Abstract
The human haptoglobin (HP) HP*2 allele contains a 1.7-kilobase (kb) intragenic duplication that arose after a unique nonhomologous recombination between the prototype HP*1 alleles. During a genetic screening of 13,000 children of survivors exposed to atomic-bomb radiation and 10,000 children of unexposed persons, two children suspected of carrying de novo mutations at the haptoglobin locus were identified (one in each group). DNA analyses of single-cell-derived colonies of Epstein–Barr virus-transformed B cells revealed that the two children were mosaics comprising HP*2/HP*2 and HP*2/HP*1 cells at a ratio of ≈3:1. We infer that the latter cells are caused by reversion of one HP*2 allele to HP*1 through an intramolecular homologous recombination between the duplicated segments of the Hp*2 allele that excised one of the segments. Because the mosaicism is substantial (≈25%), this recombination must have occurred in early embryogenesis. The frequency of finding these children and the extent of their mosaicisms corresponds to an HP*2 to HP*1 reversion rate of 8 × 10−6 per cell during development. This leads to the prediction that the HP*1 allele also will be represented, although usually at a very low frequency, in any HP2-2 person. We tested this prediction by using PCR for a single individual and found the HP*1 allele at frequencies of 4 × 10−6 and 3 × 10−6 in somatic and sperm cells. The HP*1 allele was detected by PCR in all four other HP2–2 individuals, which supports the regular but rare occurrence somatically of homologous recombination within duplicated regions in humans, in agreement with previous observations in mouse and Drosophila.
Keywords: atomic bomb survivors, mosaic, partial gene duplication, reverse mutation
Haptoglobin (HP) is an α2 serum globulin synthesized in the liver (see reviews in refs. 1 and 2) that binds free hemoglobin; the haptoglobin–hemoglobin complex prevents the elimination of hemoglobin in urine. Smithies (3), using starch gel protein electrophoresis, discovered three common phenotypes, HP1-1, HP2-1, and HP2-2, and was the first to describe their genetic cause. The variations are caused by the presence of three common autosomal alleles, HP*1F, HP*1S, and HP*2. The HP*1F and HP*1S alleles encode polypeptides of equal length, HPα1F and HPα1S, respectively, that differ by charged amino acids that make them migrate fast or slowly during starch gel electrophoresis with an acidic buffer. The third allele, HP*2, encodes an HPα2 polypeptide, which is roughly twice the length of HPα1F and HPα1S. Through protein analyses, Smithies et al. (4) suggested that the HP*2 allele contained a partial gene duplication formed by a rare and unique nonhomologous crossing-over between the HP*1F and HP*1S alleles in a heterozygote. This was later confirmed by Maeda et al. (5), who showed through DNA analyses that the length of the intragenic duplication was 1.7 kilobases (kb).
During a study of the genetic effects of atomic-bomb radiation that entailed a search for electrophoretic variation in a series of 30 proteins (6, 7), the plasma haptoglobins of 12,865 Japanese children born to exposed parents and 10,461 children born to control parents were examined by using starch gel electrophoresis. We identified haptoglobin variants in 44 children. Among these, 42 were inherited variants, but two appeared to be de novo mutants. They appeared to be variant HP2-1 heterozygotes with abnormally low amounts of HP1 protein compared with HP2, although their four parents each had the normal haptoglobin phenotype.
The present paper describes the molecular basis of these two rare mutants that proved to be extensive mosaics because of reversion of the partially duplicated HP*2 to the HP*1 prototype as a consequence of intramolecular recombination at some early stage of embryogenesis. Specifically designed PCR experiments revealed that the reversion also can be detected in somatic as well as male germ cells of an unselected HP2-2 individual at frequencies consistent with those observed in the mass screening of children.
MATERIALS AND METHODS
Samples. Among the 23,326 children screened, 12,865 children were born to parents, either one or both of whom were proximally exposed to atomic bomb radiation, and 10,461 were born to parents distally exposed or unexposed. Case I, described below, was found in the control group, and case II was found in the exposed group.
Plasma was prepared from blood drawn into a tube containing acid-citrate-dextrose (6). Genomic DNA was prepared from Epstein–Barr virus (EBV)-transformed B cell lines, white blood cells (WBCs), or sperm cells from healthy volunteers previously typed for HP phenotypes, and from family members of the present HP variants. The genomic DNA from an individual with HP2-1 modified (HP2-1M) phenotype was a gift from N. Maeda of the University of North Carolina (Chapel Hill).
HP Typing.
HP phenotypes were determined by mixing plasma with hemoglobin (20 μl of plasma and 1 μl of a fresh lysate of red blood cells, corresponding to ≈0.3% hemoglobin) followed by electrophoresis. For the first screening, HP was typed by using starch gel electrophoresis as described by Smithies (3) and modified by Weitkamp et al. (8). In suspected unusual cases, more precise typing was carried out by one-dimensional electrophoresis by using 6.2% polyacrylamide gel (6% acrylamide and 0.2% bis-acrylamide) in 40 mM Tris/200 mM glycine buffer (pH 8.5). The gels were incubated at 37°C in 150 ml of 0.2 M sodium acetate buffer (pH 4.5) containing 30% ethanol and 0.1% dimethoxybenzidine. After a 30-min incubation, HP–hemoglobin complexes were visualized by the addition of 10 ml of 5% hydrogen peroxide. Genotypes were determined by hybridization by using a hapto 6 probe (9) (a 1.4-kb PstI fragment of HP cDNA, a gift from S. Oliviero of the University of the Siena, Italy) to Southern blots of HindIII digests of genomic DNA (5).
Purification of Plasma HP.
Plasma HP was purified by properties associated with its capacity to bind hemoglobin. Human hemoglobin was prepared from red blood cell lysate after removal of cell debris by centrifugation. Sixty milligrams of hemoglobin in 0.1 M Mops buffer (pH 7.7) was mixed with 3 ml of Affi-Gel 10 (Bio-Rad) and reacted for 2 h at 4°C. The primary amino groups of hemoglobin were coupled with N-hydroxysuccinimide ester in the Affi-Gel, and stable amide bonds were formed. After the coupling reaction, free succinimide ester residues were blocked by addition of 0.5 ml of 0.1 M ethanolamine. Subsequently, the resin was washed with 30 ml of cold distilled water and suspended in 50 mM Tris⋅HCl (pH 7.8) containing 0.85% NaCl. Twenty microliters of hemoglobin-coupled Affi-Gel resin were transferred to a microcolumn, and 0.1 ml of plasma was passed through the column twice at room temperature. After extensive washing with 1 ml each of 50 mM Tris⋅HCl (pH 7.8) containing 0.5 M NaCl and distilled water, the trapped HP was eluted with 0.1 ml of 5% formic acid. The eluted HP fraction was freeze-dried.
Two-Dimensional Electrophoresis.
O’Farrell’s two-dimensional electrophoresis was carried out as described (10). About 10 μg of purified HP and 5 μg of charge standards (10) were first separated by isoelectric focusing in the presence of 9 M urea, 2% 2-mercaptoethanol, and 2% ampholine mixture (1.6% pH 3.5–10, 0.4% pH 5–7), followed by SDS/PAGE. The gels were stained with Coomassie Brilliant Blue R-250.
HP Gene Cloning.
Genomic DNA was digested completely with EcoRI and ligated with the λ EMBL4 vector. The ligated material was packaged in vitro by using Gigapack III Gold Packaging Extract (Stratagene). The resulting library was screened with a HP cDNA probe. Clones having EcoRI fragments of 11.8 kb (HP*2 allele) and 10.1 kb (HP*1 allele) were purified. The inserts were digested with HindIII and XbaI and subcloned into pUC18 plasmid vectors.
B Cell Cloning.
Single-cell-derived clonal transformation of human B lymphocytes was performed with a modification of the soft-agarose method of Yamamoto and Hinuma (11). Briefly, lymphocytes were prepared from 5 ml of peripheral blood by using Ficoll-Paque (Amersham Pharmacia). About five million lymphocytes were infected with EBV and resuspended in 0.5 ml of RPMI 1640 medium containing 15% FCS after removing the EBV source by two centrifugations. Ten milliliters of melted agarose medium consisting of 0.35% SeaPlaque Agarose (FMC) and 15% FCS in RPMI 1640 medium, and maintained at 37°C, was added to an aliquot of cell suspension and mixed quickly. A 0.8-ml portion of the mixture was added to each well of a 24-well culture plate. Cells were plated at several concentrations (1 × 105 per ml, 5 × 104 per ml, 2.5 × 104 per ml, and 1.25 × 104 per ml). After the agarose solidified, the plates were incubated at 37°C in a fully humidified CO2 incubator. Two to three weeks later, visible growing colonies, well separated from others, were isolated with a Pasteur pipette. The cells from each colony were transferred into 0.3 ml of RPMI 1640 medium containing 20% FCS, grown to optimal numbers, and harvested. Clonality of the colonies was examined by hybridization of human Ig JH fragments (12) as a probe to Southern blots of EcoRI digests of genomic DNA from each colony.
PCR To Detect Reversion from HP*2 to HP*1 Allele.
Genomic DNA either from WBC or from sperm cells of HP2-2 homozygous individuals were digested with restriction enzymes BamHI and HindIII. The digestion produced a 4.2-kb DNA fragment for the HP*2 allele and a 2.5-kb fragment for the HP*1 allele (5). The digest was size-fractionated on an agarose gel, and a gel portion expected to contain the 2.5-kb HP*1 allele was recovered. After melting the gel, the gel solution was divided into a number of tubes and subjected to PCR amplification under conditions optimized for the HP*1 allele. The primers used were CTGGGCTTTCAGGACCATA and GCTCTATTTCCCACTTTTCT. After 30 cycles of PCR amplification, the presence of the HP*1 allele was examined by Southern blot analysis with the hapto 6 probe. The reversion frequency (RF) was estimated by a Poisson formula, i.e., RF = −ln (number of tubes with the absence of HP*1 allele/average number of cells per tube).
RESULTS
Case I (Unbalanced Production of HP1 and HP2).
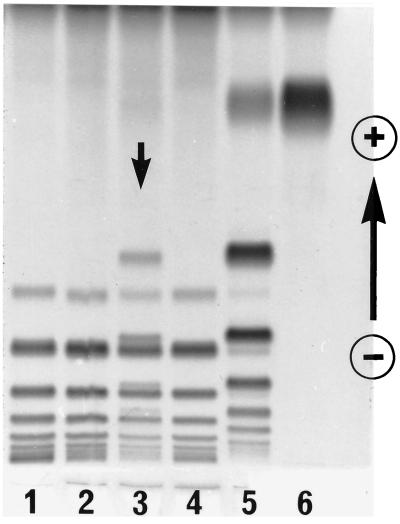
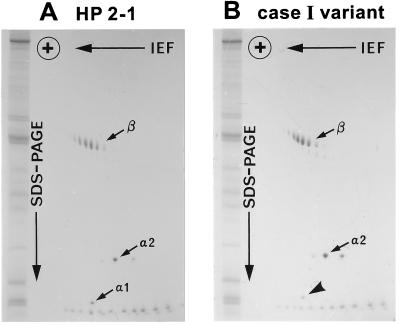
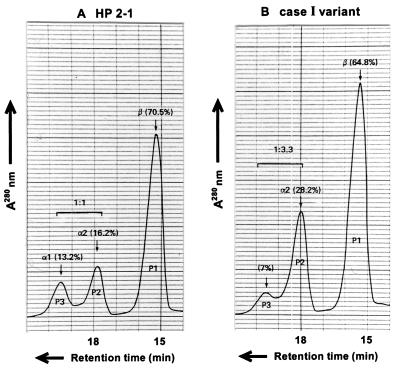
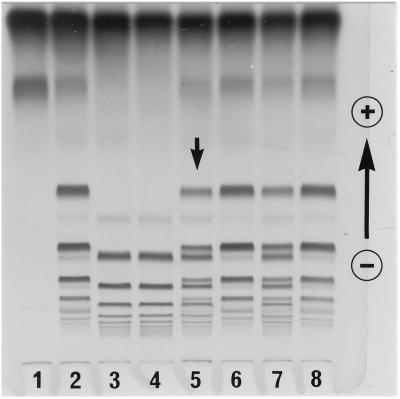
Family study of variant case I showed that the one-dimensional electrophoresis HP protein pattern of the propositus contained a set of extra bands in addition to a normal set of HP2-2 bands (Fig. 1). Both parents showed normal HP2-2 patterns. Subsequently, plasma HP from the propositus was purified by hemoglobin-affinity chromatography and analyzed by two-dimensional electrophoresis (10); the pattern of the variant (Fig. 2) was found to be similar to but not identical to that of HP2-1 heterozygotes. Specifically, in addition to a series of normal HPβ and HPα2 polypeptides spots, a weak spot was observed at the position of normal HPα1S. For further characterization, reduced and carboxymethylated HP polypeptides were separated by gel filtration with HPLC on a TSK gel 3000SW column in the presence of 8 M urea. Three kinds of polypeptides from a normal HP2-1 heterozygote were separated and eluted by size (Fig. 3A); in the figure: P1, P2, and P3 represent HPβ, HPα2, and HPα1, respectively. A similar pattern was observed in the HP variant (Fig. 3B), with the presumed mutant polypeptide (P3) eluting at the same retention time as that of HPα polypeptide, although its quantity was reduced compared with that of HPα1 in the normal HP2-1 heterozygote. Further analyses of the presumed mutant polypeptide including N-terminal amino acid sequencing and HPLC fingerprinting of trypsin digests showed that the presumed mutant polypeptide was in fact identical to normal HPα1S (data not shown).
Figure 1.
A family study of variant case I. The HP–hemoglobin complexes were separated by PAGE and stained for peroxidase activity. The plasma samples were from individuals: lane 1, HP2-2; lane 2, mother; lane 3, propositus; lane 4, father; lane 5, HP2-1; and lane 6, HP1-1. The most anodal band in each sample was the band for free hemoglobin.
Figure 2.
Analysis of partially purified HPs by two-dimensional electrophoresis. The samples are from an HP2-1 individual (A) and the case I variant (B). The gels were stained with Coomassie Brilliant Blue R-250. Note that the relative staining intensity of the spot indicated by a bold arrow in the variant HP is reduced compared with that of the α1 spot in the normal HP2-1. The series of spots seen at the bottom of each gel are the charge standards, and those seen at the left of each gel are the molecular weight standards. IEF; isoelectric focusing.
Figure 3.
Gel filtration pattern of the partially purified HP from an HP2-1 individual (A) and from the case I variant (B). The purified HP molecules after reduction with 2-mercaptoethanol and carboxymethylation with iodoacetoamide were separated on a TSK 3000SW column. The polypeptides were monitored by 280-nm UV absorbance. The polypeptide recovered from the P3 fraction of the case I HP variant was regarded to be HPα1S. The protein level of the plasma HP of the variant was in the normal range, but the molar ratio of the α1S polypeptide to the αS2 polypeptide was less than 1:3.
Proof for the Mosaicism of Case I.
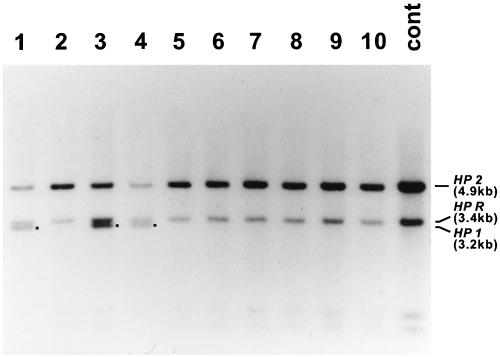
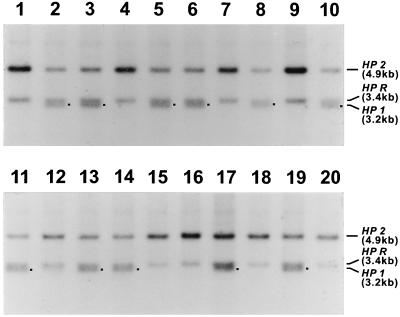
Because both parents of case I were normal HP2-2 homozygotes, we hypothesized that the HP*1 allele in the subject was generated somatically from an HP2 gene at a stage of embryogenesis early enough to result in formation of a readily detectable mosaicism. To test this hypothesis, an aliquot of blood from the case I individual was used to isolate lymphocytes, and B cell colonies were established in soft agarose after transformation by EBV. Forty one single B cell-derived colonies were isolated and examined by Southern blot; 10 of these are represented in Fig. 4. We found that 33 of 41 colonies were HP*2/HP*2 homozygotes, whereas the remaining eight were HP*2/HP*1 heterozygotes. We cloned the 3.2-kb band corresponding to the putative HP*1 allele for DNA sequencing and found that the new allele was identical to HP*1S sequence reported by Maeda et al. (5) (data not shown). The ratio of number of colonies with HP*2/HP*2 genotype to that with HP*2/HP*1 was 33:8, which strongly suggests that the somatic event generating the HP*1 allele occurred at an early stage of embryogenesis of an HP*2/HP*2 homozygote.
Figure 4.
Southern blot analysis of HP genotypes of B cell colonies. B cells from the case I variant were transformed by EBV and cloned in soft agarose. Genomic DNAs were digested with HindIII, and the Southern blots were hybridized to the hapto 6 probe (1.4-kb PstI fragment of HP cDNA). The sample in lane 11 was an HP*2/HP*2 control. The 4.9-kb band corresponded to the HP2, and the 3.4-kb band to the HP related gene (HPR) band (5). The 3.2-kb bands (shown with ■) in lanes 1, 3, and 4 were putative HP1 bands. Nucleotide sequencing of the 3.2-kb bands confirmed that they are identical with the sequence of the HP1S gene of Maeda et al. (5). A total of 41 B cell colonies were obtained for genotyping; 33 colonies were HP*2/HP*2 homozygotes, and eight were HP*2/HP*1 heterozygotes. Ten of these are represented.
Case II Was Also a Mosaic Mutant.
Our second unusual case was born to a phenotypically HP2-2 mother and an HP2-1 father (Fig. 5). We therefore analyzed the genotypes of cells from EBV-transformed B cell colonies of the case II child by Southern blot analysis as in case I. As shown in Fig. 6, nine of the 20 colonies were HP*2/HP*2 homozygotes and 11 were HP*2/HP*1 heterozygotes. This result clearly demonstrates that case II is also caused by mosaicism. Because the HP*1 allele is present in the father of the case II individual, we cannot exclude the possibility that the variant was derived from a new mutation in the HP*1 allele, although the known unique evolution of the HP*2 allele makes this unlikely. Consequently, we infer that the original genotype of the case II child was HP*2/HP*2 and that the HP*1 allele was generated from one of the two HP*2 alleles during early embryogenesis as in case I.
Figure 5.
A family study for the case II variant. HP–hemoglobin complexes were separated by a polyacrylamide gel and stained for peroxidase activity. The plasma samples were: lane 1, HP1-1; lane 2, HP2-1; lane 3, HP2-2; lane 4, mother; lane 5, propositus; lane 6, father; lane 7, mixture of equal amount of HP2-1 and HP2-2; and lane 8, HP2-1. Note that the pattern of the propositus (lane 5) was very similar to that of the mixture of HP2-1 and HP2-2 (lane 7).
Figure 6.
Southern blot analysis of the HP genotypes of B cell colonies from the case II variant. B cells were transformed by EBV and cloned in soft agarose. The genomic DNAs were digested with HindIII, and the Southern blots were hybridized to the hapto 6 probe (1.4-kb PstI fragment of HP cDNA). The 4.9-kb band corresponded to the HP2; the 3.4-kb band corresponded to the HP related gene (HPR); and the 3.2-kb band corresponded to the HP1 band (■) (5). Twenty B cell colonies were examined; nine were HP*2/HP*2 homozygotes, and 11 were HP*2/HP*1 heterozygotes.
Two Additional Cases of a Similar, but Inherited, Phenotype. In addition to the two de novo mutants, case I and case II, we identified two additional individuals (cases III and IV), with patterns in which again there were decreased amounts of HP1 relative to the HP2 protein. However, in both cases III and IV, unlike I and II, one of the parents showed one-dimensional electrophoresis protein gel patterns identical to their children. Thus, the trait was heritable.
The phenotypes of the case III and IV children match that of the HP Carlberg (HP CA) (13, 14), which resembles a mixture of HP2-2 and HP2-1 in variable proportions. Although the relative intensities of the multiple bands in the electrophoretogram seen with different HP CA individuals do not fit a simple pattern, the basic defect in HP CA appears to be a reduction of the HP1 gene product relative to that of HP2 (1). Indeed, DNA sequencing of the HP*1 allele, in case III, showed a G-to-A base substitution at nucleotide position −64 in one of the IL-6 responsive elements of the HP promoter region (15, 16). Chloramphenicol acetyltransferase assay confirmed that this substitution results in an ≈50% decreased expression compared with a normal HP*1 allele (M.K., unpublished observation). The decreased expression level was the same as that of the HP*2(−61C) allele from an individual with HP2-1M phenotype (16). In agreement with the heritable nature of the case IV phenotype, 30 EBV-transformed B cell colonies examined in case IV were all HP*2/HP*1 heterozygotes. No evidence for mosaicism was observed. We therefore conclude that these two heritable cases (III and IV) are caused by an inherited cis-acting mutation at the HP*1 allele, resulting in decreased expression of the HP1 gene and consequent imbalance of HP2 and HP1 gene products.
Frequency of the Hp*2 to Hp*1 Change in Human Somatic Cells and Male Germ Cells.
We examined the reversion frequencies in WBC and sperm in a single HP2-2 homozygous individual by a specifically designed PCR assay. Genomic DNA (20 μg, equivalent to 3 × 106 WBCs or 6 × 106 sperm cells), digested with BamHI and HindIII to produce 4.2-kb HP*2 and 2.5-kb HP*1 allele fragments, were size-fractionated. The gel portion expected to contain the HP*1 allele was subjected to PCR amplification after being divided into 60 tubes so that each tube contained DNA equivalent to approximately 5 × 104 WBCs or 1 × 105 sperm cells. Among these, 10 tubes contained HP*1 allele from the WBC DNA and 15 tubes contained HP*1 from the sperm DNA. Therefore, the HP*1 allele reversion frequencies in an Hp2-2 homozygous individual were 3.7 ± 1.2 × 10−6 in WBCs and 2.9 ± 0.7 × 10−6 in sperm, respectively.
DISCUSSION
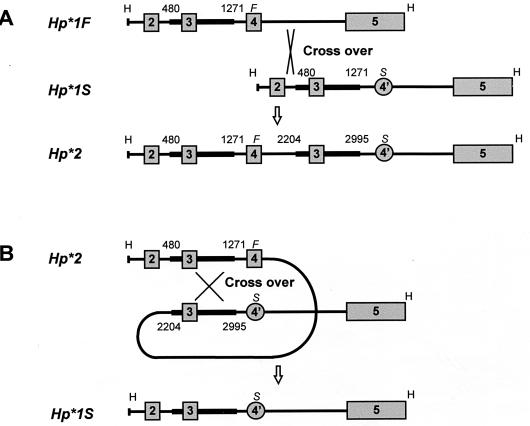
Mechanisms for Generation of HP*1 Allele from HP*2 Allele. Previous determinations of the structures of the HP*1 and HP*2 alleles revealed that the HP*2 allele contains an intragenic duplication of 1.7 kb that arose as the consequence of a unique nonhomologous recombinational event between HP1*F and HP1*S alleles (5) (Fig. 7A) sometime in the early evolution of Homo sapiens. In the somatic cells of two individuals, we found reversions from the HP*2 to the HP*1 allele that had occurred at a sufficiently early stage of embryogenesis to result in easily detectable mosaicism. Nucleotide sequence analysis confirmed the precise excision of one of the two duplicated segments from the HP*2 allele, strongly indicating that the excision was caused by a homologous recombinational event.
Figure 7.
(A) Schematic representation of formation of the human HP*2 allele by a rare nonhomologous crossing over between the fourth intron of the HP*1F allele and the second intron of the HP*1S allele in an HP*1F/HP*1S heterozygote (5). (B) Schematic representation of a postulated mechanism for the reversion mutation from HP*2 to HP*1S by homologous crossing over between the duplicated segments. The comparison of the nucleotide sequences of the HP*2 and the mutated allele (HP*1S) suggests that the intragenic homologous recombination occurred between regions 480-1271 and 2,204–2,995 of HP*2. Squares and circles represent exons. Thick bars represent homologous regions. H, HindIII site; F, HP*1F- specific region; S, HP*1S-specific region. The maps of the HindIII fragments from HP*1F, HP*1S, and HP*2 alleles are adapted from the report of Maeda et al. (5).
A priori, two mechanisms both involving homologous recombination can explain the present reversions from HP*2 to HP*1 allele in the two Hp2-2 individuals. One is unequal intermolecular somatic recombination either between homologous chromosomes or between sister chromatids. The original discovery of unequal recombination involved the Bar eye mutation in Drosophila, formed as a result of a large intrachromosomal duplication, and it was observed that Bar homozygotes occasionally give rise to a nonduplicated chromosome (reversal to the nonduplicated normal phenotype) or to a triplicated chromosome (the reciprocal product having the extreme Bar phenotype) (17). If such recombination had occurred in the present cases, one daughter cell would carry HP*1 but the other daughter cell would carry an HP*3 allele having a partial gene triplication. In our cases I and II, we see no evidence for the presence of the HP*3 allele by either protein or DNA analyses. We therefore think it more likely that the event was an intramolecular recombination resulting in the exact loss of one of the duplicated segments as illustrated in Fig. 7B. Pairing and recombination between the 1.7-kb duplicated DNA segments result in exact excision of one of the two duplicated DNA segments of the HP*2 allele. Examples of reversions from a partially duplicated allele to the wild-type allele are known in mouse pink-eyed unstable (pun) allele (18, 19) and in Drosophila white eye ivory (wi) allele (20, 21); both are believed to be caused by an intragenic homologous recombination in both germ and somatic cells.
Reversion from a partially duplicated allele to the wild-type allele by an intragenic homologous recombination in the pun homozygous mice resulting in mosaic phenotypes present in at least 1.8% of the mice (22); the frequency is much higher than the present findings at the HP*2 allele in human cells. The difference may be partly related to the length of the duplication, that is, 70 kb in pun (23) and 1.7 kb in HP*2 (5). In addition, the frequency of reverse mutation at pun was measured as spots of wild-type hairs on the mouse skin, where the pun gene is expressed, whereas reversion of HP*2, which is expressed only in the liver, was measured in nonexpressing WBC. Thus, gene expression might change the tertiary structure of the gene or nearby segments to facilitate intramolecular recombination as suggested from a study in pun homozygous embryos, in which dark melanocytes, an indicator of reversion, are more frequently observed with distance from the optic nerve (24).
May Mosaicism Explain Other HP Quantitative Variants?
Quantitative variants of human HP have been detected mainly as modification of the HP2-1 phenotype (1). Although the total amount of HP is variable, the ratio of HP1 protein to HP2 protein is normally invariant. The HP2-1M phenotype was first observed by Connell and Smithies (25) in electrophoretic examinations of purified HP from the sera of black people. The HP2-1M phenotype is common in African-Americans; about 10% of the population have the HP2-1M phenotype (26). The electrophoretic pattern of HP2-1M is similar to that of normal HP2-1 except that the slower-migrating Hp2-2 polymer bands are weaker than those of normal HP2-1 (1, 2). Although considerable variations have been observed in the intensity of the slow-migrating bands, it seems that the HP2-1M phenotype results from a reduced amount of HP2 protein compared with HP1 protein in HP2-1 heterozygotes. It has been suggested that “some kind of quantitative regulation exists, either as part of the HP2 gene itself, or closely linked to it” (1). Existence of such a regulatory element was revealed by the extensive nucleotide sequencing study of Maeda (16). She conducted direct sequencing of the HP promoter region in DNA from unrelated African-Americans and found an A-to-C substitution at nucleotide position −61 in all of 10 individuals with the HP2-1M phenotype, in two of four individuals with a possible HP2-1Mpo phenotype, but in none of 15 individuals with the ordinary HP2-1 phenotype. This finding demonstrated a strong association of the −61C mutation with decreased expression of the HP*2M allele. We found a G-to-A mutation at nucleotide position −64 of the promoter region of the HP*1CA allele in the case-III HP CA variant (heritable variant). The expression levels of both HP*1CA(−64A) and HP*2M(−61C) alleles were ≈50% of that of normal HP*1 or HP*2 allele by chloramphenicol acetyltransferase assay. Because the frequency of the HP2-1M variant is high among African-Americans (≈10%), it is likely that most cases are inherited variants of the HP*2 allele. However, any nonheritable HP2-1M variants might be the consequence of reverse mutations from HP*2 to HP*1 in a normal HP2-1 heterozygote, resulting in a phenotype indistinguishable from the HP2-1M phenotype.
When Did Reverse Mutation Take Place in the Two Mosaic Individuals? In nonmammalian zygotes, represented by amphibians and insects, cells replicate and develop into specified organs depending on the location of the cells in the eggs. For this reason, a fate map of future organs can be described on the surface of the eggs. Thus, if one of the two cells in a two-cell-stage embryo carries a new mutation, it is likely that the embryo will develop more or less into a 50:50 mosaic organism.
In mammals, however, the situation is quite different. Mammalian zygotes first need to create two major tissues, trophectoderm (extraembryonic tissue), which participates in uterine implantation, and the inner cell mass, which becomes embryo. Thus, fate maps cannot be described simply. In mice, the inner cell mass is assumed to start from about three cells randomly selected about 4 days after conception (27). The corresponding number is not known in human embryos. Consequently, the fraction of somatic cells bearing a new mutation in a mosaic individual cannot be used to indicate the origin of the mutational event. Thus, in the present two mosaic cases, it is not possible to indicate exactly when the reverse mutation from HP*2 to HP*1 took place.
Frequency of Reverse Mutation in the General Population and Somatic and Germ Cells of Hp2-2 Homozygous Individuals.
With protein analysis by electrophoresis, it is possible to detect the presence of HP1 protein at a ratio of 1:20 or higher in the presence of HP2 protein. During the study of the genetic effects of atomic-bomb radiation on 23,326 Japanese children (6, 7), we found two mosaic individuals with HP*2/HP*2 and HP*2/HP*1 cells among 13,146 children with the HP2-2 phenotype. The minimum estimate of the frequency of reverse mutation from HP*2 to HP*1 therefore must be ≈8 × 10−6 per cell [2 mutations/(13,146 × 2 alleles × 10 cells)]. We predicted that the HP*1 allele also would be present, although usually at a very low frequency that could not be detectable on protein gels, in any Hp2-2 homozygous individuals. We tested this prediction by a specifically designed PCR assay in a single individual and detected the HP*1 allele at frequencies of 4 × 10−6 and 3 × 10−6 in somatic cells and male germ cells. The frequencies were comparable with those obtained by genetic screening with protein gel electrophoresis. We also conducted additional PCR assays on WBC DNAs from four Hp2-2 homozygous volunteers and found the HP*1 allele in all of them. These results strongly support the regular but rare occurrence of homologous recombination within duplicated regions in humans, in agreement with previous observations in mouse and Drosophila.
Acknowledgments
We are grateful to Dr. K. Hiyama for her help in the family studies, Dr. D. Pierce for his statistical help, and E. Nishikori, S. Kaneoka, Y. Shimoichi, Y. Nakamoto, and J. Kaneko for technical assistance. We also thank Dr. N. Maeda for providing a genomic DNA of HP2-1 modified and Dr. S. Oliviero for providing HP cDNA probe. We thank Dr. J. V. Neel for encouragement and Dr. S. Wolff for critical reading of the manuscript. This publication is based on research performed at the Radiation Effects Research Foundation (RERF), Hiroshima and Nagasaki, in Japan. RERF is a nonprofit foundation funded equally by the Japanese Ministry of Health and Welfare and the U.S. Department of Energy through the National Academy of Sciences.
ABBREVIATIONS
- HP
haptoglobin
- EBV
Epstein–Barr virus
- WBC
white blood cell
- CA
Carlberg
- 2-1M
2-1 modified
- kb
kilobase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Giblett E R. Genetic Markers in Human Blood. Oxford: Blackwell Scientific; 1969. pp. 63–125. [Google Scholar]
- 2.Bowman B H, Kurosky A. Adv Hum Genet. 1982;12:189–261. doi: 10.1007/978-1-4615-8315-8_3. [DOI] [PubMed] [Google Scholar]
- 3.Smithies O. Biochem J. 1955;61:629–641. doi: 10.1042/bj0610629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smithies O, Connell G E, Dixon G H. Nature (London) 1962;196:232–236. doi: 10.1038/196232a0. [DOI] [PubMed] [Google Scholar]
- 5.Maeda N, Yang F, Barnett D R, Bowman B H, Smithies O. Nature (London) 1984;309:131–135. doi: 10.1038/309131a0. [DOI] [PubMed] [Google Scholar]
- 6.Neel J V, Satoh C, Goriki K, Fujita M, Takahashi N, Asakawa J, Hazama R. Proc Natl Acad Sci USA. 1986;83:389–393. doi: 10.1073/pnas.83.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neel J V, Satoh C, Goriki K, Asakawa J, Fujita M, Takahashi N, Kageoka T, Hazama R. Am J Hum Genet. 1988;42:663–676. [PMC free article] [PubMed] [Google Scholar]
- 8.Weitkamp L R, Arends T, Gallango M L, Neel J V, Schultz J, Shreffler D C. Ann Hum Genet. 1972;35:271–279. doi: 10.1111/j.1469-1809.1957.tb01401.x. [DOI] [PubMed] [Google Scholar]
- 9.Oliviero S, DeMarchi M, Bensi G, Raugei G, Carbonara A O. Hum Genet. 1985;70:66–70. doi: 10.1007/BF00389461. [DOI] [PubMed] [Google Scholar]
- 10.Asakawa J, Satoh C. Biochem Genet. 1986;24:131–148. doi: 10.1007/BF00502984. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto N, Hinuma Y. Int J Cancer. 1976;17:191–196. doi: 10.1002/ijc.2910170207. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi N, Nakai S, Honjo T. Nucleic Acids Res. 1980;24:5983–5991. doi: 10.1093/nar/8.24.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galatius-Jensen F. Acta Genet. 1958;8:248–255. doi: 10.1159/000151065. [DOI] [PubMed] [Google Scholar]
- 14.Giblett E R. Cold Spring Harb Symp Quant Biol. 1964;29:321–326. doi: 10.1101/sqb.1964.029.01.034. [DOI] [PubMed] [Google Scholar]
- 15.Oliviero S, Cortese R. EMBO J. 1989;8:1145–1151. doi: 10.1002/j.1460-2075.1989.tb03485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda N. Am J Hum Genet. 1991;49:158–166. [PMC free article] [PubMed] [Google Scholar]
- 17.Sturtevant A H, Morgan T H. Science. 1923;57:746–747. doi: 10.1126/science.57.1487.746. [DOI] [PubMed] [Google Scholar]
- 18.Gondo Y, Gardner J M, Nakatsu Y, Durham-Pierre D, Deveau S A, Kuper C, Brilliant M H. Proc Natl Acad Sci USA. 1993;90:279–301. doi: 10.1073/pnas.90.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiestl R H, Aubrecht J, Khogali F, Carls N. Proc Natl Acad Sci USA. 1997;94:4576–4581. doi: 10.1073/pnas.94.9.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins M, Rubin G M. Cell. 1982;30:71–79. doi: 10.1016/0092-8674(82)90013-7. [DOI] [PubMed] [Google Scholar]
- 21.Green M M, Todo T, Ryo H, Fujikawa K. Proc Natl Acad Sci USA. 1986;83:6667–6671. doi: 10.1073/pnas.83.18.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melvold R W. Mutat Res. 1971;12:171–174. doi: 10.1016/0027-5107(71)90138-2. [DOI] [PubMed] [Google Scholar]
- 23.Schiestl R H, Khogali F, Carls N. Science. 1994;266:1573–1576. doi: 10.1126/science.7985029. [DOI] [PubMed] [Google Scholar]
- 24.Deol M S, Truslove G M. J Embryol Exp Morphol. 1983;78:291–298. [PubMed] [Google Scholar]
- 25.Connell G E, Smithies O. Biochem J. 1959;72:115–121. doi: 10.1042/bj0720115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giblett E R. Nature (London) 1959;183:192. doi: 10.1038/183192a0. [DOI] [PubMed] [Google Scholar]
- 27.Russell L B, Russell W L. Proc Natl Acad Sci USA. 1996;93:13072–13077. doi: 10.1073/pnas.93.23.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]