res. 1994;54:1775–1781. [PubMed] [Google Scholar]
- 56.Adam T, Arpin M, Prevost M-C, Gounon P, Sansonetti P J. J Cell Biol. 1995;129:367–381. doi: 10.1083/jcb.129.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]