Abstract
The chaperonin GroEL assists protein folding by binding nonnative forms through exposed hydrophobic surfaces in an open ring and mediating productive folding in an encapsulated hydrophilic chamber formed when it binds GroES. Little is known about the topology of nonnative proteins during folding inside the GroEL–GroES cis chamber. Here, we have monitored topology employing disulfide bond formation of a secretory protein, trypsinogen (TG), that behaves in vitro as a stringent, GroEL–GroES-requiring substrate. Inside the long-lived cis chamber formed by SR1, a single-ring version of GroEL, complexed with GroES, we observed an ordered formation of disulfide bonds. First, short-range disulfides relative to the primary structure formed, both native and nonnative. Next, the two long-range native disulfides that “pin” the two β-barrel domains together formed. Notably, no long-range nonnative bonds were ever observed, suggesting that a native-like long-range topology is favored. At both this time and later, however, the formation of several medium-range nonnative bonds mapping to one of the β-barrels was observed, reflecting that the population of local nonnative structure can occur even within the cis cavity. Yet both these and the short-range nonnative bonds were ultimately “edited” to native, as evidenced by the nearly complete recovery of native TG. We conclude that folding in the GroEL–GroES cavity can favor the formation of a native-like topology, here involving the proper apposition of the two domains of TG; but it also involves an ATP-independent conformational “editing” of locally incorrect structures produced during the dwell time in the cis cavity.
Keywords: chaperonin, protein folding, topology
Although binding-active and folding-active states of the GroEL–GroES chaperonin system have been recognized and structurally characterized (1–3), the conformational states of nonnative polypeptide during folding in this system remain poorly understood. A variety of studies, including hydrogen-deuterium exchange (4–10) and, more recently, direct NMR observation (11), have reported on the conformational state of nonnative protein while bound in an open ring of GroEL indicating that it occupies an unfolded state, lacking any significant ordered secondary or tertiary structure. This finding implies that binding in an open GroEL ring may be associated with an unfolding action (12–14), which is potentially the result of multivalent binding by surrounding apical domains (15) that, in energetic terms, may be equivalent to taking the protein to the top of its folding energy landscape. The subsequent binding of ATP and GroES to the same GroEL ring as polypeptide triggers rapid release of polypeptide in <1 sec from the hydrophobic binding sites into a now-encapsulated hydrophilic chamber, where productive folding immediately commences (16, 17). In a cycling reaction, folding proceeds in this space for a period of ≈10 sec, the longest step of the reaction cycle, before a sequence of cis ATP hydrolysis immediately followed by trans ATP binding ejects GroES and substrate polypeptide into the bulk solution (17). For many “stringent” GroEL–GroES-dependent substrate protein molecules, this is not sufficient time to reach native form, and nonnative forms are released into the bulk solution (18, 19), where they can either return to GroEL for another attempt at folding, or, in the setting of a cellular compartment, be kinetically partitioned among the various chaperones and proteases that are present. As an example of such behavior in vitro, a single reaction cycle is sufficient to productively fold only a few percent of the input molecules of rhodanese or Rubisco (16, 19).
In contrast with the short dwell time in the cis cavity in the cycling reaction, cis ternary complexes formed with a single-ring version of GroEL, called SR1, and GroES are long-lived and persist essentially indefinitely, because the allosteric signal for GroES release cannot be produced (16, 17, 20). In such noncycling SR1 complexes, stringent substrate proteins like rhodanese or Rubisco proceed nearly quantitatively to the native state with the same kinetics as the cycling reaction (16, 17). This finding indicates that it is dwell time in the cis cavity that leads to the production of the native state of these substrate molecules. Consistently, if nonnative molecules are prevented during a cycling reaction from returning from the bulk solution to an open GroEL ring (and ultimately to the cis cavity), they promptly cease to reach native form (21). Thus, the exact fate of molecules folding inside the cis cavity is of central interest to understanding the folding reaction.
To date, we know little about the trajectory of cis folding. For example, the topological changes occurring in the cis cavity remain uncharted except for a FRET study that examined the behavior of a pair of fluorophores placed near the N and C termini of Rubisco, in which it was observed that the initiation of folding was associated with shortening by a few angstroms of the end-to-end distance from that observed in the GroEL-Rubisco binary complex, suggesting that some degree of compaction of the substrate occurs upon its encapsulation by GroES (22). However, questions such as whether only native topologies are assumed during cis folding or whether nonnative ones are also populated remain open. Here we have taken a step toward addressing such questions, using disulfide bond formation as a reporter of the topology of a secretory protein found to behave in vitro as a stringent substrate of GroEL.
Results
The Disulfide-Containing Secretory Protein, Trypsinogen (TG), Behaves as a Stringent, GroEL–GroES-Dependent Substrate Protein in Vitro.
Natural chaperonin substrates, which reside in the reducing compartment of the cytosol or mitochondria, do not have disulfide bonds that can be used for reporting on the acquisition of native topology. By contrast, secretory proteins in the relatively oxidizing compartment of the ER often have disulfides, and these disulfides have been used to report on folding in the classic early experiments of Anfinsen with ribonuclease (23) and in more recent experiments, for example, with BPTI (24), ribonuclease A (25, 26), and lysozyme (27, 28) (see also refs. 29–32). We uncovered a secretory protein, bovine TG, the 24-kDa proform of the monomeric enzyme trypsin, comprised of two orthogonal β-barrels and containing six disulfides in the native state (Fig. 1a), that behaved in vitro as a stringent GroEL substrate. The unfolded protein was efficiently bound by GroEL or the single-ring version, SR1, after it was diluted from a denaturant/reductant-containing mixture (see Fig. 2a), and the native form was fully recovered (Fig. 1b) after the addition of GroES, ATP, and a redox pair, reduced and oxidized glutathione, that enabled disulfide rearrangement during folding. Recovery of native TG was monitored at each time point by halting the folding reaction and then incubating with enterokinase to cleave the propeptide from TG followed by an assay for trypsin activity.
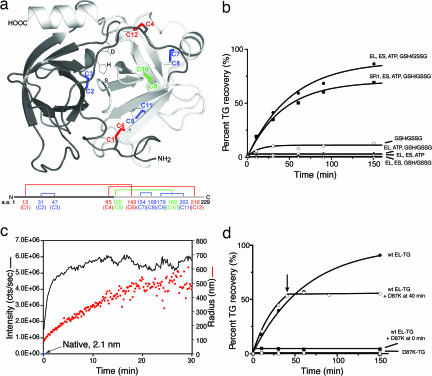
Fig. 1.
TG is a stringent GroEL–GroES-dependent substrate in vitro, folded in the cis cavity. (a) Ribbons diagram of native TG (Protein Data Bank ID code 1TGS) (39) with disulfide bonds colored as follows: red, long-range; green, medium-range C5-C10 bond; blue, short-range. These designations were based on the distance between cysteines in the primary sequence, defined as follows: long-range, >70 aa; medium-range, 40–70 aa; short-range, <40 aa. Active-site side chains of mature trypsin lying in the cleft between the two orthogonal β-barrels are represented as sticks. Only the last two residues of the propeptide (amino acids 5–6) are modeled in the figure. Underneath the ribbons diagram is a schematic illustrating the disulfide bond arrangement of native TG against the primary structure. Amino acids are numbered by taking the first residue of the propeptide (Val) as number 1. (b) TG is refolded by GroEL–GroES-ATP and by SR1–GroES-ATP, in the presence of a reduced glutathione (GSH)/oxidized glutathione (GSSG) redox pair, with similar kinetics. Binary complexes of TG and chaperonin were formed, and reactions commenced as illustrated in Fig. 2a. At the indicated points, the reaction was halted by adding chelator and IAM, and the mixture was treated with enterokinase to cleave the propeptide and assayed for trypsin activity, as described in SI Methods. Note that in the case of the SR1 reaction, a 20-min incubation at 4°C was included before enterokinase treatment to release GroES and native TG from the SR1 complex. (c) Failure of TG to renature upon direct dilution into redox buffer (without chaperonin) is associated with aggregation, as demonstrated by dynamic light scattering (see SI Methods). The intensity of scattering is shown as the black trace, and the calculated radius of the aggregates is shown in orange. (d) The addition of a GroEL “trap” molecule (D87K) that can bind but not release nonnative forms (17) is associated with immediate halt of recovery of native TG in a cycling GroEL–GroES reaction, reflecting that nonnative forms of TG are being continuously released during the GroEL–GroES reaction, as is the case with other stringent GroEL substrates (see SI Methods for details).
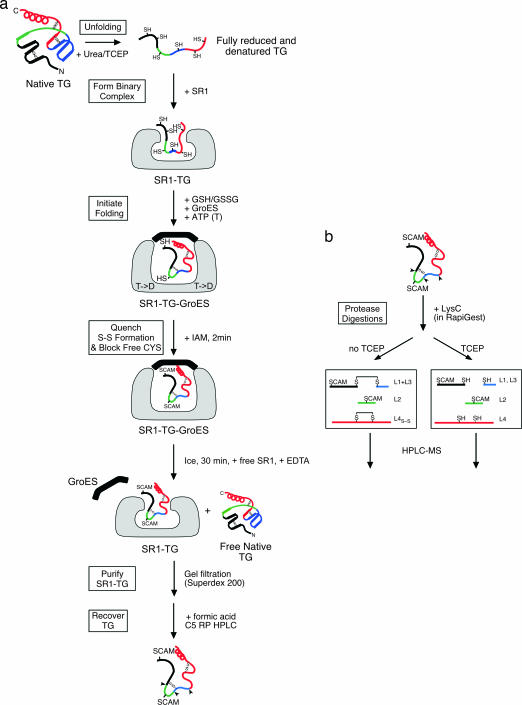
Fig. 2.
Scheme for identification of disulfides formed in TG during SR1–GroES-mediated refolding. (a) A schematized substrate representing TG is shown, with two short-range disulfides and one long-range disulfide in the native state. The polypeptide is colored black, green, blue, and red proceeding from the N to the C terminus along the primary structure. When TG is reduced and unfolded with urea and then diluted into a mixture with SR1, it forms a binary complex (second panel from the top), in which, if exposed to oxidizing conditions, only short-range disulfides were observed (SI Table 2). Starting with binary complex in fully reduced conditions in the experiments presented, folding was initiated by adding ATP, GroES, and a reduced glutathione (GHT)/oxidized glutathione (GSST) redox pair, and TG was recovered at various time points after halting the reaction with iodoacetamide by using the series of steps illustrated. SCAM, carboxyamidomethyl-cysteine. (b) The disulfides in the recovered TG were identified by using proteolysis with LysC and HPLC-MS, as diagrammed (see text). As illustrated here in a, at the point of quenching the reaction with IAM, two short-range nonnative disulfides have formed, whereas the two other thiols that are free become alkylated.
In contrast with chaperonin binding and refolding, when unfolded TG was diluted into buffer lacking GroEL (but containing the redox pair), TG misfolded (Fig. 1b) and promptly formed aggregates, as indicated by dynamic light scattering (Fig. 1c). No enzymatic activity was recovered, even when starting with unfolded TG concentrations as low as 10 nM (data not shown). These findings are consistent with previous studies that show that spontaneous refolding of reduced and denatured TG fails to produce native TG unless aggregation is prevented either by immobilizing TG on agarose (33) or by starting with unfolded TG that has been modified by the formation of mixed disulfides with glutathione (34). Aggregated TG did not form intermolecular disulfides [supporting information (SI) Fig. 5], and when examined by using proteolysis with LysC and HPLC-MS, the aggregated monomers were found to contain only short-range intramolecular disulfides (Fig. 4, right column, and SI Table 1, right column), potentially indicating a relatively disordered structure.
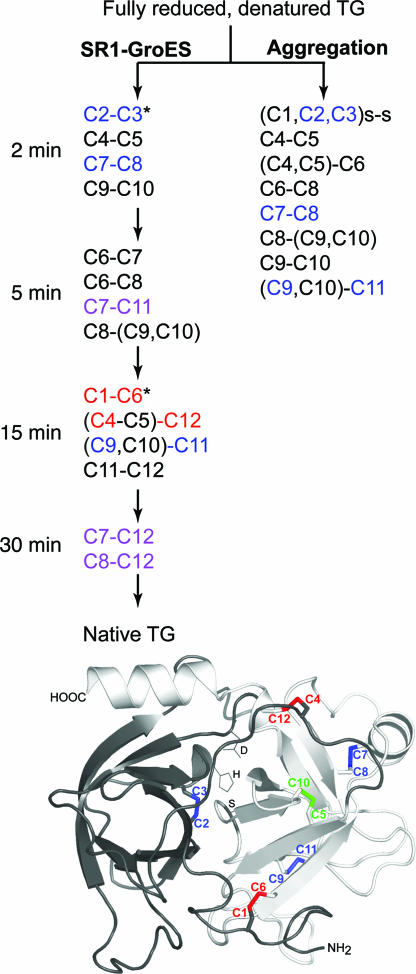
Fig. 4.
Formation of disulfides during the first 30 min of SR1–GroES-mediated folding of TG, compared with disulfides formed during spontaneous folding/aggregation. Disulfides are color-coded as follows: black, short-range nonnative disulfides; blue, short-range native; purple, medium-range nonnative; red, long-range native. Parentheses indicate peptides containing more than one cysteine, where the particular cysteine involved in the disulfide could not be identified. *, For the potentially ambiguous L2 peptide with three cysteines (C1, C2, C3), MS/MS analysis (see SI Methods) indicated that ≈70% contained the native C2-C3 disulfide. Based on this finding, it was inferred that the long-range disulfide observed later between this peptide and peptide L8 was the native C1–C6 one.
GroEL–GroES-assisted folding of TG was faithfully recapitulated by SR1, which supports productive folding of other GroEL–GroES-dependent substrates inside long-lived SR1–GroES complexes (16, 17) (the absence of the second ring abolishes the normal allosteric signal for GroES ejection). As observed for other substrates (16, 17), the same kinetics of recovery of native TG was observed with SR1–GroES as those for the cycling GroEL–GroES reaction (Fig. 1b).
Disulfide Bond Formation in Trypsinogen During Folding Inside SR1–GroES.
The topology of TG during folding in the SR1–GroES cis cavity was monitored by identifying the disulfides formed at various times after the reaction was initiated (Fig. 2). At each time point, the folding reaction was halted by adding iodoacetamide (IAM) to alkylate free cysteines in both the TG and the glutathione component of the redox pair. TG was then released from the SR1–GroES complex by using a cold-temperature incubation in the presence of EDTA that respectively dissociated GroES from SR1 (17, 19) and prevented nucleotide-dependent reassociation. The nonnative states of TG were recovered because of their stable binding by the open SR1 ring, allowing them to be separated from both newly folded native TG and GroES by gel filtration. Notably, the collective of nonnative TG conformations are efficiently captured by open chaperonin rings, as best indicated by the ability of the cycling GroEL–GroES TG folding reaction to produce nearly 100% recovery of native TG (Fig. 1b). In this context, nonnative intermediates, with virtually the same collective of disulfide bonds as those that are formed inside SR1–GroES, undergo rounds of release into the solution and rebinding by GroEL (see Fig. 1d concerning rounds of release; and for intermediates formed in the two reactions, compare Fig. 4 and SI Fig. 6).
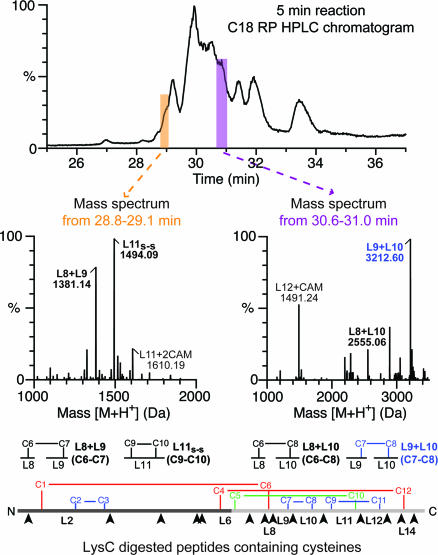
The nonnative TG intermediates recovered with SR1 were separated from it by using reversed-phase chromatography (Fig. 2a), and, to localize the disulfides in the TG intermediates, the isolated TG was subjected to LysC digestion followed by HPLC-MS (Fig. 2b), with and without reduction. As an example of this analysis, an HPLC chromatogram of the LysC digestion products prepared under nonreducing conditions from a 5 min SR1–GroES-mediated TG folding reaction is displayed in Fig. 3Top. The MaxEnt mass readouts from two retention time regions are shown in Fig. 3 Middle. A nonnative disulfide bond formed between C6 and C7 is identified in the 28.8–29.1 min profile (Fig. 3 Middle Left) as an adduct of LysC peptides 8 and 9 with a mass of 1,381.14 Da. A native disulfide bond, C7-C8, is identified in the 30.6–31.0 min profile (Fig. 3 Middle Right), as an adduct of LysC peptides 9 and 10 with a mass of 3,212.60 Da. Similar analyses carried out on entire chromatograms for the various time points produced a full accounting of disulfide-linked LysC peptides at the various times of folding (SI Table 1).
Fig. 3.
Identification of disulfides formed during SR1–GroES-mediated folding of TG. TG intermediates recovered from an SR1–GroES-mediated folding reaction that was carried out for 5 min by using the steps shown in Fig. 2a were digested with LysC, and the peptides prepared under nonreducing conditions were analyzed by using HPLC-MS with a C18 column (Top) and a Q-TOF1 mass spectrometer (see SI Methods). (Middle) MaxEnt molecular mass outputs from two different indicated periods of elution are shown, and the identified peptides are diagrammed beneath the graphs. (Bottom) Shown is a schematic of native TG and its LysC sites (arrowheads). LysC cleaves after K6, 49, 75, 95, 97, 131, 142, 145, 155, 176, 192, 206, 214, and 223, and the peptides are designated L1–L15 from the N to the C terminus. L2, for example, contains C1, C2, and C3.
At 2 and 5 min, disulfides that are predominantly short-range with respect to the primary structure were formed, both native ones and nonnative ones (shown in blue and black, respectively, in Fig. 4). By 15 min, however, long-range bonds were formed (shown in red in Fig. 4). Both of the long-range native bonds that pin the β-barrel domains together, i.e., C1-C6 and C4-C12, appeared to have been formed (shown in red in Fig. 4). The presence of C1-C6 was inferred from a disulfide linkage detected between the LysC peptide (L2) that contained C1, C2, and C3, and the peptide (L8) that contained C6, based on the presence already at an earlier time (2 min) of the native C2–C3 linkage (refer to Fig. 3 Bottom). Similarly, a disulfide linkage between the LysC peptide with C4 and C5 (L6) and the peptide containing C12 (L14) was observed. Here we were unable to clearly resolve whether it was C4 or C5 that formed the linkage with C12 (but see Disulfide Bond Formation in Trypsinogen During Folding by Cycling GroEL–GroES and SI Fig. 6 concerning resolution of this ambiguity for the GroEL–GroES reaction).
Medium-range nonnative bonds were also formed during the reaction (shown in purple in Fig. 4). In as little as 5 min, a medium-range nonnative bond was observed between C7 and C11 (shown purple in Fig. 4), mapping to the “righthand” β-barrel, and at 30 min, two additional medium-range nonnative bonds in the same β-barrel were observed. Once a disulfide bond was detected, either native or nonnative, it generally appeared in the analysis of later time-points up to the 30 min half-time of the folding reaction (SI Table 1). Notably, the fraction of individual native bonds appeared to increase with time, whereas that of nonnative ones decreased, but more precise quantitation was not possible. We note also that the medium-range native disulfide bond, C5–C10 (shown in green in Fig. 4), was not observed in SR1–GroES-produced TG intermediates at any time point, but was readily observed in native TG, and infer that it is the last disulfide to form on the pathway to the native state. Presumably, once it is formed, TG is native and no longer binds to GroEL.
Disulfide Bond Formation in Trypsinogen During Folding by Cycling GroEL–GroES.
Folding in the long-lived SR1–GroES cis cavity was compared with the physiologic folding reaction that is mediated by GroEL–GroES, where there is cycling of nonnative substrate on and off of the chaperonin, with the lifetime of polypeptide inside any given cis complex limited to ≈10 sec. Nearly the same set of disulfides was observed (SI Table 1, middle columns, and SI Fig. 6), except that the medium-range nonnative bond, C8–C12 (purple), that formed at 30 min inside SR1–GroES, was not observed. Interestingly, the order of long-range disulfide bond formation was different at GroEL–GroES. For example, the long-range bond, (C4,C5)–C12, formed at 15 min in SR1–GroES, was formed by 5 min in the GroEL–GroES reaction (where it was resolved by using MS/MS and found to be the native C4–C12 bond). Conversely, C1–C6, formed by 15 min at SR1–GroES, was not observed in the GroEL–GroES reaction until 30 min. Thus, the pathway of long-range native disulfide bond formation is different between the two reactions; yet, notably, neither produced nonnative long-range bonds. The difference in timing of long-range bond formation may relate to periodic rebinding by an open GroEL ring in the cycling reaction, which likely exerts an unfolding action on the substrate protein, effectively pulling it up the energy landscape. By contrast, the reaction inside the long-lived cis complex of SR1–GroES does not experience such an action. Consistent with such a difference of behavior is the progression of disulfide bond formation in the two reactions: at 5 min, the SR1–GroES reaction produced as many as four to six disulfide bonds, whereas the cycling GroEL–GroES reaction produced no more than four disulfides, progressing to as many as five to six disulfide bonds only by 15–30 min (SI Fig. 7).
Discussion
Correct Formation of Long-Range Topology in the Cis Folding Chamber.
In summary, we observed here that when TG was folded either inside the cis cavity of the noncycling SR1–GroES or in the cycling GroEL–GroES reaction, the only long-range disulfide bonds formed were native ones (shown in red in Fig. 4 and SI Fig. 6). In particular, long-range nonnative bonds were not observed. These bonds could potentially have formed either between C4 or C12 at the “top” of TG and C1 or C6 at the “bottom” (four possible bonds) or between C2 or C3 in the “left” barrel and thiols in the “right” one (16 possible bonds). The consequence of such native bond selection is that the two β-barrels of trypsinogen are properly apposed (see Fig. 1a) and bring together the two “halves” of the active site pocket. This apposition apparently does not occur in the absence of the chaperonin system–intermolecular interactions that lead to wholesale aggregation supervene (by 30 sec; see Fig. 1c). By contrast, in isolation in the cis cavity, such intermolecular interaction cannot occur, and TG has the opportunity to form long-range native topology.
Nonnative Short-Range Structures Are Formed but Are “Edited” to Native During the Cis Folding Reaction.
Although the only long-range disulfides observed were native ones, most of the possible nonnative disulfides of short range and a few specific disulfides of medium range were observed, reflecting that nonnative contacts are formed on a local basis within the domains of the substrate protein. In the case of the medium-range disulfides, whether they represent particular off-pathway steps or, in some cases, productive on-pathway intermediates cannot be distinguished.
A process of conformational rearrangement of nonnative states and progressive native structure formation during SR1–GroES-mediated refolding would indicate that the cis cavity, at least at the level of local structure, can support conformational editing. Notably, this occurs without resorting to any directly associated nucleotide binding or hydrolysis. In particular, the energy of ATP binding has already been expended at the step of ATP/GroES binding that initially forms the cis cavity (35), and the energy of hydrolysis is expended with the single turnover occurring at SR1–GroES within ≈10 sec of formation of the complex (20). This finding implies, as revealed particularly by the many-minute lifetime but productive character of the SR1–GroES cis cavity, that there must be relatively low transition state barriers inside the cis cavity between the misfolded forms observed and properly folded ones, which do not require much input of energy to cross. The basis for such behavior seems unclear. For example, simple confinement by nearby chaperonin walls does not easily explain a facile crossing of a barrier between off-pathway and on-pathway states. Notably, multiplication of the GroEL C-terminal “tails,” as recently designed to suggest tight confinement as a supportive mechanism for folding within the cis cavity (36), was without any significant enhancing effect on the rate of TG folding (0.024 sec−1 for wild-type, 0.027 sec−1 for duplicated tails, and no recovery of native TG for triplicated and quadruplicated tails). Thus, it remains to be demonstrated how/whether the cis cavity might actively support a role in reversing nonnative bonds and favoring formation of native ones. Perhaps thermal fluctuations alone are sufficient to surmount these barriers.
Methods
Proteins.
Wild-type and mutant (SR1, D87K) GroELs and wild-type GroES were expressed in Escherichia coli and purified as described in refs. 16, 17, 20, and 35. TG from bovine pancreas (Sigma–Aldrich, St. Louis, MO) was purified by using chromatography on a MonoQ column (Amersham, Piscataway, NJ) to obtain the β form of trypsinogen (37). Purified TG was dialyzed against 1 mM HCl containing 1 mM benzamidine (Sigma) to inhibit autoactivation and then against 1 mM HCl. See SI Methods for details of storage and activity assay.
GroEL–GroES-Mediated Folding of TG.
TG was denatured and fully reduced by incubation in 6 M urea, 15 mM tris(2-carboxyethyl)phosphine hydrochloride, and 50 mM tris (pH 7.4) for 30 min at 20°C. GroEL-TG binary complexes were formed by 100-fold dilution of denatured TG into a refolding buffer [50 mM Tris, pH 8.0/50 mM KCl/10 mM MgCl2/1 mM tris(2-carboxyethyl)phosphine hydrochloride] that contained a 2-fold molar excess of GroEL. Refolding was initiated by adding GroES (2-fold molar excess over GroEL), a redox buffer consisting of 4.5 mM glutathione and 1.5 mM oxidized glutathione (final concentrations), and ATP to give a final concentration of 5 mM. At various times, the reaction was quenched by adding EDTA to give a final concentration of 20 mM, and free cysteines were modified by adding 100 mM IAM to form carboxyamidomethyl-cysteine, which prevented further disulfide exchange and trapped disulfide folding intermediates. This modification occurs in <1 sec (38) and is quantitative, because no free cysteines were detected by using mass spectrometry. SR1–GroES-mediated refolding was carried out identically, with the addition of a final incubation, after adding IAM, at 4°C for 20 min to release GroES from the complex. TG recovery was measured by using enterokinase treatment and trypsin assay (see SI Methods) either immediately (for wild-type) or after the 4°C incubation (for SR1). No specific steps were taken to exclude calcium ions from the refolding reaction, but the concentration must be well below millimolar, because both chaperonin and denatured TG had been extensively dialyzed. In addition, adding calcium up to a final concentration of 10 mM to the spontaneous folding reaction had no effect on the recovery of native TG.
Identification of Folding Intermediates During Chaperonin-Mediated Folding.
In the case of the GroEL–GroES reaction, folding was initiated as described in GroEL–GroES-Mediated Folding of TG and was quenched at various times by adding EDTA and IAM. In the case of SR1–GroES, free SR1 (2-fold molar excess over TG) was added to the reaction mixture after quenching to ensure binding of nonnative TG species by open chaperonin rings, and the sample was incubated at 4°C for 30 min to release GroES and native TG. Binary complexes of GroEL or SR1 and nonnative TG intermediates were separated from free GroES and free native TG by using gel filtration chromatography on Superdex 200 (Amersham). TG intermediates were separated from GroEL subunits by using RP-HPLC after dissociation by acidification, and disulfide-linked peptides were identified by using HPLC-MS after LysC digestion as described in SI Methods.
Supplementary Material
Acknowledgments
We thank Shawn Cao of Wyatt Technology Corp. for assistance with light-scattering experiments. This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Abbreviations
- TG
trypsinogen
- IAM
iodoacetamide.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610989104/DC1.
References
- 1.Horwich AL, Farr GW, Fenton WA. Chem Rev. 2006;106:1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 3.Thirumalai D, Lorimer GH. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 4.Zahn R, Spitzfaden C, Ottiger M, Wüthrich K, Plückthun A. Nature. 1994;368:261–265. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]
- 5.Robinson CV, Gross M, Eyles SJ, Ewbank JJ, Mayhew M, Hartl FU, Dobson CM, Radford SE. Nature. 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- 6.Groβ M, Robinson CV, Mayhew M, Hartl FU, Radford SE. Protein Sci. 1996;5:2506–2513. doi: 10.1002/pro.5560051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg MS, Zhang J, Sondek S, Matthews CR, Fox RO, Horwich AL. Proc Natl Acad Sci USA. 1997;94:1080–1085. doi: 10.1073/pnas.94.4.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gervasoni P, Gehrig P, Plückthun A. J Mol Biol. 1998;275:663–675. doi: 10.1006/jmbi.1997.1481. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Walter S, Horwich AL, Smith DL. Nat Struct Biol. 2001;8:721–728. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]
- 10.Park ES, Fenton WA, Horwich AL. FEBS Lett. 2005;579:1183–1186. doi: 10.1016/j.febslet.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wüthrich K. Proc Natl Acad Sci USA. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahn R, Plückthun A. J Mol Biol. 1994;242:165–174. doi: 10.1006/jmbi.1994.1567. [DOI] [PubMed] [Google Scholar]
- 13.Walter S, Lorimer GH, Schmid FX. Proc Natl Acad Sci USA. 1996;93:9425–9430. doi: 10.1073/pnas.93.18.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zahn R, Perrett S, Fersht AR. J Mol Biol. 1996;261:43–61. doi: 10.1006/jmbi.1996.0440. [DOI] [PubMed] [Google Scholar]
- 15.Farr G, Furtak K, Rowland MC, Ranson NA, Saibil HR, Kirchhausen T, Horwich AL. Cell. 2000;100:561–573. doi: 10.1016/s0092-8674(00)80692-3. [DOI] [PubMed] [Google Scholar]
- 16.Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 17.Rye HS, Burston SG, Fenton WA, Beechem JM, Xu Z, Sigler PB, Horwich AL. Nature. 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 18.Weissman JS, Kashi Y, Fenton WA, Horwich AL. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 19.Todd MJ, Viitanen PV, Lorimer GH. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 20.Weissman JS, Hohl CM, Kovalenko O, Chen S, Braig K, Saibil HR, Fenton WA, Horwich AL. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 21.Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- 22.Lin Z, Rye HS. Mol Cell. 2004;16:23–34. doi: 10.1016/j.molcel.2004.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anfinsen CB, Haber E, Sela M, White FH. Proc Natl Acad Sci USA. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissman JS, Kim PS. Science. 1991;253:1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- 25.Rothwarf DM, Li YJ, Scheraga HA. Biochemistry. 1998;37:3767–3776. doi: 10.1021/bi972823f. [DOI] [PubMed] [Google Scholar]
- 26.Narayan M, Welker E, Wedemeyer WJ, Scheraga HA. Acc Chem Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- 27.Roux P, Ruoppolo M, Chaffotte AF, Goldberg ME. Protein Sci. 1999;8:2751–2760. doi: 10.1110/ps.8.12.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Berg B, Chung EW, Robinson CV, Dobson CM. J Mol Biol. 1999;290:781–796. doi: 10.1006/jmbi.1999.2915. [DOI] [PubMed] [Google Scholar]
- 29.Maskos K, Huber-Wunderlich M, Glockshuber R. J Mol Biol. 2003;325:495–513. doi: 10.1016/s0022-2836(02)01248-2. [DOI] [PubMed] [Google Scholar]
- 30.Molinari M, Helenius A. Methods Enzymol. 2002;348:35–42. doi: 10.1016/s0076-6879(02)48623-5. [DOI] [PubMed] [Google Scholar]
- 31.Jansens A, van Duijn E, Braakman I. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- 32.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JCA. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 33.Sinha NK, Light A. J Biol Chem. 1975;250:8624–8629. [PubMed] [Google Scholar]
- 34.Odorzynski TW, Light A. J Biol Chem. 1979;254:4291–4295. [PubMed] [Google Scholar]
- 35.Chaudhry C, Farr GW, Todd MJ, Rye HS, Brunger AT, Adams PD, Horwich AL, Sigler PB. EMBO J. 2003;2:4877–4887. doi: 10.1093/emboj/cdg477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 37.Higaki JN, Light A. Anal Biochem. 1985;148:111–120. doi: 10.1016/0003-2697(85)90635-9. [DOI] [PubMed] [Google Scholar]
- 38.Creighton TE. J Mol Biol. 1975;96:767–776. doi: 10.1016/0022-2836(75)90151-5. [DOI] [PubMed] [Google Scholar]
- 39.Bolognesi M, Gatti G, Menegatti E, Guarneri M, Marquart M, Papamobos E, Huber R. J Mol Biol. 1982;162:839–868. doi: 10.1016/0022-2836(82)90550-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.