Abstract
We investigated the ubiquitously expressed hsp70-associating protein Hap46, which is also called RAP46 and is homologous to BAG-1, for activities independent of hsp70 interactions. We observed in vitro binding to various DNA fragments but detected no apparent sequence specificity. Deletion of the amino-terminal decapeptide, which contains two clusters of three basic amino acids each, abolished the DNA-binding ability of Hap46. Similarly, exchange of either of these positively charged clusters for three alanines resulted in loss of DNA binding. Using a fusion of Hap46 and green fluorescent protein, we found preferential accumulation in cell nuclei on heat stress as compared with unstressed cells. The repressive effect of heat shock on overall transcriptional activity in human DU145 carcinoma cells was largely prevented when Hap46 was overexpressed by transfection. Such overproduction of Hap46 also resulted in enhanced expression of specific reporter gene constructs and in increased levels of mRNAs specific for hsp70 and hsp40 after temperature stress. In vitro transcription with nuclear extracts was stimulated greatly by Hap46. Like DNA binding, transcriptional enhancement required amino-terminally located basic amino acid residues but not the carboxyl-terminal portion of Hap46 known to participate in hsp70 interaction. Our results show that Hap46 is a bifunctional protein that can interact with both hsp70s and DNA, employing different portions of the molecule. They also suggest that Hap46 is involved in temperature-sensitive regulation of transcription, acting as a general transcriptional activator.
The ubiquitously expressed mammalian protein Hap46 of an apparent molecular mass of 46 kDa was discovered originally as a receptor-associating protein by screening a human cDNA library with glucocorticoid receptors expressed by the baculovirus system in Sf9 insect cells (1). We observed similar interactions with other members of the large family of nuclear receptors (1). The murine homologue of Hap46, BAG-1, which differs significantly at the amino-terminal end, was detected by the same approach, except that either Bcl-2 or a receptor tyrosine kinase, again produced by use of baculoviruses, was used as bait (2, 3). This apparent convergence is easily explained by the properties of Hap46 and BAG-1, which interact directly and specifically with members of the 70-kDa heat-shock protein (hsp) family (4–6), including hsp70 of insect origin (5). Similarly, the constitutively expressed, cognate form hsc70 is a direct binding partner for Hap46 or BAG-1. To signify this biochemical fact, we introduced the designation “hsp70/hsc70-associating protein” Hap46 (5, 7). BAG-1 binds to hsc70 as a monomer with a 1:1 stoichiometry (8), and ternary complexes of Hap46 and BAG-1 with hsc70 and a variety of unrelated proteins, including various transcription factors, are generated by in vitro incubations (5, 9). Hap46 and BAG-1 were found to interact directly with the amino-terminal ATP-binding domain of hsp70/hsc70 (4–7), whereas the other proteins associate with the carboxyl-terminally located substrate-binding domain of hsp70/hsc70 (7, 9), particularly if they are in a misfolded state. Interaction of Hap46 with the ATP-binding domain of hsp70s most likely causes conformational alterations that then lead to increased ATP binding and enhanced ATPase activity of the molecular chaperone (6, 9, 10).
Various forms of Hap46 and BAG-1 have been detected that differ in molecular size because of different start sites for translational initiation (11). Recently, a family of Hap46-related proteins, all of which interact with hsp70/hsc70 and contain a conserved region of roughly 50 amino acid residues near their carboxyl termini, has been described (12). In fact, the carboxyl-terminal portions of Hap46 and BAG-1 have been shown by deletion studies to be required for binding to hsp70s (Fig. 1) and were found to be sufficient for stimulating the ATPase activity of hsc70 (4, 10, 13). Therefore, we searched for additional interactions and other functions of Hap46. The amino-terminal domain was of particular interest, because it contains clusters of basic amino acid residues followed by glutamic acid-rich repeats (1). Because Hap46 was discovered originally in the context of our studies on steroid hormone receptors (1), which are specific transcription factors, and more recently was found to influence the activities of several nuclear receptors in different systems (14–16), we were especially interested in checking for transcriptional effects. In this study, we demonstrate enhancement of in vitro transcription by Hap46. We also observed stimulation of transcription in heat-stressed cells overexpressing Hap46 pertaining to the synthesis of total mRNA as well as expression of a specific reporter gene. These effects are discussed in view of our finding that Hap46 has DNA-binding ability.
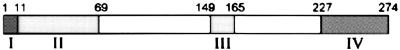
Figure 1.
The domain structure of Hap46 is shown as a bar diagram. (I) Positively charged sequence MKKKTRRRST required for DNA-binding (residues 1–10). (II) Acidic hexapeptide repeats (residues 11–68). (III) Putative nuclear localization sequence (residues 149–164). (IV) hsp70/hsc70-interacting region (residues 227–274).
MATERIALS AND METHODS
Vectors and Protein Expression.
Chloramphenicol acetyltransferase (CAT) encoding vector pcDNA3/CAT was from Invitrogen. Human Hap46 and mutant Hap46ΔC47 (residues 1–227) cDNAs were inserted into HindIII and EcoRI sites of pcDNA3.1/HisA (Invitrogen), from which the polyhistidine coding sequences had been removed by HindIII digest. Mutant Hap46ΔC47 was created by PCR with primers 5′-GGGTAAGCTTGGCGCTCGCAGGCCGCGG-3′ and 5′-CCGAATTCATGGCAGGATCAGTGTGTC-3′. Fusion constructs of Hap46 with the green fluorescent protein (GFP; ref. 17) were obtained by inserting Hap46 cDNA into HindIII and PstI sites of pEGFP-N1 (CLONTECH). Fusion proteins of glutathione S-transferase (GST) with Hap46 and Hap46 mutants were made by inserting the corresponding fragments into BamHI and EcoRI sites of pGEX-2T (Amersham Pharmacia). GST-Hap46ΔC47 was obtained by PCR with primers 5′-ATGGATCCATGAAGAAGAAAACCCGGCGC-3′ and 5′-CCGAATTCATGGCAGGATCAGTGTGTC-3′. GST-Hap46 K(2-4)→A was obtained with primers 5′-CCGCGTGGATCCATGACGACGACGACCCGGCGCCGCTCG-3′ and 5′-TGGAATTCTGCTACACCTCACTCGGC-3′, and GST-Hap46 R(6-8)→A was obtained with primers 5′-GTGGATCCATGAAGAAGAAAACCCCGCCGCCGTCGACCCGGAGCGAG-3′ and 5′-TGGAATTCTGCTACACCTCACTCGGC-3′. All PCR constructs were verified by sequencing. GST-Hap46ΔN10 (residues 11–274) was constructed by inserting a truncated cDNA fragment obtained directly from a cDNA library (1) into the EcoRI site of pGEX-2T. GST fusion proteins expressed in Escherichia coli were purified in PBS/1% Triton X-100 on GSH-Sepharose (Amersham Pharmacia), cleaved with thrombin (18), and checked for purity by SDS/PAGE (Fig. 2). As judged by Coomassie blue or silver staining, proteins were at least 95% pure; however, some specific degradation products showed up, which, in immunoblots, stained specifically for Hap46 (see below). The amino-terminal deletant Hap46ΔN10 consistently migrated faster than the wild-type protein (Fig. 2, lane 2 vs. 1) because of deletion of highly charged residues.
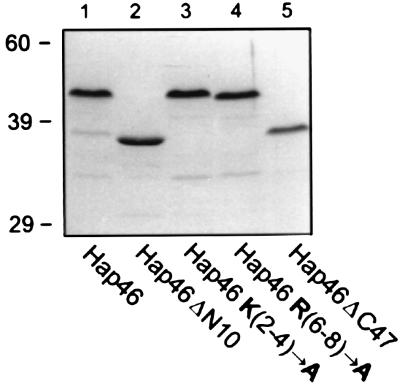
Figure 2.
Bacterially expressed Hap46. GST fusion proteins of wild-type and mutant Hap46 were expressed in E. coli, purified by affinity chromatography, released by thrombin cleavage, and analyzed by SDS/PAGE. Full-length Hap46 (lane 1), Hap46ΔN10 (lane 2), Hap46 K(2-4)→A (lane 3), Hap46 R(6-8)→A (lane 4), and Hap46ΔC47 (lane 5) were used. Staining was performed with Coomassie blue R250.
Cell Culture and Transfections.
DU145 cells from the American Type Culture Collection (no. HTB-81) were grown in RPMI medium 1640 with 10% (vol/vol) FCS. Viability was ascertained by trypan blue exclusion. Transfections were performed with DOSPER reagent (15 μl; Roche Molecular Biochemicals), Hap46 expression or control vectors (4 μg each), and pcDNA3/CAT (1 μg) per 50-mm plate. After 20 h, cells were either exposed to 42°C for 2 h or left at 37°C. After further incubation at 37°C for 3 h, cells were harvested for CAT assays (18) or RNA isolation. For metabolic labeling, cells were treated as above, but immediately before heat-shock, they received phosphate-free medium with 200 μCi [33P]phosphoric acid (4,000 Ci/mmol; ICN) per 50-mm plate.
RNA Analysis.
33P-labeled poly(A)+ RNA was isolated on oligo(dT) affinity matrix (Oligotex, Qiagen, Chatsworth, CA) and either quantitated by counting or analyzed by electrophoresis in 1% agarose-formaldehyde gels. RNA was then transferred to nylon filters (18) and subjected to autoradiography. Northern hybridizations with the same filters were performed with [32P]dCTP-labeled cDNAs for human hsp70 and hsp40 under high-stringency conditions (18).
Protein-Interaction Experiments.
Interaction of GST-Hap46 fusion protein with hsc70, purified to homogeneity from bovine brain (19), occurred in 20 mM Hepes buffer, pH 7.4/150 mM NaCl/10 mM MgCl2/0.1% Nonidet P-40/1 mM DTT as described (5). GSH-Sepharose was washed extensively with PBS/0.3% Tween 20. Bound protein was eluted with SDS sample buffer and analyzed by SDS/PAGE and immunoblotting.
Immunoblotting.
SDS/PAGE and transfer to Immobilon-P membranes were carried out as described (1). Hsc70 was detected by antibody N27F3-4 (StressGen Biotechnologies, Victoria, Canada), and Hap46 was detected by monoclonal antibody KS-6C8 (20). Staining was performed with peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reagent (Amersham Pharmacia).
Electrophoretic Mobility-Shift Assays.
A 125-bp fragment of phage λ-DNA (HindIII digest) and a 283-bp fragment of pcDNA3/CAT (NaeI digest; mostly sequences from the neomycin resistance gene) were 5′ end labeled with [32P]ATP (3,000 Ci/mmol; ICN) and subjected to mobility-shift assays (18) in 20 mM Hepes, pH 7.4/100 mM KCl/10 mM MgCl2/1 mM DTT/4% (vol/vol) glycerol. Routine assays (30 μl) contained 2 μg of protein and 50,000 cpm of DNA (0.1 ng) and were incubated for 30 min at 25°C; nonlabeled bulk DNA was omitted. Recombinant wild-type and mutant Hap46 proteins were used throughout after proteolytic removal of amino-terminal GST tags, as described above; hsc70 (4 μg) was added as indicated.
In Vitro Transcription.
A 1.2-kilobase DNA fragment [Promega, cytomegalovirus (CMV) Positive Control DNA] containing the CMV promoter was used as template in transcriptions in vitro with HeLa nuclear extract (Promega; 70 μg of protein per 25-μl assay); ATP, CTP, and UTP (final concentrations 400 μM each); GTP (16 μM); and 5 μCi of [32P]GTP (3,000 Ci/mmol; ICN). Hap46, Hap46ΔN10, and hsc70 (5 μg each per 25 μl) were added as indicated. Incubation occurred in 20 mM Hepes buffer, pH 7.9/100 mM KCl/3 mM MgCl2/0.2 mM EDTA/0.5 mM DTT/20% (vol/vol) glycerol for 60 min at 30°C. Hap46 and Hap46ΔN10 were generated from GST fusion proteins as described above. Incorporated radioactivity was quantitated by counting, and RNA was analyzed on 6% denaturing polyacrylamide gels.
RESULTS
Hap46 Is a DNA-Binding Protein. Because binding to hsp70s involves the carboxyl-terminal domain of Hap46 (Fig. 1), we wondered whether it might interact with other cellular components, possibly by making use of amino-terminal sequences. In search of directly interacting proteins, we employed two independent approaches: screening a human liver cDNA expression library in λgt11 with carboxyl-terminally truncated Hap46ΔC47 (residues 1–227) and interaction experiments with cell extracts and truncated Hap46ΔC47 fused to GST and immobilized on GSH-Sepharose. In neither experimental setup did we obtain evidence of proteins other than hsp70/hsc70 that were able to interact directly with Hap46 (data not shown).
We then asked whether Hap46 might have nucleic acid-binding ability. To this end, we used DNA of differing origins, most notably a 283-bp fragment derived from a plasmid and a 125-bp fragment of phage λ-DNA origin, both in radiolabeled form. When these were incubated with Hap46 and subjected to electrophoretic mobility-shift assays, we indeed observed binding even under high-salt conditions, as evidenced by an upshift of labeled DNA (Fig. 3 A and B, lanes 2). This mobility shift is indeed caused by Hap46, because the recombinant protein used in these assays was at least 95% pure (Fig. 2, lane 1) and because control material from bacteria solely expressing the GST protein did not show a similar effect (data not shown). In other experiments, we used calf thymus DNA adsorbed to cellulose and incubated with in vitro synthesized, [35S]methionine-labeled Hap46. We observed significant retention of Hap46 on this matrix (data not shown). DNA binding was also confirmed by standard nitrocellulose-filter binding assays with HindIII fragments of λ-DNA. Together, these data show that Hap46 is a DNA-binding protein. When we used different amounts of Hap46 and a fixed amount of labeled DNA in mobility-shift assays, we observed progressive retardation of the shifted DNA with increasing protein levels (Fig. 3C). This result suggests that one DNA molecule can simultaneously interact with several Hap46 copies, which, however, might rapidly dissociate and reassociate with the DNA strand during the course of electrophoresis.
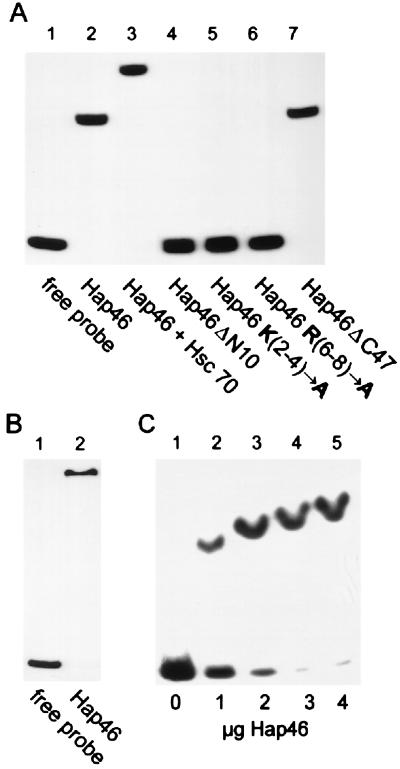
Figure 3.
Hap46 binding to DNA. (A) A radiolabeled 283-bp fragment of pcDNA3/CAT was either employed as such (lane 1) or used for electrophoretic mobility-shift assays with full-length Hap46 (lane 2), Hap46ΔN10 (lane 4), Hap46 K(2-4)→A (lane 5), Hap46 R(6-8)→A (lane 6), and Hap46ΔC47 (lane 7). In the experiment shown in lane 3, Hap46 was used in combination with hsc70. Analysis was by gel electrophoresis and autoradiography. (B) A 125-bp fragment from phage λ-DNA was subjected to the same assay with full-length Hap46 added (lane 2) or not (lane 1). (C) The 283-bp DNA fragment was used for gel-shift assays with either no protein added (lane 1) or with Hap46 at 1 μg (lane 2), 2 μg (lane 3), 3 μg (lane 4), or 4 μg (lane 5) per 30-μl assay.
We also checked deletion mutants of Hap46 for DNA binding. The carboxyl-terminally truncated version Hap46ΔC47, which no longer binds to hsp70s, was found to interact efficiently with DNA (Fig. 3A, lane 7). By contrast, deletion of the amino-terminal decapeptide, resulting in Hap46ΔN10, completely abolished DNA binding (Fig. 3A, lane 4), whereas hsc70 binding was not affected at all (data not shown). To delineate the importance of the basic amino acid residues contained within this decapeptide further, we exchanged the clusters of three lysines (positions 2 to 4; Fig. 1) and three arginines (positions 6 to 8) with three alanines each. We found that neither of these mutant Hap46 proteins was able to interact with DNA (Fig. 3A, lanes 5 and 6), suggesting that both clusters of positively charged residues contribute to DNA-binding ability. These results further prove that the gel shift produced by our full-length Hap46 preparation (Fig. 3A, lane 2) is not caused by some contaminant.
These data clearly show that DNA binding involves the amino-terminal but not the carboxyl-terminal portion of Hap46. Consistent with this notion, ternary complexes can be formed in which Hap46 interacts simultaneously with hsc70 and DNA, as evidenced by a supershift obtained on exposing the 283-bp DNA fragment to Hap46 and hsc70 (Fig. 3A, lane 3). Because interaction of Hap46 with hsp70s and with DNA involves distinct regions of the molecule, we wondered about the relative stability of these interactions. When we bound hsc70 to the above GST-Hap46 fusion protein attached to GSH-Sepharose, we found that roughly half of the hsc70 readily dissociated from the matrix when the temperature was raised from 37° to 42°C (Fig. 4, lane 2). Controls showed that GST-Hap46 was not released from the matrix by this temperature upshift (data not shown). Electrophoretic mobility-shift assays at elevated temperatures, however, showed that complexes of DNA and Hap46 are stable up to at least 42°C (data not shown).
Figure 4.
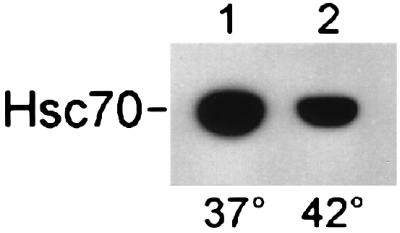
Temperature effect on the interaction of Hap46 with hsc70. The GST-Hap46 fusion protein (5 μg) was incubated with hsc70 (1 μg) and GSH-Sepharose for 1 h at 37°C followed by 1 h at either 37° (lane 1) or 42°C (lane 2). Bound protein was analyzed by immunoblotting with hsc70-specific antibody N27F3-4. Quantitation was performed by densitometric scanning and showed that, in the control experiment (lane 1), hsc70 had been retained quantitatively on the matrix.
Intracellular Localization of Hap46.
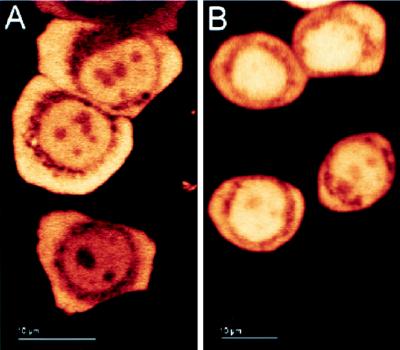
Previously, Hap46 has been found to contain a putative nuclear localization sequence (ref. 1; Fig. 1). Because it is indeed a DNA-binding protein, one would expect it to colocalize, at least partially, with cell nuclei. We therefore checked for compartmentalization by use of Hap46 cDNA fused to the 5′ end of the cDNA encoding GFP (17). Human HeLa and DU145 cells as well as murine 3T3 Swiss cells were transfected with this fusion construct and were analyzed in situ by confocal laser-scanning microscopy. We consistently found the fluorescence signal roughly equally distributed between nucleus and cytoplasm in cells kept at 37°C, as shown in Fig. 5A for HeLa. When we exposed transfectants of these cell lines to a 42°C heat stress for 2–4 h, we observed preferential nuclear accumulation of Hap46-GFP, as shown for HeLa cells in Fig. 5B. Interestingly, both unstressed and heat-treated cells showed a perinuclear ring of endoplasmic reticulum that was almost devoid of Hap46-GFP fluorescence.
Figure 5.
Cellular distribution of Hap46. HeLa cells were transfected with expression vector encoding the Hap46-GFP fusion protein. Cells were kept overnight at 37°C and analyzed either immediately by confocal laser-scanning microscopy (A) or after exposure to 42°C for 2 h (B). Light areas in photographs denote high fluorescence intensity. Different cells show different levels of transfection. (Bars = 10 μm.)
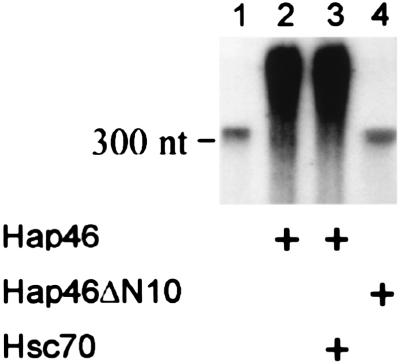
Hap46 Stimulates in Vitro Transcription. We next wanted to find out whether Hap46 is able to affect transcriptional activities. We checked for this by use of a standard in vitro transcription assay with nuclear extracts. As shown in Fig. 6 (lane 2 vs. 1), addition of Hap46 to this system dramatically stimulated the production of RNA (10- to 20-fold). The standard template used here was a DNA fragment of roughly 1,200 bp containing the CMV promoter. On stimulated transcription, RNA molecules were significantly larger in size than the direct run-off transcript of 363 nt observed in the absence of Hap46. The largest product is 1.2 kilobases in size and is thus the result of end-to-end transcription, suggesting that this in vitro transcription occurs independently of the CMV promoter.
Figure 6.
Hap46 stimulates in vitro transcription. Transcription assays were done with [32P]GTP and nuclear extracts from HeLa cells. Labeled RNA was analyzed by gel electrophoresis and autoradiography. Transcription reactions occurred in the absence (lane 1) or presence of full-length Hap46 (lanes 2 and 3) or mutant Hap46ΔN10 (lane 4) with a CMV promoter carrying DNA template. In the experiment shown in lane 3, Hap46 was used in combination with hsc70. The position of a 300-nt RNA is indicated.
We wondered whether hsp70s would affect transcription enhanced by Hap46 and carried out an assay in which we used wild-type HAP46 together with additional hsc70. However, we did not observe any difference (Fig. 6, lane 3 vs. 2; evaluated by counting). We also investigated the above amino-terminal truncation Hap46ΔN10. Clearly, this mutant no longer stimulated in vitro transcription (Fig. 6, lane 4), suggesting that the amino-terminal portion of Hap46 is involved in transcriptional activation.
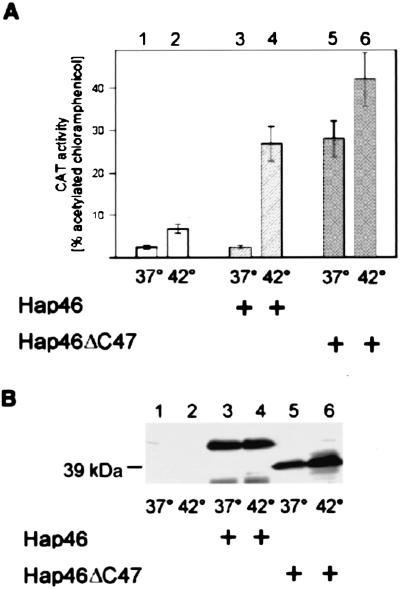
Hap46 Stimulates Transcription in Intact Cells. To check whether Hap46 similarly affects transcription in vivo, we employed several cell lines and achieved at least a 10-fold overexpression of Hap46 by transient transfection with the cDNA. In one line of experiments, we used CAT as an indicator gene and cotransfected the cells with a corresponding construct containing the CMV promoter. Fig. 7 shows experiments with human prostate carcinoma cells DU145. CAT expression in control cells was enhanced roughly 2-fold by 42°C heat-shock conditions as compared with 37°C conditions (Fig. 7A, column 2 vs. 1), as was to be expected from the temperature sensitivity of the CMV promoter (21). Most significantly, overexpression of Hap46 resulted in a large stimulation of reporter gene expression after 42°C heat treatment (Fig. 7A, column 4 vs. 2). Data from three independent experiments showed a roughly 5-fold increase in CAT activity on Hap46 overexpression after 42°C thermal stress relative to heat treatment without overexpression of Hap46. Increased activity was also obtained when we used the carboxyl-terminal truncation Hap46ΔC47 (Fig. 7A, columns 5 and 6). Interestingly, this deletion mutant gave rise to rather high levels of expression at 37°C, i.e., with no heat stress. Similar experiments with HeLa and COS-7 cells produced similar results (data not shown).
Figure 7.
Hap46 stimulates CAT reporter gene expression under heat stress. DU145 cells were transfected with expression vectors encoding CAT alone (lanes 1 and 2), CAT in combination with full-length Hap46 (lanes 3 and 4), or mutant Hap46ΔC47 (lanes 5 and 6). Cells were exposed to heat stress at 42°C for 2 h, as indicated. (A) CAT activity was measured in cell extracts by standard procedures, and turnover is expressed in percentage of acetylated chloramphenicol (18). Data show averages from three independent experiments with error bars indicating maximum deviations. (B) Aliquots of cell extracts, each containing 50 μg of total protein, were analyzed by immunoblotting with Hap46-specific antibody KS-6C8. Endogenous Hap46 contained in untransfected DU145 cells (lanes 1 and 2) did not show up at the short exposure times used here to detect overexpressed wild-type Hap46 and Hap46ΔC47. The position of the 39-kDa marker protein (rabbit muscle aldolase) is indicated.
In control immunoblotting experiments, we checked the levels of Hap46 expression in control DU145 cells and in transfectants (Fig. 7B). Untransfected DU145 cells do contain endogenous Hap46; however, detection of these low levels of endogenous Hap46 requires significantly longer exposure times than those used here. Full-length Hap46 (Fig. 7B, lanes 3 and 4) as well as Hap46ΔC47 (Fig. 7B, lanes 5 and 6) of the proper sizes were detected. Hap46 expression after transfection (lanes 3 and 4) was roughly 10- to 20-fold above the endogenous level, as judged from control immunoblots with increasing amounts of Hap46. Moreover, protein expression was largely independent of heat treatment (lanes 4 and 6 vs. 3 and 5). The amino-terminally truncated protein Hap46ΔN10 turned out to be unstable in DU145 cells (data not shown); consequently, it made no sense to look for effects on CAT expression.
We also used CAT constructs containing glucocorticoid response elements and the herpes simplex I thymidine kinase promoter in conjunction with glucocorticoid receptors expressed from the respective cDNA. With the steroid triamcinolone acetonide present throughout, transfection with Hap46 cDNA yielded no further CAT induction at 37°C beyond that produced by the hormone itself. CAT activity was also not affected in the absence of exogenous Hap46 on exposing cells to 42°C thermal stress. However, on overexpression of Hap46, we observed a roughly 3-fold stimulation of CAT activity after 42°C heat treatment (data not shown). This result shows that Hap46 is just as effective when the reporter is under hormonal control and that the temperature effect of Hap46 is independent of the promoter type.
To search in a more comprehensive way for transcriptional activation in cells, we metabolically labeled DU145 cells with [33P]phosphate after transfection with Hap46 cDNA. Poly(A)+ RNA was then isolated and analyzed by gel electrophoresis and autoradiography (Fig. 8A). Heat shock at 42°C for 2 h caused a drastic decrease in message levels in untransfected cells (Fig. 8, lane 2 vs. 1). Quantitation by counting incorporated radiolabel revealed a 5- to 10-fold reduction in mRNAs on thermal stress. Strikingly, this inhibition was almost completely reversed on overexpression of Hap46 (Fig. 8, lane 4 vs. 2), suggesting that, in these cells, Hap46 exerts a protective effect against heat stress on the level of transcription.
Figure 8.
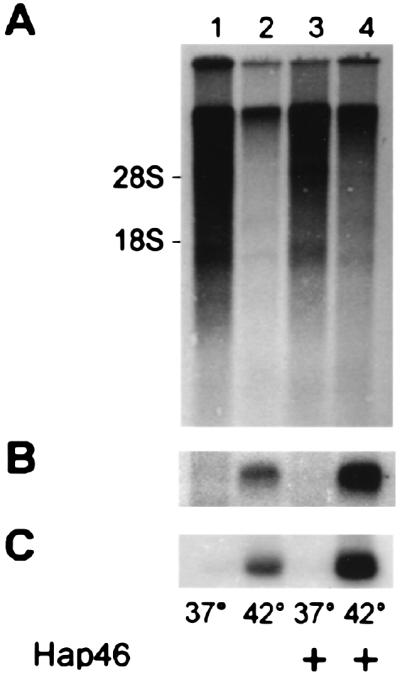
Hap46 affects cellular mRNA levels. (A) DU145 cells were either transfected with Hap46 cDNA (lanes 3 and 4) or not (lanes 1 and 2) and exposed to heat stress at 42°C for 2 h, as indicated. Metabolic labeling with [33P]phosphoric acid was performed immediately before heat treatment. Poly(A)+ RNA was isolated, run on gels, transferred to filters, and detected by autoradiography on a 16-h exposure. (B) The same filters were submitted to Northern hybridization with 32P-labeled hsp70 cDNA. (C) Northern hybridization with 32P-labeled hsp40 cDNA was performed after stripping the filters (18). RNA was detected by autoradiography. Positions of 28S and 18S rRNAs are shown.
When we submitted the above mRNA preparations to Northern analysis, checking for hsp70 and hsp40 specific messages, we found, as to be expected, induction with temperature upshift (Fig. 8 B and C, lane 2 vs. 1). At increased Hap46 levels, induction of hsp70 and hsp40 mRNA was even more prevalent (Fig. 8 B and C, lanes 4 vs. 2). Quantitative evaluation gave a roughly 5-fold enhancement of these mRNAs by Hap46 after 42°C treatment as compared with heat shock without overexpression of Hap46. Taken together, these results identify Hap46 as a factor that is able to stimulate transcriptional activities within cells, particularly under conditions of thermal stress.
These observations suggest that overexpression of Hap46 might make cells less susceptible to the deleterious effects of heat, as was observed with DU145 cells. Full-length Hap46 caused a significantly increased survival rate after heat stress at 42°C (Fig. 9, column 2 vs. 1).
Figure 9.
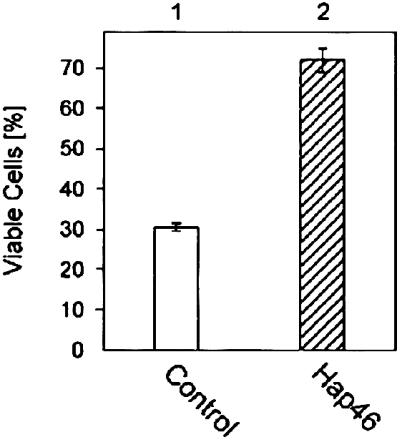
Hap46 protects cells against temperature stress. DU145 cells were transfected with a Hap46 expression vector (column 2) or a control plasmid (column 1). On the day after transfection, cells were exposed to a 2-h heat shock at 42°C and subsequently returned to 37°C. After another 48 h, viable cells were counted. Data show averages from two independent experiments with error bars indicating deviations.
DISCUSSION
The results presented here clearly show that Hap46 is a DNA-binding protein. We employed a variety of DNA fragments of different origins and found Hap46 to interact with all of them, suggesting that Hap46 has general DNA-binding ability with multiple Hap46 molecules binding per DNA; however, we cannot exclude specific interaction with rather short and abundant sequence motifs. DNA binding involves the amino-terminal portion of Hap46, because truncation of a stretch of 10 amino-terminal residues completely abolished the DNA-binding potential (Fig. 3A). This amino-terminal decapeptide contains six positively charged amino acids, which are arranged in two clusters of three basic residues each (Fig. 1). Both of these clusters are involved in the interaction with DNA, because substitution of either one of them by three alanines selectively disrupted DNA binding (Fig. 3A). Thus, the amino-terminal portion of Hap46 is clearly required for DNA binding but, of course, may not be sufficient. Further experiments are needed to delineate the complete DNA-binding domain of Hap46. Despite a stretch of basic residues at the amino terminus, Hap46 is altogether a negatively charged protein containing a rather high proportion of acidic amino acids (1) and having a calculated pI of 4.85. Thus, we are not dealing simply with ionic interactions of proteins like cytochrome c or lysozyme with the polyanionic macromolecule DNA. When we carried out preliminary gel-mobility assays with Hap46 carrying GST at its amino terminus, we observed that this fusion protein interacted much less efficiently than Hap46 itself, suggesting that the amino-terminal end of Hap46 needs to be accessible for DNA binding to occur.
Although Hap46 consists of 274 amino acids (1), the murine homologue BAG-1 contains only 219 residues, (2) which, however, show about 80% identity to residues 61–274 of the Hap46 sequence. With BAG-1 significantly differing from Hap46 in the amino-terminal portion, we would expect BAG-1 not to have the DNA-binding properties described in the present paper for Hap46.
Consistent with DNA-binding ability, we found that Hap46 is localized—in part—in cell nuclei, as detected by use of the fusion with GFP. A similar result has also been obtained by specific immunohistochemical staining, and the extent of nuclear localization may depend on the cell type and state of differentiation (20). Most importantly, we found the Hap46-GFP fusion protein to accumulate preferentially within the nuclear compartment on applying heat stress to cells (Fig. 5B). Increased expression of Hap46 has been observed in several tumor cell types (20, 22). Possibly, this overexpression helps neoplastic cells to survive under the stressful conditions of cell culture or within solid tumors (see below), thus favoring selection of cells with high levels of expression.
Taken together, our results suggest that Hap46 might be able to interact with the genome directly. Thus, we checked for effects on transcription and found that overexpression of Hap46 indeed resulted in stimulated expression of reporter gene constructs and of endogenous hsp70 and hsp40 genes on exposing the cells to heat stress (Figs. 7A and 8 B and C). Moreover, the level of total mRNAs was found to be increased, and the inhibitory effect of heat stress was compensated by overproduction of Hap46 (Fig. 8A). These observations are consistent with our finding that addition of bacterially expressed Hap46 to a standard in vitro transcription system with nuclear extracts caused a dramatic stimulation of RNA production (Fig. 6). Thus, Hap46 acts as a transcriptional activator in this in vitro system employing DNA not complexed with histones. In intact cells, however, transcriptional enhancement by Hap46 involves proper promoters and results in transcripts of normal lengths (Fig. 8). The effect of Hap46 in intact cells may in fact come about by remodeling chromatin structure, possibly in concert with hsp70s (see below).
The experiments presented here lead us to conclude that Hap46 is a bifunctional protein and may exert pleiotropic actions. The amino-terminal portion of Hap46 is involved in the effects described in this report. On the other hand, the carboxyl-terminal domain of Hap46 is required for interaction with hsp70s (Fig. 1) and modulates their chaperoning activity (4, 5, 9, 10). Interaction with a promiscuous variety of partner proteins, particularly if they are at least partially misfolded, has been detected in vitro. Most, if not all, of these interactions are mediated by hsp70s. It is not at all clear at present whether similar complexes also exist in vivo; however, it has been proposed that Hap46 may play a role in initiating the degradation of such misfolded proteins, which are generated by heat stress (5). In principle, the stress protective effect of Hap46 either could involve such degradative mechanisms or could be caused by compensating the effect of thermal stress on the transcriptional level, as suggested by the present experiments.
In the cellular context under normal physiological conditions, Hap46 probably exists in complexes with hsp70s. With Hap46 binding to DNA, it may either function in transcriptional activation independently of hsp70s or carry hsp70 to the transcriptional machinery where the complex of both proteins could be functionally active. The results of our experiments with truncated versions of Hap46 strongly favor the former possibility, because the carboxyl-terminal portion of Hap46 that is needed for interaction with hsp70s is not required for the stimulation of transcriptional activity (Fig. 7A). Thus, temperature-induced dissociation of Hap46–hsp70 complexes, which occurs readily in vitro (Fig. 4), may well occur in vivo under thermal stress. Even if hsp70 is induced on heat shock, as has long been known (23), Hap46 released from the complex may then get to function as a general transcriptional activator and initiator protein.
The bifunctional character of Hap46, interacting with both hsp70s and DNA, is reminiscent of HSF1, NF-κB, and the Wilm’s tumor suppressor protein WT1. These proteins are transcriptionally active and have been shown to associate with hsp70 (24–26). Complexes with the molecular chaperone seem to be involved in their respective cellular regulations.
Consistent with previous data (4), we observed that increased expression of Hap46 can make cells more resistant to thermal stress (Fig. 9), although this resistance may depend on cell type. We suppose that this increase in heat tolerance is accomplished, not only through enhanced expression of molecular chaperones like hsp70, but mostly by compensating the shut-down of general transcriptional activity on heat stress (Fig. 8A). From our experiments with full-length and truncated variants of Hap46 (Figs. 6 and 7A), it becomes clear that the amino-terminal portion of Hap46, which contains a series of positively charged amino acid residues, is essential for the transcriptional effects presented here. Further elucidation of the molecular mechanism involved in the stress-protective effect of Hap46 will be a challenge.
Acknowledgments
We thank Dr. J. C. Reed (Burnham Institute, La Jolla, CA) for monoclonal antibody KS-6C8, Dr. E. Günther (Universität Göttingen, Göttingen, Germany) for a human hsp70 DNA probe, Dr. Y. Minami (Tokyo Metropolitan Institute of Medical Science, Tokyo) for human hsp40 cDNA, Dr. A. Schüssler (Universität Heidelberg) for help with the confocal laser-scanning microscope, Mr. R. Streicher (Leica) for support in electronic evaluation of the microscopic data, and Mr. B. Segnitz for purified bovine hsc70 and for help with photographs. This work was supported by the Deutsche Forschungsgemeinschaft.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- CMV
cytomegalovirus
- GFP
green fluorescent protein
- GST
glutathione S-transferase
Note Added in Proof
In preliminary electrophoretic mobility-shift assays with DNA fragments of 18-22 bp, we observed no significant Hap46 binding. This observation supports the view that multiple molecules of Hap46 bind to DNA to produce stable complexes.
References
- 1.Zeiner M, Gehring U. Proc Natl Acad Sci USA. 1995;92:11465–11469. doi: 10.1073/pnas.92.25.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 3.Bardelli A, Longati P, Albero D, Goruppi S, Schneider C, Ponzetto C, Comoglio P M. EMBO J. 1996;15:6205–6212. [PMC free article] [PubMed] [Google Scholar]
- 4.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeiner M, Gebauer M, Gehring U. EMBO J. 1997;16:5483–5490. doi: 10.1093/emboj/16.18.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Höhfeld J, Jentsch S. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebauer M, Zeiner M, Gehring U. FEBS Lett. 1997;417:109–113. doi: 10.1016/s0014-5793(97)01267-2. [DOI] [PubMed] [Google Scholar]
- 8.Stuart J K, Myszka D G, Joss L, Mitchell R S, McDonald S M, Xie Z, Takayama S, Reed J C, Ely K R. J Biol Chem. 1998;273:22506–22514. doi: 10.1074/jbc.273.35.22506. [DOI] [PubMed] [Google Scholar]
- 9.Bimston D, Song J, Winchester D, Takayama S, Reed J C, Morimoto R I. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebauer M, Zeiner M, Gehring U. Mol Cell Biol. 1998;18:6238–6244. doi: 10.1128/mcb.18.11.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Packham G, Brimmel M, Cleveland J L. Biochem J. 1997;328:807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takayama S, Xie Z, Reed J C. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 13.Lüders J, Demand J, Schönfelder S, Frien M, Zimmermann R, Höhfeld J. Biol Chem. 1998;379:1217–1226. doi: 10.1515/bchm.1998.379.10.1217. [DOI] [PubMed] [Google Scholar]
- 14.Froesch B A, Takayama S, Reed J. J Biol Chem. 1998;273:11660–11666. doi: 10.1074/jbc.273.19.11660. [DOI] [PubMed] [Google Scholar]
- 15.Kullmann M, Schneikert J, Moll J, Heck S, Zeiner M, Gehring U, Cato A C B. J Biol Chem. 1998;273:14620–14625. doi: 10.1074/jbc.273.23.14620. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Takayama S, Zheng Y, Froesch B, Chen G-q, Zhang X, Reed J C, Zhang X-k. J Biol Chem. 1998;273:16985–16992. doi: 10.1074/jbc.273.27.16985. [DOI] [PubMed] [Google Scholar]
- 17.Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji F I. Proc Natl Acad Sci USA. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 19.Chappell T G, Konforti B B, Schmid S L, Rothman J E. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 20.Takayama S, Krajewski S, Krajewska M, Kitada S, Zapata J M, Kochel K, Knee D, Scudiero D, Tudor G, Miller G J, et al. Cancer Res. 1998;58:3116–3131. [PubMed] [Google Scholar]
- 21.Bruening W, Giasson B, Mushynski W, Durham H D. Nucleic Acids Res. 1998;26:486–489. doi: 10.1093/nar/26.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Hao Y, Tang S C, Pater A. Exp Cell Res. 1999;247:200–207. doi: 10.1006/excr.1998.4349. [DOI] [PubMed] [Google Scholar]
- 23.Pelham H R B. Cell. 1986;46:959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- 24.Guzhova I V, Darieva Z A, Melo A R, Margulis B A. Cell Stress Chaperones. 1997;2:132–139. doi: 10.1379/1466-1268(1997)002<0132:msphiw>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Mosser D D, Morimoto R I. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maheswaran S, Englert C, Zheng G, Lee S B, Wong J, Harkin D P, Bean J, Ezzell R, Garvin A J, McCluskey R T, et al. Genes Dev. 1998;12:1108–1120. doi: 10.1101/gad.12.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]