Abstract
The bacterial RecA protein has been the most intensively studied enzyme in homologous genetic recombination. The core of RecA is structurally homologous to that of the F1-ATPase and helicases. Like the F1-ATPase and ring helicases, RecA forms a hexameric ring. The human Dmc1 (hDmc1) protein, a meiosis-specific recombinase, is homologous to RecA. We show that hDmc1 forms octameric rings. Unlike RecA and Rad51, however, hDmc1 protein does not form helical filaments. The hDmc1 ring binds DNA in the central channel, as do the ring helicases, which is likely to represent the active form of the protein. These observations indicate that the conservation of the RecA-like ring structure extends from bacteria to humans, and that some RecA homologs may form both rings and filaments, whereas others may function only as rings.
The Escherichia coli RecA protein has been intensively studied for many years by using genetic, biochemical, and biophysical techniques (1–4). It has been hoped that insight gained into how RecA functions to catalyze homologous genetic recombination might be helpful in understanding such eukaryotic processes as meiosis. The RecA protein forms an unusual nucleoprotein filament on DNA, in which the DNA is both extensively stretched and untwisted (5–7). The yeast (8) and human (9) Rad51 proteins, which are homologous to RecA, form very similar nucleoprotein structures. The more distantly related bacteriophage T4 UvsX protein assembles in a filament with nearly identical helical parameters (10), suggesting that the structural properties of this filament are important to function and have been conserved over large evolutionary distances.
RAD51 in yeast is involved in DNA recombination and repair (11), but is not an essential gene. However, disruption of RAD51 in mice leads to embryonic lethality (12), suggestive of a role in general DNA metabolism and genome stability. Another eukaryotic RecA homolog is the Dmc1 protein. DMC1 (disrupted meiotic cDNA) is a meiosis-specific gene in yeast and animals, and disruption of it leads to meiotic arrest in both yeast (13) and mice (14), and accumulation of double-strand breaks with 3′ ends (12). In at least one plant, Arabidopsis, however, Dmc1 is induced during mitosis (15). It has been shown in vitro that yeast (16) and human (17, 18) Rad51 proteins can promote an ATP-dependent strand-exchange reaction. Human Dmc1 (hDmc1) protein also can catalyze such reactions in vitro, but with much lower activity (19).
The homology between RecA and its eukaryotic homologs is contained within the nucleotide-binding core. The nucleotide-binding core of the hexameric F1-ATPase is structurally homologous to the core of the RecA protein (20), even though these proteins have vastly different functions and substrates (the loops of RecA that bind DNA are topologically similar to the loops in the F1-ATPase that bind the γ-subunit, a coiled-coil protein). In addition, the conserved motifs present in DNA and RNA helicases have been shown to be part of the same core found in RecA for three helicases, which is likely to be true for all helicases (21, 22). Because helicases act in many aspects of recombination, replication, transcription, and repair of DNA (23), they have a greater functional similarity to RecA than does the F1-ATPase. Some of the most intensively studied helicases function as hexameric rings, including the E. coli transcription-termination rho protein (24), E. coli DnaB (25), bacteriophage T7 gp4 (26), simian virus 40 large T antigen (27), and the papilloma virus E1 protein (28). Interestingly, RecA forms a hexameric ring that appears to be a structural homolog of the hexameric helicases, both at the level of tertiary and quaternary structure (29). Rings of human Rad51 protein also have been observed (30), but neither function nor DNA binding have yet been demonstrated for this form of either the RecA or Rad51 proteins. It has been assumed that the only active form of the RecA protein and its homologs is the helical nucleoprotein filament.
MATERIALS AND METHODS
hDmc1 Preparation.
Three different preparations of hDmc1 protein were analyzed. In one, the protein was prepared as described (19), and ≈10% of the protein exhibited proteolytic degradation. The other two preparations involved his-tagged hDmc1 and showed no proteolysis. The purification of hDmc1 protein after overexpression in E. coli will be described elsewhere (J.Y.M., A. A. Davies, N. Hajibagheri, E. Van Dyck, F. E. Benson, A. Z. Stasiak, A. Stasiak, and S.C.W., unpublished work; Z.L. and C.M.R., unpublished work).
DNA Substrates.
Single-stranded (ss) M13 mp18 was purchased from New England Biolabs, double-stranded (ds) M13 mp18 from Boehringer Mannheim and φX174 (replicative form) from Sigma. φX174 fragments with blunt ends were produced by digesting 10 μg of form I φX174 with HpaI (10 units/μl, Promega). DNA then was loaded on a low melting point agarose gel and subjected to electrophoresis. The resulting 1,264- and 3,710-bp bands were excised and extracted. For dsDNA with 3′ ss tails of increasing length, 25 μg form I φX174 was cut with PstI (13 units/μl, IBI), followed by λ exonuclease digestion (5 units/μl, Boehringer Mannheim) for either 40 sec, 5 min, or 10 min at 37°C. Tailed duplex DNA was loaded on a low melting point agarose gel, subjected to electrophoresis, excised, and extracted. For dsDNA with 5′ ss tails, dsM13 mp18 was cut with EcoRI (12 units/μl, Promega), followed by exonuclease III digestion (200 units/μl, Promega). DNA was loaded on a low melting point agarose gel, subjected to electrophoresis, excised, and extracted.
Ring Formation.
Isolated rings were formed by incubating 1.0 μM hDmc1 for 10 min at 37°C with 1 mM MgCl2, 1 mM DTT, 2 mM ATP-γ-S, and 25 mM Pipes, pH 7. For clusters of rings, 1.0 μM hDmc1 was incubated for 10 min at 37°C with 11 μM heat-denatured partially duplex φX174, 1 mM MgCl2, 1 mM DTT, and 25 mM Pipes, pH 7. For long stacks of rings, 1 μM hDmc1 was incubated for 10 min at 37°C with 10 or 11 μM partially duplex φX174, 1 mM MgCl2, 1 mM DTT, and 25 mM Pipes, pH 7 in the presence or absence of 2 mM ATP-γ-S.
Electron Microscopy and Image Analysis. Images were recorded on film at a magnification of ×30,000 with a JEOL 1200 EXII electron microscope, and negatives were densitometered by using a Leaf45 scanner (at 4 Å/pixel). Most computer processing used the spider software (31). The three-dimensional reconstruction of hDmc1 was generated from a set of 16,875 images, collected from two preparations that did not show any rings with 7-fold symmetry. A top view was generated by ranking the images on the strength of the 8-fold rotational power, by using a reference-free alignment (32) of the 4,000 highest images. An additional 41 classes were generated by using a K-means algorithm that grouped the images according to similarity clusters (33), after first separating out those images that corresponded to side views obtained on DNA. Nine of the final 42 classes corresponded to tilts of 80° or larger, based on the final angular assignments, whereas all of the other classes were assigned tilt angles of 40° or less. Each class contained at least 100 images. A starting model was generated by back projection using only one averaged side view and the top view, which served to generate initial angular assignments for the other classes. The process was iterated more than 20 times, as new three-dimensional reconstructions were used to generate the angular assignments by using cross-correlation functions between the observed classes and projections of the reconstruction. The process converged quickly, as a stable solution appeared within the first few iterations.
Given the limited number of views, it is not possible to estimate the resolution of the reconstruction by dividing the views into two groups, generating two independent reconstructions, and analyzing such statistics as Fourier ring correlations. For tilts about the 8-fold symmetry axis of the ring, only two views 22.5° apart are needed to generate an isotropic 27-Å resolution reconstruction. Because the side views of the ring fell into only two characteristic classes, separated by ≈22°, we expect that the resolution of the reconstruction is ≈27 Å.
RESULTS
We find that the meiosis-specific recombinase hDmc1 protein forms octameric rings (Figs. 1 and 2). The rings are ≈140 Å in diameter, with a central hole that is ≈25 Å in diameter. As with the hexameric helicase rings formed by T7 gp4 (34) and E. coli DnaB (35), these rings in projection have a clearly chiral or pin-wheel appearance. Ring formation does not require ATP, Mg2+, or the presence of DNA. To exclude the possibility that ring formation in the absence of exogenous DNA was the result of a contaminant DNA in the protein preparation, the samples were incubated with Bal 31, a ds exonuclease and a ss endonuclease. Although we cannot exclude the possibility that trace quantities of polynucleotide still existed, the presence of numerous rings after this incubation suggests that a DNA substrate is not necessary for ring formation.
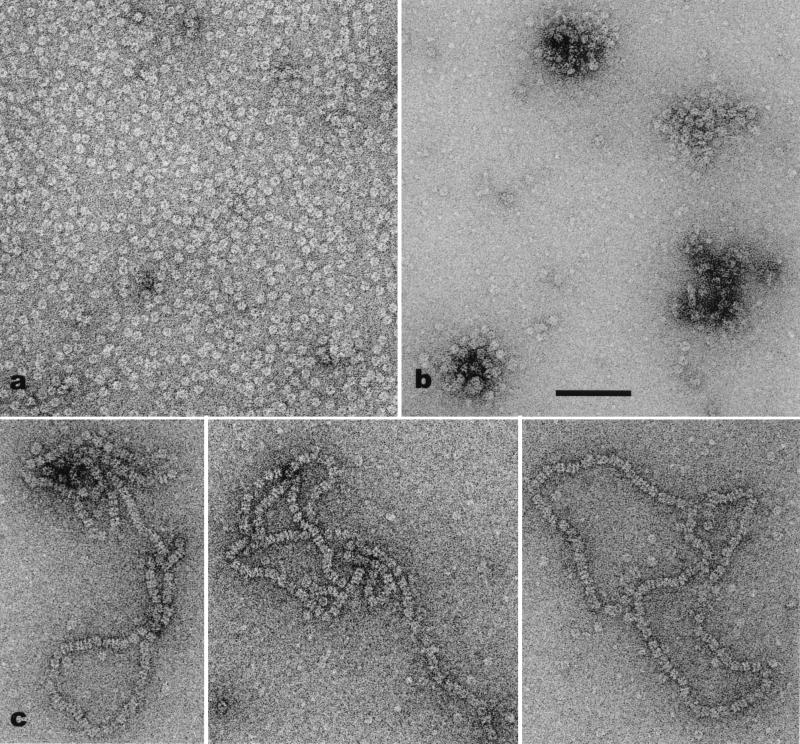
Figure 1.
Electron micrographs of hDmc1. (a) Forming rings in the absence of DNA. (b) Clusters of rings with ssDNA. (c) Stacks of double rings on partially duplex φX174. In the absence of DNA (a) the rings appear to be uniformly distributed, with very little aggregation. In contrast, when ssDNA is added, the rings frequently are clumped together. Four such aggregates can be seen in b. The formation of the long stacks of rings (c) seems to require a ss-dsDNA junction. (Scale bar = 1,000 Å.)
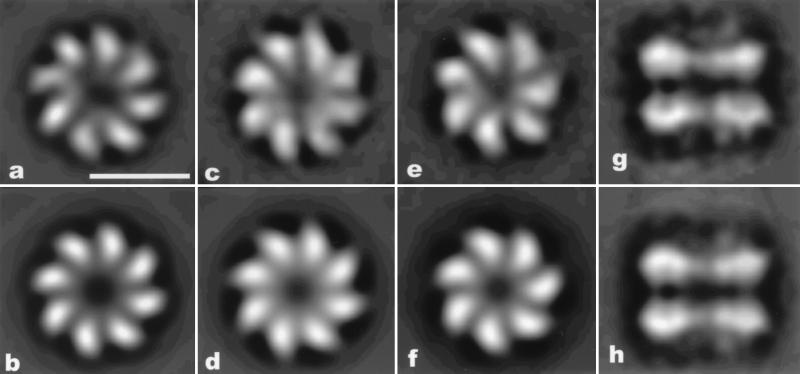
Figure 2.
(a) Average of 3,000 top 8-fold rings of a set of 13,646 rings from a single hDmc1 preparation. A symmetrized version of this average is shown in b. In one preparation of hDmc1 ≈10% of the protein existed as a large proteolytic fragment. From this preparation 15,948 rings were collected. (c) An average of 2,000 rings with the highest 8-fold symmetry is shown. (e) An average of 3,000 rings with the highest 7-fold symmetry is shown. (d and f) Eight-fold and 7-fold symmetrized versions, respectively, of these averages are shown. (g) An average of 605 double rings found on dsDNA substrates having ssDNA tails. The rings are spaced apart by ≈55 Å. The bipolar symmetry (each ring is facing in the opposite direction) of these structures can be seen by the similarity between the raw average (g) and a symmetrized version of this average (with mirror symmetry imposed) in h. (Scale bar in a is 100 Å.)
One preparation of hDmc1 protein, where a small fraction of the protein was observed to exist as a large proteolytic fragment (19), gave rise to approximately equal numbers of rings with seven subunits and with eight subunits (Fig. 2 c–f). Two other preparations, where there was no apparent proteolytic degradation, did not yield any significant number of rings with seven subunits, as judged by rotational power analysis. Because the fraction of the protein that was proteolytically degraded was much less than 50%, and probably closer to 10%, this finding suggests that a single truncated subunit in a ring can change the symmetry of the ring from 8-fold to 7-fold (if subunits randomly coassemble, we would expect from binomial statistics that 50% of the rings would have eight intact subunits if 8% of the protein were degraded).
Taking advantage of the different projections offered by both the side views and “tilted” top views, a three-dimensional reconstruction was generated from 42 different image classes. Only one of these classes, the top view projected down the 8-fold axis (Fig. 2b), had symmetry imposed. The resulting reconstruction is shown in Fig. 3.
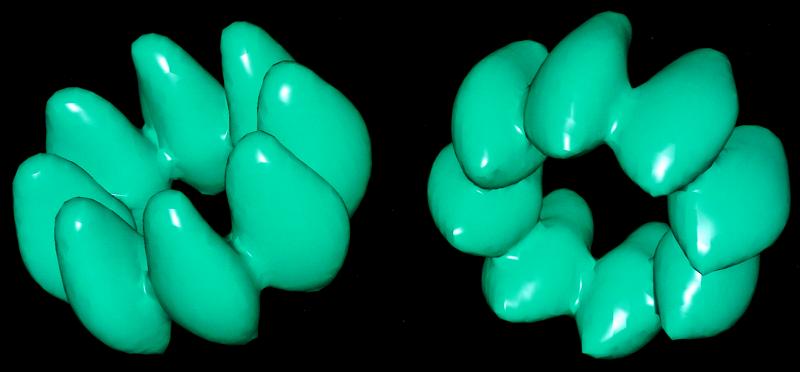
Figure 3.
A three-dimensional reconstruction of the octameric hDmc1 ring is shown in two different views. The ring binds DNA in the central channel. The reconstruction was generated from a total of 16,875 images of rings, which were averaged into 42 different classes. The surface of the reconstruction has been chosen to enclose 100% of the expected molecular volume, assuming a partial specific volume for protein of 0.75 cm3/gr.
In contrast to RecA and Rad51, under none of the conditions used did we see helical filaments formed by hDmc1. These conditions involved different DNA substrates (ssDNA, dsDNA, dsDNA with ssDNA tails, partially denatured dsDNA, etc.), different nucleotide cofactors (ATP and ATP-γ-S), and different Mg2+ concentrations. Can hDmc1 be forming very short helical filaments that might be mistaken for small stacks of rings? This possibility seems unlikely, as we analyzed 705 double rings, and none of the image classes corresponded to a helical turn in projection. This result argues that the form of the protein responsible for the observed DNA protection (19) must be the octameric ring.
In the absence of DNA the hDmc1 rings were distributed on the electron microscopy grids as single particles, whereas with DNA they frequently were arranged in higher-order structures. With ssDNA they displayed a tendency to form clusters of various sizes (Fig. 1b), and with dsDNA with either 3′ or 5′ tails they assembled pairwise in long stacks (Fig. 1c). No long stacks of rings were observed with dsDNA containing blunt ends. This finding suggests that the stacks are nucleated at the ss-dsNA interface, and several observations suggest that they extend over the duplex DNA. The stacks appeared to cover a significant portion of the molecules, under conditions where the ssDNA tails will be fairly short. The bipolar nature of the double rings (Fig. 2 g and h) is also consistent with the bipolar symmetry of the dsDNA, similar to the mode of binding observed for RuvB to dsDNA (36). When dsDNA containing a ssDNA tail was heat-denatured, a few stacks of rings were detected, but the very low number of stacks was probably caused by incomplete denaturation of the DNA. High energy nucleotide cofactors or Mg2+ had no effect on the assembly of long stacks of hDmc1 rings.
The coaxial arrangement of rings bound to DNA substrates (Fig. 1c) shows that the path of the DNA is through the central channel of the ring. If the DNA were bound to the sides of the rings, one would expect to see a staggered arrangement of rings. Examination of many electron micrographs failed to find any such staggered arrays of rings. The possibility that the coaxial stacks are caused solely by protein–protein interactions can be excluded for two reasons: the spacing between rings is variable, and long stacks are not seen in the absence of a DNA substrate. This binding of DNA in the central channel is the same mode shown for RuvB (36), T7 gp4 (34, 37), DnaB (38), and simian virus 40 large T antigen (39), all of which are part of a superfamily of RecA-related proteins.
DISCUSSION
It has been suggested that the Dmc1 protein will have the same secondary and tertiary structure as the E. coli RecA protein (40). Is the failure of Dmc1 to form helical filaments surprising? Not if one considers the family of proteins that include actin, HSP70, and hexokinase, which all share a common tertiary structure and appear to have evolved from a common ancestral protein (41). Within this family, only actin forms a helical filament. The highly conserved helical parameters present in the RecA (5, 7), UvsX (10), and Rad51 (8) filaments may be a large constraint on the evolutionary sequence divergence of these proteins. This constraint may be enhanced by the fact that RecA (29), yeast Rad51 (X.Y., T. Ogawa, and E.H.E., unpublished work), and human Rad51 (30) proteins also form rings. The possibility that hDmc1 does not form helical filaments, and the resulting elimination of the constraint imposed by the helical architecture on the conservation of protein residues, may be consistent with the phylogenetic observation that DMC1 genes consistently evolve faster than recA and RAD51 genes (42).
Although it appears from our observations that the form of Dmc1 protein responsible for binding DNA is an octameric ring, we have no knowledge of whether Dmc1 functions in vitro and in vivo as individual rings or as extended stacks of rings. Bacteriophage T7 gp4 protein forms extended stacks of rings on ssDNA in vitro (37), but it is likely that the protein acts at a replication fork as a single ring (34). Further, we cannot exclude the possibility that an accessory protein or cofactor would cause the protein to form helical filaments. However, our observations do suggest that under the conditions used in vitro, the active form of the Dmc1 protein is an octameric ring.
Our results suggest that other eukaryotic RecA homologs also may function as rings, rather than as helical filaments. Further, the conservation of a ring structure from the bacterial RecA protein to the human Rad51 protein is most likely caused by a functional DNA-binding property of these structures. Although the manner in which the Dmc1 rings function in homologous recombination is not yet known, it is probable that, like the hexameric helicases and RuvB, they will use the energy of ATP hydrolysis to either move along a DNA substrate or pump DNA.
Acknowledgments
This work was supported by National Institutes of Health GM35269 (E.H.E.) and GM55304 (C.M.R), Human Frontier Sciences Program RG335 (E.H.E.), and the Imperial Cancer Research Fund (S.C.W.).
ABBREVIATIONS
- hDmc1
human Dmc1
- ss
single-stranded
- ds
double-stranded
References
- 1.West S C. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 2.Radding C M. J Biol Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 3.Egelman E H, Stasiak A. Micron. 1993;24:309–324. [Google Scholar]
- 4.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stasiak A, DiCapua E, Koller T. J Mol Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 6.Nishinaka T, Ito Y, Yokoyama S, Shibata T. Proc Natl Acad Sci USA. 1997;94:6623–6628. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stasiak A, DiCapua E. Nature (London) 1982;229:185–186. [Google Scholar]
- 8.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 9.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Egelman E H. J Mol Biol. 1993;232:1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- 11.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 12.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 14.Pittman D L, Cobb J, Schimenti K J, Wilson L A, Cooper D M, Brignull E, Handel M A, Schimenti J C. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 15.Doutriaux M P, Couteau F, Bergounioux C, White C. Mol Gen Genet. 1998;257:283–291. doi: 10.1007/s004380050649. [DOI] [PubMed] [Google Scholar]
- 16.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 17.Baumann P, Benson F E, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 18.Gupta R C, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Golub E I, Gupta R, Radding C M. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abrahams J P, Leslie A G, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 21.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Nature (London) 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 22.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 23.Matson S W, Bean D W, George J W. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 24.Gogol E P, Seifried S E, von Hippel P H. J Mol Biol. 1991;221:1127–1138. doi: 10.1016/0022-2836(91)90923-t. [DOI] [PubMed] [Google Scholar]
- 25.Reha-Krantz L J, Hurwitz J. J Biol Chem. 1978;253:4043–4050. [PubMed] [Google Scholar]
- 26.Patel S S, Hingorani M M. J Biol Chem. 1993;268:10668–10675. [PubMed] [Google Scholar]
- 27.Mastrangelo I A, Hough P V C, Wall J S, Dobson M, Dean F B, Hurwitz J. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 28.Fouts E T, Yu X, Egelman E H, Botchan M R. J Biol Chem. 1999;274:4447–4458. doi: 10.1074/jbc.274.7.4447. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Egelman E H. Nat Struct Biol. 1997;4:101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 30.Baumann P, Benson F E, Hajibagheri N, West S C. Mutat Res. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 31.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 32.Penczek P A, Radermacher M, Frank J. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 33.Penczek P A, Zhu J, Frank J. Ultramicroscopy. 1996;63:205–218. doi: 10.1016/0304-3991(96)00037-x. [DOI] [PubMed] [Google Scholar]
- 34.Yu X, Hingorani M M, Patel S S, Egelman E H. Nat Struct Biol. 1996;3:740–743. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Jezewska M J, Bujalowski W, Egelman E H. J Mol Biol. 1996;259:7–14. doi: 10.1006/jmbi.1996.0297. [DOI] [PubMed] [Google Scholar]
- 36.Stasiak A, Tsaneva I R, West S C, Benson C J B, Yu X, Egelman E H. Proc Natl Acad Sci USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egelman E H, Yu X, Wild R, Hingorani M M, Patel S S. Proc Natl Acad Sci USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jezewska M J, Rajendran S, Bujalowska D, Bujalowski W. J Biol Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 39.Dean F B, Borowiec J A, Eki T, Hurwitz J. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 40.Story R M, Bishop D K, Kleckner N, Steitz T A. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- 41.Flaherty K M, McKay D B, Kabsch W, Holmes K C. Proc Natl Acad Sci USA. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stassen N Y, Logsdon J M, Jr, Vora G J, Offenberg H H, Palmer J D, Zolan M E. Curr Genet. 1997;31:144–157. doi: 10.1007/s002940050189. [DOI] [PubMed] [Google Scholar]