Abstract
The lack of myostatin promotes growth of skeletal muscle, and blockade of its activity has been proposed as a treatment for various muscle-wasting disorders. Here, we have examined two independent mouse lines that harbor mutations in the myostatin gene, constitutive null (Mstn−/−) and compact (Berlin High Line, BEHc/c). We report that, despite a larger muscle mass relative to age-matched wild types, there was no increase in maximum tetanic force generation, but that when expressed as a function of muscle size (specific force), muscles of myostatin-deficient mice were weaker than wild-type muscles. In addition, Mstn−/− muscle contracted and relaxed faster during a single twitch and had a marked increase in the number of type IIb fibers relative to wild-type controls. This change was also accompanied by a significant increase in type IIB fibers containing tubular aggregates. Moreover, the ratio of mitochondrial DNA to nuclear DNA and mitochondria number were decreased in myostatin-deficient muscle, suggesting a mitochondrial depletion. Overall, our results suggest that lack of myostatin compromises force production in association with loss of oxidative characteristics of skeletal muscle.
Keywords: dystrophy, histology, mitochondria, physiology
Lack of myostatin function results in the excessive growth of skeletal muscle, demonstrating the existence of a powerful mechanism to control muscle size in normal individuals (1). The myostatin gene encodes a member of the TGF-β family of signaling molecules and has been highly conserved throughout vertebrate evolution (2). This finding, together with the extremely rare incidence of spontaneous mutations within the gene (3, 4), points to biological advantage and associated evolutionary constraints on muscle size by this pathway. In some respects, this is paradoxical, because muscularity has been positively associated with vigor and reproductive fitness. Such views may have neglected a critical evaluation of the functional aspects of muscle hypertrophy induced by the absence of myostatin. Doubts of this sort are indirectly supported by the observation that this increase in muscle mass is not accompanied by a proportionate increase in muscle force (5, 6). Furthermore, cattle with hereditary muscular hypertrophy (double-muscled cattle), many of which have been shown to harbor mutations in the myostatin (Mstn) gene, are actually prone to muscle damage after mild exercise (7–10). It is reported, however, that myostatin-deficient mice do not suffer from muscle fiber damage when subjected to brief periods of exercise (11).
Targeted inactivation or antibody blocking of myostatin in the dystrophin-deficient mdx mice, a model of Duchenne muscular dystrophy, demonstrated that dystrophic muscle could indeed be stimulated to grow (5, 12, 13). The effect on force development in these animal models differed depending on the methods of myostatin blockade. Whereas specific force output in mdx mice remained decreased after treatment with anti-myostatin antibody, treatment with stabilized myostatin propeptide resulted in an increase of specific force output (5, 12, 13). On a discordant note, targeted inactivation of myostatin in the dyw/dyw mouse, the animal model for merosin-deficient congenital muscular dystrophy, revealed no improvement of the muscle pathology and actually increased postnatal mortality (14).
Despite these uncertainties, the use of specific antibodies to block myostatin has now been proposed as a new therapeutic strategy to stimulate muscle growth, and a multicenter clinical trial on muscular dystrophy patients is in process. In this report, we analyzed the functional and structural characteristics of muscle from myostatin-null mice. Importantly, the main body of our study has been performed on the same line originally used to describe the myostatin knockout phenotype and to confer the beneficial value of blocking myostatin function in muscle-wasting disorders (1, 5).
Results
Decreased Specific Force Generation in the Myostatin Knockout Mouse.
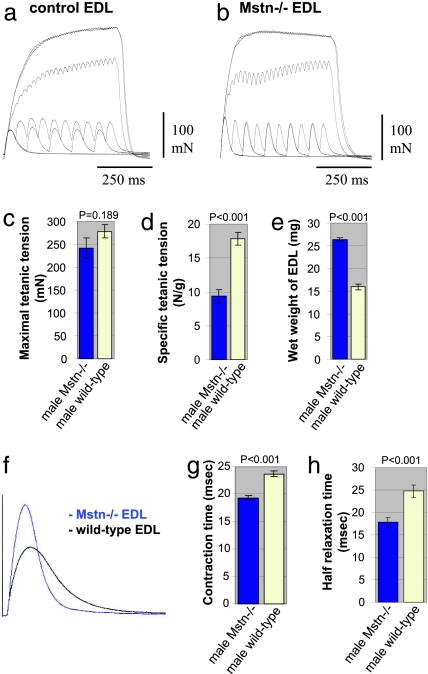
To examine the relationship between muscle size and improved function in Mstn−/− mice, we first performed a series of tests of force generation. The maximal isometric twitch force (Pt) and tetanic force (Po) of extensor digitorum longus (EDL) muscles of 7-month-old male Mstn−/− and age-matched C57BL/6 wild-type mice are presented in Table 1 and Fig. 1. Maximal Po was reached at 100 Hz in both Mstn−/− and C57BL/6 wild-type muscles and did not increase with stimulation at higher frequencies. We therefore compared values obtained at 100 Hz from wild-type and Mstn−/− mice (Fig. 1 a and b). Despite the increased muscle bulk in the myostatin-deficient animals, maximal Po was similar to that of wild-type mice of the same genetic background (P = 0.189; Table 1; Fig. 1 c and e). Moreover, when maximal Po was normalized for muscle weight (specific force), we found that Mstn−/− male mice generated only 53% of the force developed by wild-type animals (P < 0.001; Table 1; Fig. 1d). Similarly, normalization of Po for the cross-sectional area also revealed a reduction in specific force generation (P = 0.010; Table 1).
Table 1.
Physiological and morphometric properties of Mstn−/− and C57BL/6 wild-type EDL muscles
| Gender (age, months) | Mstn−/− EDL | C57BL/6 wild-type EDL | P* | |
|---|---|---|---|---|
| Maximal tetanic tension, mN | Male (7) | 242 ± 22 (14) | 278 ± 14 (12) | 0.189 |
| Male (2) | 164 ± 4 (3) | 166 ± 1 (4) | 0.5 | |
| Female (10) | 251 ± 18 (6) | 297 ± 12 (4) | 0.106 | |
| Specific tetanic tension, N/g | Male (7) | 9.4 ± 0.99 (13) | 17.8 ± 0.92 (9) | <0.001 |
| Male (2) | 8.9 ± 0.63 (3) | 17.2 ± 0.99 (4) | 0.001 | |
| Female (10) | 11.0 ± 0.92 (6) | 27.2 ± 1.45 (4) | <0.001 | |
| Specific tetanic tension, N/mm2 | Male (7) | 80.4 ± 14.5 (5) | 135.2 ± 7.8 (5) | 0.010 |
| Maximal twitch force, mN | Male (7) | 65.6 ± 5.46 (14) | 57.5 ± 2.81 (12) | 0.204 |
| Twitch/tetanus ratio | Male (7) | 0.275 ± 0.035 (14) | 0.21 ± 0.03 (12) | <0.001 |
| Contraction time, ms | Male (7) | 19.4 ± 0.33 (14) | 23.7 ± 0.56 (12) | <0.001 |
| Relaxation time, ms | Male (7) | 17.8 ± 1.10 (14) | 24.8 ± 1.40 (12) | 0.001 |
| Wet weight, mg | Male (7) | 26.4 ± 0.40 (13) | 15.9 ± 0.68 (9) | <0.001 |
| Male (2) | 18.6 ± 0.90 (3) | 9.7 ± 0.48 (4) | 0.003 | |
| Female (10) | 23.1 ± 0.79 (6) | 10.9 ± 0.21 (6) | <0.001 | |
| CSA, mm2 | Male (7) | 3.17 ± 0.35 (5) | 2.07 ± 0.06 (5) | 0.035 |
| Mitochondria number per unit area | Male (2) | 30.3 ± 0.55 (4) | 41.3 ± 0.69 (4) | <0.001 |
Values are given as means together with mean standard error, and number of muscles examined is given in parentheses. For statistical analysis, the unpaired t test (*) was used. P < 0.05 was considered significant.
Fig. 1.
Physiological properties and size of EDL muscles from male adult Mstn−/− mice compared with age-matched C57BL/6 wild types. (a and b) Records of superimposed isometric contraction in response to direct muscle stimulation at 1, 10, 20, 50, 100, and 200 Hz are shown for EDL muscles from wild-type (a) and Mstn−/− (b) mice. (c) Maximal Po of EDL muscles from Mstn−/− mice (blue column) compared with wild types (yellow column) (P = 0.189). (d) Maximal Po expressed as a function of muscle weight of EDL muscles from Mstn−/− mice (blue column) compared with wild types (yellow column) (P < 0.001). (e) Wet weight of EDL muscles that were measured after force testing from Mstn−/− mice (blue column) compared with wild types (yellow column) (P < 0.001). (f) Records of isometric twitch contraction of EDL muscles from a Mstn−/− mouse (blue trace) and a wild-type mouse (black trace). (g) Contraction times after a single twitch stimulation of EDL muscles from Mstn−/− mice (blue column) compared with wild types (yellow column) (P < 0.001). (h) Relaxation times after a single twitch stimulation of EDL muscles from Mstn−/− mice (blue column) compared with wild types (yellow column) (P < 0.001).
The weight of 7-month-old male Mstn−/− EDL muscle was increased by 66% over wild-type values (P < 0.001; Table 1; Fig. 1e), and the total cross-sectional area of the Mstn−/− EDL exceeded control values by 53% (P = 0.035; Table 1).
We also performed contraction measurements on EDL muscles from 10-month-old female Mstn−/− mice and found a similar maximal Po (P = 0.106) and a marked reduction in specific force (P < 0.001) despite a >2-fold higher muscle mass (P < 0.001) than in age-matched female wild types (Table 1). Maximum and specific tetanic force was similar in male and female Mstn−/− mice.
Force measurements were performed on EDL muscles from young animals (2 months of age) and again gave similar maximal Po for the two genotypes (P = 0.5; Table 1). The wet weight of homozygous EDL muscle was increased to 191% compared with wild types (P = 0.003; Table 1), thus resulting in a decreased specific force for homozygous muscle (P = 0.001; Table 1), confirming data obtained from older mice. Reduction in specific force was 52% for 2-month-old and 53% for 7-month-old Mstn−/− male mice compared with age-matched male wild-type mice.
Additionally, muscles of the Mstn−/− EDL showed a decay of tension during tetanic stimulation at 100 and 200 Hz, whereas in the wild-type EDL, there was an increase in tension over the stimulation period (Fig. 1 a and b). Moreover, the maximal twitch force was slightly higher in the Mstn−/− than the wild-type EDL (P = 0.204), but the maximal tetanic force was slightly lower than the wild-type EDL (Table 1; Fig. 1f), resulting in a significant alteration of the twitch/tetanus ratio of 0.27 for Mstn−/− compared with 0.21 for wild-type EDL (Table 1).
Male Mstn−/− EDL muscles also showed significantly shorter time to peak tension (contraction time) (P < 0.001) and shorter time to half relaxation than wild-type muscles (P = 0.001; Table 1; Fig. 1 f–h). Similar results were also observed in female Mstn−/− vs. wild-type muscles (data not shown).
Decreased Specific Force Generation in the compact Mouse.
Essentially similar results were obtained from force measurements on skeletal muscle from the compact (Berlin High Line, BEH(C/C)) mouse that displays hypermuscularity but differs from Mstn−/− in both its genetic background and the nature of the mutation in the myostatin gene. We found a slightly higher maximal Po in EDL muscles from the 2-month-old homozygous compact females (BEH(C/C)), albeit statistically not significant (P = 0.64), which converted to a significant reduction in specific force (P < 0.025), allowing for the 2-fold higher muscle mass (P < 0.05) compared with EDL muscles from their wild-type controls [Table 2 and supporting information (SI) Fig. 5]. Thus, both Mstn−/− and compact mice have significantly smaller specific force values as compared with their wild-type littermates.
Table 2.
Physiological properties of EDL muscles from 2-month-old female homozygous compact (BEHC/C) and wild- type (BEH+/+) mice
| C/C | +/+ | P* | |
|---|---|---|---|
| Maximal tetanic tension, mN | 254 ± 32 (3) | 216 ± 8.6 (3) | 0.64 |
| Specific tetanic tension, N/g | 11.9 ± 2.1 (3) | 19.5 ± 1.0 (3) | 0.025 |
| Wet weight, mg | 21.7 ± 1.0 (3) | 11.2 ± 1.0 (3) | <0.005 |
Values are given as means together with mean standard error; the number of muscles examined is given in parentheses. For statistical analysis, the unpaired t test (*) was used. P < 0.05 was considered significant.
Lack of Myostatin Predisposes to Differentiation to Type IIB Fibers.
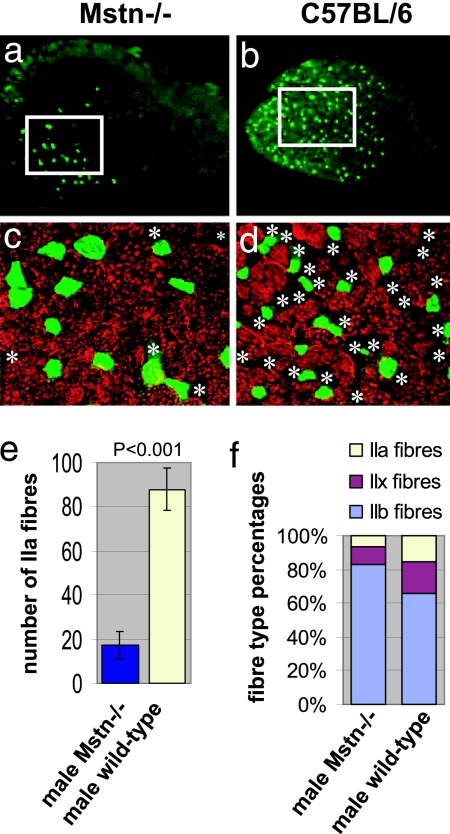
Functional studies revealed a shortening in contraction and relaxation time in Mstn−/− muscle. We therefore investigated the fiber-type distribution of the EDL muscle (Fig. 2), finding very few type I fibers (slow fibers) in either Mstn−/− or C57BL/6 wild-type muscles (data not shown). The total number of IIa fibers on midbelly sections was five times lower in the Mstn−/− EDLs than in wild-type mice (P < 0.001; Fig. 2e). IIa fibers were generally segregated in the deeper region of the muscle. Therefore, we determined the fiber-type distribution at this site (Fig. 2 a–d). We counted IIa, IIb, and non-IIa/non-IIb fibers in this region and expressed the distribution as percentage of relative frequency. Fibers negative for both isoforms were considered nonhybrid IIx fibers. We found a reduction in IIa and IIx fibers and a concomitant increase in IIb fibers in Mstn−/− EDL muscles (Fig. 2f). This shift in the fiber-type distribution in the myostatin-deficient EDL muscle is consistent with the shortening of contraction and relaxation time.
Fig. 2.
Muscle fiber-type profiles in EDL muscles from male Mstn−/−- and C57BL/6 wild-type mice. (a and b) Transverse sections of whole EDL (5× objective). The green fluorescence in a shows expression of MHC IIa in only a few fibers of Mstn−/− EDL compared with C57BL/6 wild-type EDL shown in b. IIa fibers can be seen to predominate in the deep muscle regions of the muscles, which are shown on the left side of the images. The white frames show areas of deep muscle region where the fiber type distribution was analyzed. The regions outlined in a and b are shown enlarged in c and d and show an overlay of staining for MHC IIa (green) and MHC IIb (red) by using ×20 objective. Asterisks depict non-IIa/non-IIb fibers. (e) Total number of IIa fibers in the whole EDL muscle sections from Mstn−/− mice (blue column) compared with wild types (yellow column) (P < 0.001). (f) Histogram showing the fiber-type distribution in the deep region of the EDL muscles. Fibers expressing MHC IIa and IIb and fibers negative for IIa/IIb were counted. Non-IIa/non-IIb fibers were considered as IIx fibers because there were hardly any fibers expressing slow MHC.
Oxidative Enzymes Are Decreased in Mstn−/− Muscle.
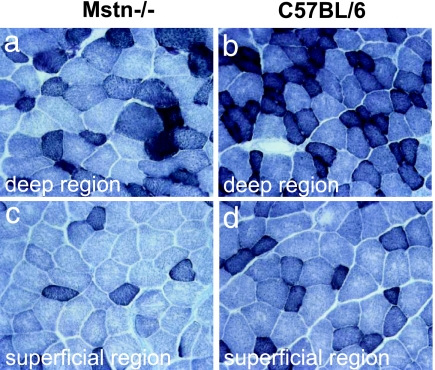
In view of the fiber-type bias toward fast glycolytic type IIb fibers, suggesting a concomitant decrease in mitochondrial activity, we performed staining for the mitochondrial enzymes succinate dehydrogenase (SDH), cytochrome oxidase (COX), and NADH reductase on sections of EDL muscles. Staining for SDH activity revealed markedly fewer strongly stained fibers (high activity) and an increase in a larger proportion of very pale fibers (low activity) in Mstn−/− than in wild-type mice (Fig. 3). Similarly, fibers from Mstn−/− EDL muscles showed a lower activity for COX and NADH paralleling that for SDH (data not shown).
Fig. 3.
Oxidative properties of adult EDL muscles from 7-month-old male Mstn−/− mice relative to age-matched C57BL/6 wild types. SDH staining of Mstn−/− EDL (a and c) and of control EDL (b and d) in the deep (a and b, respectively) and superficial (c and d, respectively) regions. Darkly stained fibers contain high SDH activity, and pale-stained fibers contain low SDH activity.
However, SDH activity in muscle from Mstn−/− mice seemed generally lower compared with control muscle and was difficult to explain solely by a fiber-type switch toward fast glycolytic fibers (Fig. 3).
mtDNA Depletion and Decreased Mitochondria Number in Mstn−/− Muscle.
The above results suggested a decreased number of mitochondria in myostatin-deficient muscle. We therefore determined the ratio of the number of mtDNA (MT-CO1) copies per single-copy nuclear gene (Ndufv1) by quantitative real-time PCR with gene-specific probes (SI Fig. 6). This assay has been controlled on rho0-cells, which are devoid of mtDNA, and did not yield a signal for the MT-CO1 probe (data not shown), thus excluding the possibility of amplification of nuclear pseudogenes. Half the copy number of the nuclear autosomal single-copy gene can thus be equaled with the number of myonuclei. In wild-type muscle, despite a different fiber-type composition of EDL (fast muscle) and soleus (slow muscle), the mtDNA/myonucleus ratios were similar between these two muscles (209.8 and 200.2, respectively; P = 0.754), suggesting that different fiber types contain about same number of mitochondria per myonucleus. Interestingly, we found a significantly lower mtDNA/myonucleus ratio in muscle from Mstn−/− mice compared with wild-type muscle (from 205.0 to 104.8; P = 0.007, if EDL and soleus were taken together). The decrease was more pronounced in EDL muscle (from average 209.8 to 75.2 P = 0.014) as compared with soleus muscle (from 200.2 to 134.4, P = 0.027), thus suggesting a mitochondrial depletion in muscle from Mstn−/− mice independent of fiber-type composition.
We additionally determined the number of mitochondria in muscle fibers of the EDL muscle from 2-month-old male Mstn−/− mice and male C57BL/6 wild-type mice by analyzing electron micrographs taken at the midbelly region. We found that the myofibers from male Mstn−/− mice, on average, had 30.3 mitochondria per unit area compared with 41.3 mitochondria for wild-type mice (P < 0.001; 455 fibers from four EDL muscles from Mstn−/− mice; 401 fibers from four EDL muscles from C57BL/6 wild-type mice; Table 1), thus confirming mitochondrial depletion in muscle from Mstn−/− mice.
Muscle from Mstn−/− Mice Accumulates Tubular Aggregates.
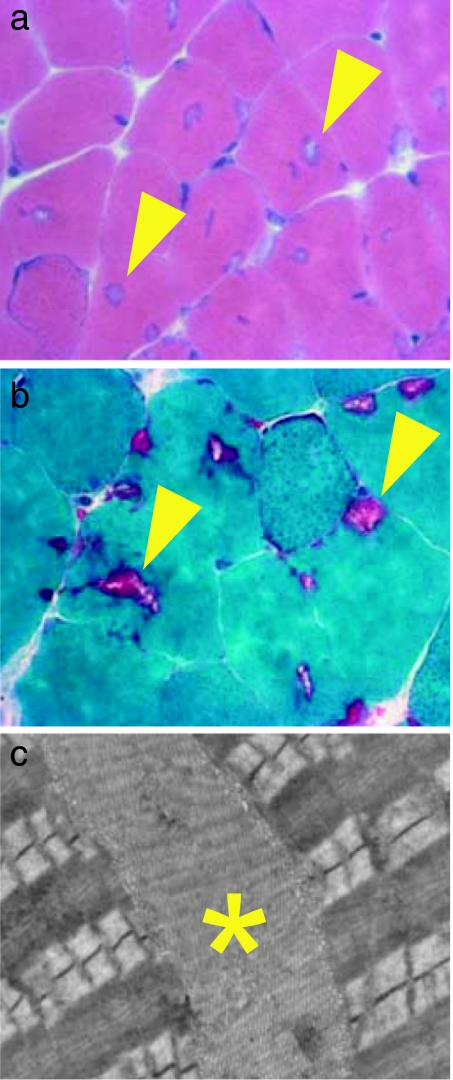
Histological examination by H&E and Gomori's trichrome stains revealed many cytoplasmic inclusions in the EDL of both male (7 months old; Fig. 4a and b) and female (10 months old) Mstn−/− mice. These were rare in age-matched C57BL/6 wild-type males and were never detected in wild-type females (SI Fig. 7). We also found similar inclusions in the tibialis anterior and gastrocnemius muscles, both of which contain a high proportion of type IIb fibers, but not in the soleus muscle (data not shown). Cytoplasmic inclusions were found predominantly in superficial regions of these muscles, which contain mainly IIb fibers. Electron microscopy revealed them to be a membranous accumulation of tubules and saccular dilatations, which defined them as tubular aggregates (Fig. 4c).
Fig. 4.
Histological analysis of tubular aggregates from adult EDL muscles of Mstn−/− mice. (a) Transverse sections of the EDL muscles from a 7-month-old male Mstn−/− mouse after staining with H&E showing the presence of numerous cytoplasmic inclusions (arrowheads) in many muscle fibers. (b) Transverse sections of the EDL from a 7-month-old male Mstn−/− mouse after Gomori's trichrome staining showing cytoplasmic inclusions in purple color (arrowheads). (c) Electron micrograph taken from an EDL of a 7-month-old male Mstn−/− mouse showing stacks of tubular and saccular formations (∗), which define tubular aggregates, between the contractile apparatus.
Cytoplasmic inclusions were not present in the EDL muscles from 2-month-old Mstn−/− mice, suggesting that they accumulated over time. Moreover, cytoplasmic inclusions were not present in EDL muscles from 2-month-old compact mice (BEHC/C) examined in this study (data not shown).
Immunolabeling showed that these inclusions accumulated SERCA1, the calcium-release channel of the sarcoplasmic reticulum of fast muscle fibers (SI Fig. 7). However, they were largely devoid of the dihydroxypyridine receptor (DHPR) of the transverse tubules desmin or myosin heavy chain, when stained with an antibody that detects all forms of MHC (data not shown). Triple staining for MHC type IIb, SERCA 1, and laminin-γ1 confirmed the presence of cytoplasmic inclusions in IIb fibers only (SI Fig. 7 and data not shown). Other abnormalities such as fiber necrosis or centrally located nuclei were rarely seen in the muscle of Mstn−/− or wild-type mice.
The frequency of tubular aggregates showed no correlation with specific force generation in the Mstn−/− mice (data not shown).
Discussion
In this study, we have investigated the functional and cellular characteristics of enlarged skeletal muscle from mouse lines with mutations in the myostatin gene (Mstn−/− and BEHC/C). In accord with other reports, we found that a deficiency in myostatin resulted in increased muscle mass (1, 6, 15–18), but that this increase was not accompanied by a proportionate increase in force generation. It follows that specific tetanic tension was significantly decreased when normalized for muscle size, which was observed in both mouse lines. These findings clearly indicate that the increase in muscle mass of myostatin mutant animals confers no strength advantage over wild-type controls.
Although previous studies on the Mstn−/− mouse strain and on wild-type mice that were treated with antimyostatin antibodies revealed an increase in grip strength (5, 6, 11), the increase in force was disproportionately low when compared with the increase in muscle mass, suggesting a decrease in specific force generation. In light of our results, it is surprising that a blockade of myostatin in the mdx mouse, the animal model of Duchenne muscular dystrophy actually increased the total as well as specific force output (5, 12, 13).
A relevant comparison may be made with another form of inhibition of the myostatin-signaling pathway resulting in a similar physiological phenotype to the myostatin mutants. Overexpression of ski in MSVski transgenic mice resulted in Type IIb fiber hypertrophy accompanied by a 30% decrease in specific force generation (19). Ski negatively regulates Smad phosphorylation (20–22), thereby inhibiting signaling of TGF-β-like factors, such as myostatin. These findings contrast with muscle-specific overexpression of insulin-like growth factor 1, where fiber hypertrophy is accompanied by increased maximum force generation and maintained specific force levels (23). One may suggest that massive hypertrophy simply could alter the angle of pull on the muscle fibers during contraction, thereby reducing the ability to generate a higher force output. However, that hypertrophy increases force output after overexpression of IGF-1, but not after myostatin knockout, points to additional factors that cause the force problems in a lack of myostatin.
In agreement with previous findings on the Mstn−/− mouse line (24), we found a predominance of type IIb fibers in the EDL muscle from Mstn−/− mice together with a marked reduction in types IIa and IIx fibers, as well as preliminary evidence of a similar IIb fiber predominance for tibialis anterior and gastrocnemius muscles. We also noted a substantial deficit in the activity of oxidative enzymes in myostatin-deficient EDL muscle, in accord with the high proportion of fast glycolytic fibers that, in general, contain less mitochondria. However, the extent of loss of activity of oxidative enzymes is difficult to explain solely by a fiber-type switch toward fast glycolytic fibers. We found that the mtDNA copy number per myonucleus in muscles from Mstn−/− mice was lower than that seen even in fast glycolytic fibers of wild-type muscle, and we found a decreased number of mitochondria in muscles from Mstn−/− mice. We conclude that these findings truly reflect a mitochondrial depletion that cannot be explained by a simple switch of fiber types, and our data suggest a marked diminution in mitochondria per unit volume of cytoplasm.
This finding, together with previous reports of reduced capillary density in myostatin-deficient muscle (18), prompts the idea that myostatin may function to optimize aerobic metabolism in skeletal muscle. It has been reported that blockade of mitochondrial respiration in skeletal muscle resulted in decreased tetanic force generation (25, 26), and in patients with mitochondrial depletion syndrome, muscle weakness is one leading clinical feature (26, 27). Thus, the low specific force in hypertrophic muscle due to lack of myostatin, may be attributable to the associated mitochondrial depletion.
Our results do not specify whether the altered fiber-type composition in muscle of Mstn−/− mice arises during development, or whether fibers convert later in life. Nor is it evident whether they represent a direct effect on the muscle or whether the firing patterns of the innervating motorneurons are implicated (28), a topic for further investigation. Interestingly, a similar bias toward glycolytic myofiber phenotype has also been found in cattle with hereditary muscle hypertrophy associated with mutation of the myostatin gene (29). This predominance of fast glycolytic fibers may well explain the increase in contraction speed and shortening in relaxation time in myostatin-deficient muscle, but, given the normal association of IIb fibers with high force production, it is surprising that maximal stimulation hardly changed force production. Thus, Mstn−/− muscle does not seem to benefit from the high tetanic force generation to be expected of glycolytic fibers, and the mitochondrial depletion would suggest increased fatigability and exercise intolerance, a mark of derangement rather than orderly change in the differentiation process.
This view is sustained by the observed accumulation with age of tubular aggregates composed of sarcoplasmic reticulum components and located exclusively in type IIb fibers in Mstn−/− muscle. This feature has not been reported (1, 5, 6, 11). Although their presence may be associated with the source of the physiological problems, our data do not implicate tubular aggregates as the primary cause of reduced force generation. Thus, both young mice of Mstn−/− and compact mice (BEHC/C) genotypes exhibited low specific force production before they had developed tubular aggregates, and female Mstn−/− mice produced far fewer tubular aggregates than males but exhibited a low specific force output very similar to that of males. Moreover, within the male group, there was no correlation between the number of myofibers containing cytoplasmic inclusions and specific force output. Again, comparison is invited with the MSVski transgenic mouse model of fiber hypertrophy, which also exhibits a decrease of specific force generation in combination with increased and aggregated sarcoplasmic reticulum (30). It seems likely that aberrations of physiological function of sarcoplasmic reticular organization both arise, perhaps independently, as a result of physiological stresses associated with dysregulated fiber hypertrophy. One such explanation that springs to mind is that the disturbances may reflect different aspects of problems in calcium handling. The decay in tension during tetanic stimulation together with the increased maximum isomeric twitch force (Pt)/Po ratio and the proliferation of sarcoplasmic reticulum may both be mechanistically linked to an altered calcium homeostasis in myostatin-deficient muscle. This hypothesis fits well, too, with the dearth of mitochondria, which can act as fast calcium sinks in skeletal muscle (31) as well as providing ATP for calcium uptake into the sarcoplasmic reticulum; their lack in Mstn−/− might well stimulate a compensatory formation of the sarcoplasmic reticulum. Intriguingly, small numbers of tubular aggregates are found in muscles of older male wild-type mice of the reference wild type used here and of various inbred and outbred laboratory mouse strains (32, 33) but not in wild-type females, which correlates with the higher level of myostatin in wild-type female than male muscle (34).
Our twitch-force testings revealed an increase in contraction speed and shortening in relaxation time in myostatin-deficient muscle, which may sufficiently be explained by the fiber-type conversion toward fast glycolytic fibers observed in myostatin-deficient muscle. Alternatively, differences in the connective tissue between Mstn−/− and wild-type mice may lead to a more rapid uptake of the series elastic component and may act as an additional contributing factor to induce the more rapid twitch kinetics. Additionally, the expanded sarcoplasmic reticulum that morphologically became evident in the form of tubular aggregates might be responsible for a more rapid release and reuptake of calcium, thus accounting for the differences in twitch kinetics. However, its exact role remains to be determined.
Current interest in myostatin centers on the potential of therapeutic blockade of this cytokine to increase the size and strength of muscle as a means of counteracting muscle weakness associated with sarcopenia and muscular dystrophy. Although a number of studies have demonstrated some benefit from this strategy in mice carrying the mdx muscular dystrophy, the basis of the clinical improvements observed is unclear, and the experimental regimes have been fairly short term. Nonetheless, human trials of myostatin-blocking agents are being conducted currently. Our data on nonmyopathic mice, while not directly contradicting previous work on the mdx mouse, do raise questions as to the precise biological effects of myostatin blockade and suggest an urgent need for further elucidation of the mechanisms of muscle growth and differentiation that are affected by such treatments.
Materials and Methods
Animals.
Mstn−/+ founder breeding pairs on a C57BL/6 background were a kind gift of Se-Jin Lee (Johns Hopkins University, Baltimore, MD) (1). Myostatin knockout and wild-type mice (C57BL/6) were bred in the animal facilities of Royal Veterinary College. After the establishment of the colony, mice were screened and shown to be free of common rodent infectious diseases. Studies were performed on 2- and 7-month-old male Mstn−/− and male C57BL/6 mice and 10-month-old female Mstn−/− and female C57BL/6 mice unless otherwise stated. All investigations on animals carried out for this study had prior approval of the institutional ethics approval committee (Royal Veterinary College). Animals were kept under guidelines set out by the Home Office (United Kingdom). Animals were kept under license PPL/70/5218 (Home Office, United Kingdom).
Additionally, mice from a subline of the BEH have been used (35). The BEH line is homozygous for the compact mutation (36) and is coded as MstnCompt−dl1ABC (37, 38). Mice from the BEH line were crossed with mice from the Berlin Low line (BEL) to introduce the wild type back into the BEH mice, to allow segregation of the wild-type and the compact allele in this subline, coded BEHC/+ (35). Heterozygous mice from BEHC/+ have been repeatedly backcrossed to mice from the BEH line to reestablish the high growth background of this line, which had been long-term selected for high growth (35). The mice used here were sampled after completion of five backcross generations (including F1), which had on average 98.44% of genetic background of the BEH line reestablished. Heterozygous mice of this generation were mated inter se to produce the experimental animals for this study [all three genotypes: homozygous compact (BEHC/C), heterozygous (BEHC/+), and homozygous for wild type (BEH+/+)].
Physiological Studies.
All functional studies were performed in vitro on EDL muscles. The time to reach peak tension (contraction time) and the half-relaxation time (time between maximum and half-maximum force during the relaxation phase of the twitch, t½r) were determined. The maximal Po was determined from the plateau of the frequency-force relationship. For detailed information, see SI Table 3.
Relative Quantification of mtDNA Copy Numbers.
Four male Mstn−/− and five male C57BL/6 control mice were killed by cervical dislocation at 7 weeks. The EDL and soleus muscles of the right leg were immediately explanted in toto and flash-frozen in liquid nitrogen. DNA was extracted, and quantifications of the mtDNA encoded gene MT-CO1 (GenBank accession no. NC_001807) and of the single-copy nuclear gene Ndufv1 (GenBank accession no. NM_133666) were performed by using TaqMan chemistry (Applied Biosystems, Weiterstadt, Germany). A detailed protocol can be viewed in SI Text.
Histology.
Each EDL muscle was weighed and then mounted in OCT (VWR, Poole, U.K.) and frozen in melting isopentane cooled in liquid nitrogen. Ten-micrometer transverse sections from the midbelly region were cut on a cryostat. Serial sections were stained for H&E, modified Gomori's trichrome, SDH, NADH–tetrazolium, and cytochrome oxidase.
Immunohistochemistry.
A list of antibodies and concentrations used is in SI Text. Double staining was performed to depict expression of myosin heavy chain (MHC) IIa and IIb. Triple staining was performed to depict expression of MHC IIb, laminin-γ1, and SERCA1 or DHPR. For detailed immunohistochemistry protocols, see SI Table 3.
Electron Microscopy.
Samples (1 mm3) were initially fixed in 4% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 1.5–2 h, followed by washing in buffer for 30 min and fixation for 1 h in 1% osmium tetroxide in 0.1 M cacodylate buffer. Samples were then dehydrated and infiltrated with propylene oxide followed by embedding in araldite. Semithin (1 μm) and ultrathin (70-nm) sections were cut by using a Leica Ultracut (Leica, Bensheim, Germany). Sections were stained with uranyl acetate and contrasted with lead citrate according to standard protocols and viewed on Jeol JEM-1011 (JEOL, Tokyo, Japan). The mitochondria number was determined by using a point-counting method on micrographs taken at ×3000 magnification according to previously published methods (39, 40).
Image Analysis.
The total cross-sectional area was determined from H&E-stained transverse sections from the midbelly region of EDL muscles by using imaging software (Leica QWin).
Number of Experiments.
The number of each experiment is listed in SI Text.
Supplementary Material
Acknowledgments
We thank Elaine Shervill, Helen Smith, Lucy Feng, and Angelika Zwirner for excellent technical assistance. We are indebted to Prof. Se-Jin Lee for providing founder mice for our Mstn−/− colony and to Dr. Simon Hughes (King's College London) for providing antibodies. This work was funded by MDA USA (Grant MDA3870, to H.A.), the Wellcome Trust (Grants 066195 and 078649, to R.M. and K.P.), and l'Association Monégasque Contre les Myopathies and Duchenne Parent Project France (M.S.).
Abbreviations
- EDL
extensor digitorum longus
- Po
maximal tetanic tension
- SDH
succinate dehydrogenase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at http://www.pnas.org/cgi/content/full/0604893104/DC1.
References
- 1.McPherron AC, Lawler AM, Lee SJ. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee SJ. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- 3.Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. N Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 4.Ferrell RE, Conte V, Lawrence EC, Roth SM, Hagberg JM, Hurley BF. Genomics. 1999;62:203–207. doi: 10.1006/geno.1999.5984. [DOI] [PubMed] [Google Scholar]
- 5.Wagner KR, McPherron AC, Winik N, Lee SJ. Ann Neurol. 2002;52:832–836. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- 6.Whittemore LA, Song K, Li X, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A, et al. Biochem Biophys Res Commun. 2003;300:965–971. doi: 10.1016/s0006-291x(02)02953-4. [DOI] [PubMed] [Google Scholar]
- 7.Holmes JH, Ashmore CR, Robinson DW. J Anim Sci. 1973;36:684–694. doi: 10.2527/jas1973.364684x. [DOI] [PubMed] [Google Scholar]
- 8.McPherron AC, Lee SJ. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, et al. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 10.Marchitelli C, Savarese MC, Crisa A, Nardone A, Marsan PA, Valentini A. Mamm Genome. 2003;14:392–395. doi: 10.1007/s00335-002-2176-5. [DOI] [PubMed] [Google Scholar]
- 11.Wagner KR, Liu X, Chang X, Allen RE. Proc Natl Acad Sci USA. 2005;102:2519–2524. doi: 10.1073/pnas.0408729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- 13.Bogdanovich S, Perkins KJ, Krag TO, Whittemore LA, Khurana TS. FASEB J. 2005;19:543–549. doi: 10.1096/fj.04-2796com. [DOI] [PubMed] [Google Scholar]
- 14.Li ZF, Shelton GD, Engvall E. Am J Pathol. 2005;166:491–497. doi: 10.1016/S0002-9440(10)62271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Hadhazy M, Wehling M, Tidball JG, McNally EM. FEBS Lett. 2000;474:71–75. doi: 10.1016/s0014-5793(00)01570-2. [DOI] [PubMed] [Google Scholar]
- 16.Grobet L, Pirottin D, Farnir F, Poncelet D, Royo LJ, Brouwers B, Christians E, Desmecht D, Coignoul F, Kahn R, Georges M. Genesis. 2003;35:227–238. doi: 10.1002/gene.10188. [DOI] [PubMed] [Google Scholar]
- 17.Wolfman NM, McPherron AC, Pappano WN, Davies MV, Song K, Tomkinson KN, Wright JF, Zhao L, Sebald SM, Greenspan DS, Lee SJ. Proc Natl Acad Sci USA. 2003;100:15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehfeldt C, Ott G, Gerrard DE, Varga L, Schlote W, Williams JL, Renne U, Bunger L. J Muscle Res Cell Motil. 2005;26:103–112. doi: 10.1007/s10974-005-1099-7. [DOI] [PubMed] [Google Scholar]
- 19.Charge SB, Brack AS, Hughes SM. Am J Physiol. 2002;283:C1228–C1241. doi: 10.1152/ajpcell.00206.2002. [DOI] [PubMed] [Google Scholar]
- 20.Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Liu X, Eaton EN, Lane WS, Lodish HF, Weinberg RA. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- 23.Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N. Nat Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- 24.Girgenrath S, Song K, Whittemore LA. Muscle Nerve. 2004;31:34–40. doi: 10.1002/mus.20175. [DOI] [PubMed] [Google Scholar]
- 25.Zhang SJ, Bruton JD, Katz A, Westerblad H. J Physiol. 2006;572:551–559. doi: 10.1113/jphysiol.2005.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancuso M, Salviati L, Sacconi S, Otaegui D, Camano P, Marina A, Bacman S, Moraes CT, Carlo JR, Garcia M, et al. Neurology. 2002;59:1197–1202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- 27.Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, Schon EA, DiMauro S. Am J Hum Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- 28.Pette D, Vrbova G. Muscle Nerve. 1999;22:666–677. doi: 10.1002/(sici)1097-4598(199906)22:6<666::aid-mus3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 29.Wegner J, Albrecht E, Fiedler I, Teuscher F, Papstein HJ, Ender K. J Anim Sci. 2000;78:1485–1496. doi: 10.2527/2000.7861485x. [DOI] [PubMed] [Google Scholar]
- 30.Bruusgaard JC, Brack AS, Hughes SM, Gundersen K. Acta Physiol Scand. 2005;185:141–149. doi: 10.1111/j.1365-201X.2005.01462.x. [DOI] [PubMed] [Google Scholar]
- 31.Andrade FH, McMullen CA, Rumbaut RE. Invest Ophthalmol Visual Sci. 2005;46:4541–4547. doi: 10.1167/iovs.05-0809. [DOI] [PubMed] [Google Scholar]
- 32.Agbulut O, Destombes J, Thiesson D, Butler-Browne G. Histochem Cell Biol. 2000;114:477–481. doi: 10.1007/s004180000211. [DOI] [PubMed] [Google Scholar]
- 33.Chevessier F, Marty I, Paturneau-Jouas M, Hantai D, Verdiere-Sahuque M. Neuromuscul Disord. 2004;14:208–216. doi: 10.1016/j.nmd.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 34.McMahon CD, Popovic L, Jeanplong F, Oldham JM, Kirk SP, Osepchook CC, Wong KW, Sharma M, Kambadur R, Bass JJ. Am J Physiol. 2003;284:E377–E381. doi: 10.1152/ajpendo.00282.2002. [DOI] [PubMed] [Google Scholar]
- 35.Bunger L, Laidlaw A, Bulfield G, Eisen EJ, Medrano JF, Bradford GE, Pirchner F, Renne U, Schlote W, Hill WG. Mamm Genome. 2001;12:678–686. doi: 10.1007/s00335001-3018-6. [DOI] [PubMed] [Google Scholar]
- 36.Bunger L, Ott G, Varga L, Schlote W, Rehfeldt C, Renne U, Williams JL, Hill WG. Genet Res. 2004;84:161–173. doi: 10.1017/s0016672304007165. [DOI] [PubMed] [Google Scholar]
- 37.Szabo G, Dallmann G, Muller G, Patthy L, Soller M, Varga L. Mamm Genome. 1998;9:671–672. doi: 10.1007/s003359900843. [DOI] [PubMed] [Google Scholar]
- 38.Varga L, Szabo G, Darvasi A, Muller G, Sass M, Soller M. Genetics. 1997;147:755–764. doi: 10.1093/genetics/147.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weibel ER. Stereological Methods. London: Academic; 1979. [Google Scholar]
- 40.Weibel ER. Stereological Methods. London: Academic; 1980. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.