Abstract
Internal ribosome entry site (IRES) RNAs from the hepatitis C virus (HCV) and classical swine fever virus (CSFV) coordinate cap-independent assembly of eukaryotic 48S initiation complexes, consisting of the 40S ribosomal subunit, eukaryotic initiation factor (eIF) 3 and the eIF2/GTP/Met-tRNAiMet ternary complex. Here, we report that these IRESes also play a functional role during 80S ribosome assembly downstream of 48S complex formation, in promoting eIF5-induced GTP hydrolysis and eIF2/GDP release from the initiation complex. We show that this function is encoded in their independently folded IRES domain II and that it depends both on its characteristic bent conformation and two conserved RNA motifs, an apical hairpin loop and a loop E. Our data suggest a general mode of subunit joining in HCV and HCV-like IRESes.
Keywords: eukaryotic initiation factors, HCV, IRES RNA, RNA structure, translation initiation
Introduction
In higher eukaryotes, there are at least two distinct pathways known to lead to the assembly of 80S ribosomes. The first pathway requires a 5′-capped mRNA and the full complement of canonical eIFs (Sachs et al, 1997; Kapp and Lorsch, 2004; Merrick, 2004). First, a 43S particle, comprising the 40S subunit, eIF1, 1A, 3 and the eIF2/GTP/Met-tRNAiMet ternary complex, is recruited to the 5′-cap structure of the mRNA through interactions with the 5′-cap binding complex eIF4F. The 43S particle then scans the mRNA, assisted by the helicase, eIF4A and its cofactor eIF4B, to locate the initiation codon and convert into a 48S initiation complex (Sachs et al, 1997). During scanning, the binding of eIF1A in the ribosomal A site and eIF1 near the platform is believed to modulate 40S conformation, which in turn ensures proper start codon selection (Pestova et al, 1998a; Lomakin et al, 2003; Algire et al, 2005; Maag et al, 2005). After AUG start codon recognition and codon–anticodon base pairing between the mRNA and Met-tRNAiMet in the ribosomal P site triggers eIF5-mediated hydrolysis of GTP bound to eIF2 and subsequent dissociation of eIF2/GDP from the 48S complex (Unbehaun et al, 2004; Algire et al, 2005). Finally, eIF5B mediates release of the remaining eIFs and the joining of the 60S subunit in another GTP-dependent process to form 80S ribosomes (Pestova et al, 2000; Unbehaun et al, 2004).
In contrast, in the second pathway, 48S complexes are formed without a 5′-cap structure or scanning, and only a subset of the canonical eIFs is required. This process is mediated by IRES elements found within the 5′ UTR of some cellular mRNAs and several viral RNAs, such as the genomic RNA of HCV (Sachs et al, 1997; Hellen and Sarnow, 2001; Stoneley and Willis, 2004). HCV IRES-mediated initiation is driven by the high-affinity IRES–40S interaction, which promotes stable binding of eIF3 and the eIF2/GTP/Met-tRNAiMet ternary complex to form a 48S particle with established codon-anticodon base pairing in the P site (Pestova et al, 1998b; Ji et al, 2004; Otto and Puglisi, 2004). The assembly of active 80S ribosomes still seems to require eIF5, eIF5B, GTP and 60S subunits (Pestova et al, 1998b), but the detailed pathway of 80S assembly during HCV IRES-mediated initiation of translation is still unknown.
The secondary structure of the HCV IRES RNA displays two major domains, II and III (Brown et al, 1992; Kieft et al, 1999), which contain structural elements crucial for initiation of translation (Figure 1A) (Pestova et al, 1998b; Ji et al, 2004; Otto and Puglisi, 2004). The global domain organisation and several RNA structural motifs in these domains are conserved among related viruses from the Flaviviridae family, such as CSFV, the bovine viral diarrhea virus (BVDV) and GB virus B (GBV-B) (Pestova et al, 1998b; Honda et al, 1999; Pestova and Hellen, 1999). Functional roles of individual IRES domains have been studied in the HCV IRES RNA. Mutations in the basal part of domain III abolish 40S subunit binding (Kieft et al, 2001), whereas mutations in the apical loops of domain III affect recruitment of eIF3 and subsequently of the ternary complex, and thereby all block the assembly of 48S complexes (Ji et al, 2004; Otto and Puglisi, 2004). Domain II, in contrast, is not required for 40S subunit binding (Kieft et al, 2001; Otto et al, 2002) and its deletion does not alter recruitment of eIF2, Met-tRNAiMet and eIF3, but reduces the translational activity up to five-fold by blocking 80S formation (Pestova et al, 1998b; Kolupaeva et al, 2000; Ji et al, 2004; Otto and Puglisi, 2004). Primer extension inhibition reactions, so-called toeprinting analysis, performed on domain II-deleted binary HCV IRES-40S complexes, displayed significantly weaker toeprints as compared with wild-type (wt) HCV IRES, suggesting that the HCV ORF is less stably bound to the 40S subunit without domain II (Kolupaeva et al, 2000). Moreover, 48S complexes assembled onto HCV IRES RNA lacking domain II consistently displayed weaker toeprints, suggesting a defect in codon–anticodon base pairing and therefore in proper 48S formation, which would subsequently impair 80S ribosome assembly (Pestova et al, 1998b; Ji et al, 2004; Otto and Puglisi, 2004). The cryo-electron microscopy (cryo-EM) analyses of the 40S subunit and binary HCV IRES–40S complexes revealed a significant modulation of 40S conformation upon binding of the IRES RNA (Spahn et al, 2001). The larger domain III adopts an elongated conformation within the binary HCV IRES–40S complex, positioning itself on the 40S platform while changing 40S conformation. Domain II forms an independently folded module within the IRES RNA with an overall L-shape both free in solution (Lukavsky et al, 2003) and when bound to the 40S subunit (Spahn et al, 2001). Direct interaction of domain II with the 40S subunit has been evidenced by protection of domain II from RNAse digestion and chemical footprinting in the context of a binary HCV IRES–40S complex (Kieft et al, 2001; Lytle et al, 2002) and a domain II-dependent crosslink to the ribosomal head protein S5 (rpS5), which is located in the E site of the 40S subunit (Kolupaeva et al, 2000; Fukushi et al, 2001). Correspondingly, the cryo-EM structure showed that the apical hairpin loop and loop E motif of domain II interact with the ribosomal head and platform near the E site, thereby closing the mRNA cleft (Spahn et al, 2001). This domain II-dependent conformational change of the 40S subunit together with the aforementioned toeprinting experiments suggested a role for domain II in proper positioning of the mRNA in the ribosomal P site (Kolupaeva et al, 2000; Spahn et al, 2001), but to date, the precise functional role of the conserved IRES domain II remains unclear.
Figure 1.
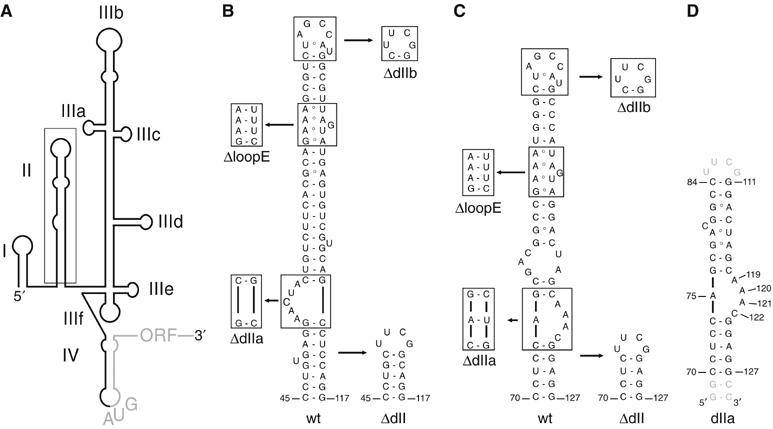
Secondary structures of the HCV and CSFV IRES domain II. (A) Secondary structure model of the HCV IRES RNA with individual domains (I–IV) indicated. The AUG start codon and the HCV ORF are shown in grey. (B) Secondary structure of the wt HCV IRES domain II. The domain II mutants used in this study are indicated and boxed. Numbering according to ref.(Pestova et al, 1998b). (C) Sequence of the wt CSFV IRES domain II. The domain II mutants used in this study are indicated and boxed. Numbering according to (B). (D) RNA construct used for the NMR structure determination of the CSFV domain IIa (nt 70–84 and 111–127). Nucleotides added to improve transcription efficiency and promote hairpin formation are shown in grey.
Here, we demonstrate that domain II of the HCV and CSFV IRESes are controlling GTP hydrolysis and eIF2 release and are therefore actively involved in 80S ribosome formation downstream of 48S assembly. The solution structure of CSFV IRES domain IIa determined by NMR spectroscopy reveals that a bent domain II conformation is a conserved structural feature of HCV-like IRESes. Mutations within domain II, such as deletion of the entire domain or its conserved structural motifs, as well as alteration of its overall shape, impair eIF2 release and thereby stall 80S ribosome assembly at the 48S stage. Our results propose a structurally encoded functional role of domain II during subunit joining, which is shared among HCV-like IRESes.
Results
Deletion of HCV IRES domain II blocks 80S assembly after 48S complex formation
To investigate the functional role of domain II in the IRES-mediated initiation pathway, a deletion mutant of the entire domain II (ΔdII) was designed in which nucleotides 48–114 of the HCV IRES RNA were replaced by a stable hairpin (Figure 1B). Assembly of eukaryotic 48S and 80S complexes in rabbit reticulocyte lysate (RRL) onto the wt and ΔdII HCV IRES RNA was monitored by toeprint and sucrose density gradient analyses (Anthony and Merrick, 1992). The non-hydrolysable GTP analog, GMPPNP, was used in the assembly reactions to inhibit eIF2 release and accumulate fully assembled 48S complexes (Merrick, 1979). Toeprint analysis of the wt 48S complex shows two expected bands of similar intensity at positions +16 and +17 from the adenine (+1) in the AUG start codon (Pestova et al, 1998b; Otto and Puglisi, 2004), reflecting correct codon–anticodon base pairing within the ribosomal P site (Figure 2A, lane 1). The ΔdII 48S complex displays a similar toeprint pattern, but with decreased intensity as compared with that of wt (Figure 2A, lane 3). This lower intensity of the toeprint signal does not seem to reflect a decreased overall stability of the ΔdII 48S complex under the assay conditions, as upstream toeprints in the pseudoknot and eIF3-binding region, as well as the full-length cDNA product display very similar band intensities for both wt and ΔdII 48S complexes (Figure 2A, lanes 1 and 3).
Figure 2.
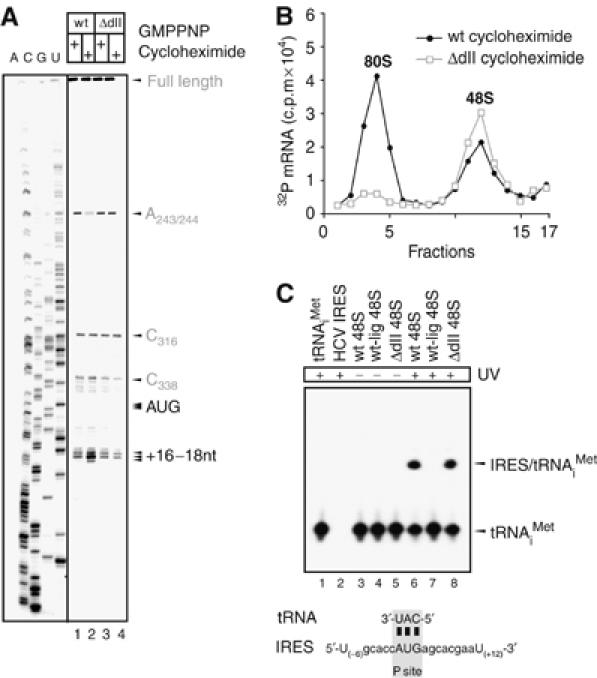
HCV IRES domain II is required for 80S ribosome assembly. (A) Toeprinting analysis of wt and ΔdII HCV IRES RNAs in RRL. Ribosomal particles assembled in RRL using wt or ΔdII IRES RNA in the presence of GMPPNP or cycloheximide, were analysed by primer extension inhibition (lanes 1–4). Arrows indicate positions of the AUG start codon and the toeprints. A dideoxynucleotide sequence generated with the same primer is shown on the left. Upstream toeprints correspond to an extension stop in the pseudoknot (C316) and in the eIF3-binding region (A243/244). A significantly weaker eIF3 toeprint (A243/244) is only observed upon 80S ribosome formation using wt HCV IRES (lane 2), but not with the ΔdII HCV IRES RNA (lane 4). (B) Sucrose density gradient analyses of wt and ΔdII mutant HCV IRES ribosomal complexes. Complexes were formed in RRL preincubated with cycloheximide to block elongation step and to trap 80S ribosomes. The positions of ribosomal complexes are indicated above appropriate peaks. (C) Detection of codon–anticodon base pairing within HCV IRES 48S complexes. Northern blot analysis of total RNA extracted from wt, ΔdII and wt-lig IRES 48S complexes before or after UV irradiation using a probe specific for the 3′-end of tRNAiMet (lanes 3–9). HCV IRES RNA and tRNAiMet were loaded as controls (lanes 1–2). Positions of free tRNAiMet and the 4-thiouridine-mediated IRES/tRNAiMet crosslinks are indicated on the right. The figure below the Northern blot depicts the sequence of the HCV mRNA surrounding the AUG start codon placed in the ribosomal P site base paired to Met-tRNAiMet. The closest uridine residues upstream and downstream of the AUG start codon are numbered relative to the A[+1] of the AUG start codon.
Assembly reactions in the presence of cycloheximide, which inhibits the elongation process, were used to accumulate fully assembled 80S ribosomes (Godchaux et al, 1967). The toeprint analysis of wt 80S complexes displays an additional band at position +18 and an intensified middle band corresponding to position +17 (Figure 2A, lane 2), indicative of correct P-site placement of Met-tRNAiMet within 80S ribosomes (Dmitriev et al, 2003; Otto and Puglisi, 2004). No equivalent changes evidencing proper 80S ribosome formation were detected when ΔdII IRES ribosomal complexes were analysed (Figure 2A, lanes 4). In addition, sucrose density gradient analysis showed the formation of 48S and 80S complexes with wt IRES, whereas almost no 80S ribosome formation was detected with the ΔdII IRES (Figure 2B). In agreement with previous studies, deletion of domain II seems to affect 48S formation to some extent (reflected in the decreased 48S toeprint) and inhibits 80S ribosome formation (Pestova et al, 1998b; Ji et al, 2004; Otto and Puglisi, 2004).
The toeprinting experiments demonstrate that the AUG start codon is placed in the ribosomal P site both in the wt and the ΔdII 48S complex, but the weaker toeprint signal observed with the ΔdII 48S complex might indicate that codon–anticodon base pairing is less stable without domain II. We therefore investigated the extent of direct contact between the AUG start codon and Met-tRNAiMet using 4-thiouridine-mediated crosslinking (Stade et al, 1989). This technique yields ‘zero-length' crosslinks upon UV irradiation, reflecting direct contacts between interaction partners (Stade et al, 1989). Accordingly, the presence of a signal would be indicative of stable base pairing between the AUG start codon (4-thiouridine labeled) and Met-tRNAiMet (unlabeled), in contrast to the former toeprinting assay which monitors codon–anticodon base pairing more indirectly by observing mRNA positioning on the 40S ribosomal subunit. 48S initiation complexes assembled in RRL onto either 4-thiouridine-labeled wt or ΔdII IRES RNA were isolated using the StrepoTag-based affinity purification method and further purified by sucrose density gradient sedimentation (Locker et al, 2006). Met-tRNAiMet was detected by Northern blotting with a 32P-labeled probe specific for tRNAiMet (Figure 2C, lanes 1 and 2). In both 48S complexes, Met-tRNAiMet was present at the same level, supporting that domain II is not required for the recruitment of the ternary complex (Figure 2C, lanes 3 and 5). Upon UV irradiation, both wt and ΔdII IRES RNAs crosslinked with equal efficiency to Met-tRNAiMet, demonstrating that stable codon–anticodon base pairing occurs independent of domain II (Figure 2C, lanes 6 and 8). The observed crosslinks are unlikely to arise from any other than the 4-thiouridine incorporated in the AUG start codon, as toeprinting experiments showed that the AUG start codon resides in the ribosomal P site and therefore no other 4-thiouridine residue would be close enough to the anticodon of Met-tRNAiMet to yield the observed ‘zero-length' crosslinks (Figure 2C). To provide further evidence that the crosslinks arise solely from a direct interaction between Met-tRNAiMet and the HCV AUG start codon, a ligated wt IRES RNA was prepared in which only domain II is 4-thiouridine-labeled (wt-lig), but not domain III, the AUG start codon or the HCV ORF, and was used to assemble 48S complexes (Figure 2C, lane 4). The absence of a crosslink in this wt-lig IRES 48S complex invigorates that the observed crosslinks above reflect codon-anticodon base pairing (Figure 2C, lane 7). In addition, UV-irradiated wt-lig 48S complexes were also assayed by reverse transcription to detect crosslink-dependent stops of primer extension within the HCV ORF, which would be indicative of previously proposed domain II-ORF interactions (Spahn et al, 2001). However, no crosslink-induced stops could be detected, suggesting that uracil bases in the apical loop of domain II do not interact directly with the HCV ORF (data not shown). Our results show that domain II is not essential for the assembly of 48S complexes with stable codon–anticodon base pairing in the P site and indicate that this domain is required for the 80S ribosome assembly process downstream of 48S complex formation (Figure 2). To identify the role of IRES domain II during the 48S to 80S ribosome transition, we further characterised each step in the IRES-driven 80S formation.
HCV IRES domain II mediates eIF2 release during subunit joining
During canonical initiation of translation, the GTPase-activating protein (GAP), eIF5, binds to 48S complexes and eIF2, which in turn enables hydrolysis of eIF2-bound GTP and subsequent release of eIF2/GDP upon start codon recognition (Chakrabarti and Maitra, 1991; Chaudhuri et al, 1994; Das et al, 2001; Unbehaun et al, 2004). To test whether deletion of domain II impairs the recruitment of eIF5 to 48S complexes, wt or ΔdII HCV IRES 48S complexes were assembled in RRL in the presence of GMPPNP as before. 48S particles were affinity-purified and the amount of eIF5 in the complexes was analysed by quantitative immunoblotting using serial dilutions of recombinant eIF5 over a linear concentration range from 10 to 1200 fMol (Figure 3A, lanes 1–4). Both wt and ΔdII HCV IRES 48S complexes (500 fMol) contained less than 10 fMol native eIF5 (Figure 3, lane 5), which was only clearly detectable upon overexposure of the Western blot (data not shown). About 1% of both wt and ΔdII 48S particles contained native eIF5. To obtain additional evidence for domain II-independent eIF5 recruitment, both 48S assembly reactions (wt and ΔdII) were also performed in RRL supplemented with recombinant eIF5 to compensate for the naturally low abundance of native eIF5 in RRL (Pestova et al, 2000). Equivalent amounts of eIF5 were detected within both wt and ΔdII 48S complexes (Figure 3A), demonstrating that efficient recruitment of eIF5 to 48S complexes is domain II-independent. This suggests that domain II is required for subsequent steps in the 80S assembly.
Figure 3.

Domain II mediates release of eIF2 from HCV 48S complexes during 80S assembly. (A) Recruitment of eIF5 to HCV IRES 48S complexes is unaffected by deletion of domain II. Quantitative immunoblotting analysis of purified wt and ΔdII IRES 48S complexes assembled in the presence and absence of recombinant eIF5 using antibodies specific to eIF5. Dilutions of recombinant eIF5 (10–1200 fMol) and 500 fMol of 48S complexes were loaded as indicated and resolved on 12% NuPAGE gel. Native eIF5 in 48S complexes assembled without addition of recombinant eIF5 could only be detected upon overexposure of the immunoblots (not shown) and was estimated to be bound to 1% of both wt and ΔdII 48S complexes. Other band intensities were quantified using the ImageQuant software, and relative levels of eIF5 were normalised to that of the 1200 fMol eIF5 intensity. A graphical representation of the relative intensities is displayed on the right. All error bars are standard error of the mean. Using a response curve analysis, the following eIF5 concentrations within 48S complexes assembled in the presence of recombinant eIF5 were determined: 470±20 fMol for wt and 445±16 fMol for ΔdII HCV IRES. (B) Analysis of eIF5-mediated eIF release from HCV IRES 48S complexes. Detection of eIF2 and eIF3 by immunoblotting with antibodies against eIF2α and eIF3-p66, respectively, before (48S) and after treatment of wt or ΔdII IRES 48S complexes with GTP and eIF5 (bound or released). (C) Sucrose density gradient analyses of wt HCV IRES 48S complexes before and after incubation with GTP and eIF5. The unchanged position of 48S ribosomal complexes indicates that the particles stay intact after eIF2 release, and that eIF3 remains bound to the particle. The first fractions have been omitted for clarity. (D) Analysis of eIF5B-mediated eIF3 release from HCV IRES 48S complexes. Detection of bound or released eIF3 by immunoblotting with antibody against eIF3-p66 after incubation of wt IRES 48S complexes with different combinations of GTP, eIF5, eIF5B and 60S subunits as indicated. (E) Sucrose density gradient analysis of wt IRES 48S complexes before and after incubation with GTP, eIF5, 60S subunits, and with or without eIF5B. The positions of ribosomal complexes are indicated above appropriate peaks. The first fractions have been omitted for clarity.
Following eIF5-mediated GTP hydrolysis, eIFs need to be displaced from the 40S subunit to allow joining of the large 60S ribosomal subunit. To identify which eIFs are released from HCV 48S complexes upon eIF5-induced GTP hydrolysis and the possible involvement of domain II in this step, we monitored the release of eIF2 and eIF3 by analysing the presence of eIF2 and eIF3 in liquid and solid fractions of wt and ΔdII 48S particles following centrifugation. Excess amounts of GTP and eIF5 were added to the assembly mixtures to ensure that exchange of GMPPNP to GTP within the 48S particles and eIF5 binding are not rate limiting (see Supplementary data and Supplementary Figure 1). In the wt 48S complex, eIF5-induced GTP hydrolysis leads to nearly complete dissociation of eIF2 from the 48S complex, whereas eIF3 remains bound to the particle (Figure 3B, wt). Sucrose density gradient analysis shows that incubation of 48S particles with GTP and eIF5 does not significantly alter the migration of 48S particles which further supports that eIF3, a 700 kDa multisubunit protein complex, remains bound (Figure 3C). The extent of eIF2 release from ΔdII 48S complexes, on the contrary, was significantly lower than that from wt particles, whereas eIF3 stayed bound as in the case of the wt (Figure 3B, ΔdII). This clearly demonstrates that domain II promotes the release of eIF2 from HCV IRES 48S complexes.
Upon eIF5-mediated hydrolysis of eIF2-bound GTP, only eIF2/GDP is released, but eIF3 stays bound to the 48S particle. In the canonical initiation pathway, eIF3 dissociates during 60S subunit joining mediated by eIF5B in another GTP-dependent process (Pestova et al, 2000; Unbehaun et al, 2004). To investigate the release of eIF3 from HCV IRES 48S complexes, similar release experiments were carried out to monitor the presence of eIF3 in supernatant and pellet fractions of 80S assembly reaction mixtures. Purified wt 48S complexes were incubated with excess of GTP and eIF5 to assure eIF2 release in the presence or absence of eIF5B and 60S ribosomal subunits. Subsequently, the mixtures were fractionated by centrifugation and eIF3 was detected by immunoblotting. Incubation of the 48S complex with only GTP and eIF5 did not result in a significant release of eIF3 (Figure 3D, lane 1 and B, wt). Neither eIF5B nor 60S subunits added singly to the 48S complex in the presence of GTP and eIF5 enhanced eIF3 release (Figure 3D, lanes 2 and 3). On the other hand, when both eIF5B and 60S subunits were added together with GTP and eIF5, a significant displacement of eIF3 from the 48S complex was observed (Figure 3D, lane 4). When ΔdII 48S particles were analysed, the eIF3 release also required GTP, eIF5, eIF5B and 60S subunits, but the detected level of eIF3 release was significantly reduced (data not shown) due to impaired release of eIF2/GDP from ΔdII 48S particles (Figure 3B, ΔdII), which is a prerequisite for eIF3 release. However, when the amount of released eIF3 was normalised to the amount of released eIF2 in both wt and ΔdII cases, the extent of eIF3 release was unaffected by deletion of domain II (data not shown). This seems to indicate that release of eIF3 is domain II-independent. Additionally, 80S ribosome formation was monitored by sucrose density gradient analysis using purified 48S particles assembled onto 32P-labeled wt HCV IRES RNA and incubated with 60S ribosomal subunits in the presence or absence of eIF5B. In agreement with the results from the previous eIF3 release experiments, a peak signifying the formation of 80S ribosomes appeared only upon addition of GTP, eIF5, 60S ribosomal subunits and eIF5B (Figure 3E). Altogether this suggests that eIF3 is released during 60S subunit joining in an eIF5B-dependent manner.
The bent domain II conformation is conserved in the CSFV IRES
Our data show that HCV IRES domain II mediates eIF2 release during 80S formation, and we raised the question which RNA structural motifs are required for this function. The solution structure of HCV IRES domain II revealed an independently folded IRES domain, containing an apical, dynamic hairpin loop and a loop E motif and an asymmetric internal loop in the basal domain IIa, which is responsible for an overall bent conformation (Lukavsky et al, 2003). Previous mutational analyses of both the apical hairpin loop and loop E motif showed that these structural elements contain crucial residues for translational activity (Reynolds et al, 1996; Odreman-Macchioli et al, 2001; Kalliampakou et al, 2002). Although these apical structural motifs display high conservation in primary sequence in related flaviviruses such as CSFV and BVDV, their basal domain IIa is distinct from the HCV IRES (Honda et al, 1999) (Figure 1B and C). To test whether CSFV IRES domain IIa still adopts a bent conformation similar to HCV IRES domain IIa, the structure of this CSFV IRES domain was determined by solution NMR spectroscopy (Figure 1D). The structure determination employed standard homo- and heteronuclear NMR techniques (Lukavsky and Puglisi, 2001) and utilised torsion angle and nuclear overhauser effect (NOE)-derived distance restraints in combination with angular restraints derived from residual dipolar couplings (RDC) to improve both local and global precision of the structure (Lukavsky and Puglisi, 2005). The final ensemble of 11 structures displays a root-mean-square deviation (r.m.s.d.) for all heavy atoms of 0.95 Å, and the asymmetric, internal loop is also very well defined (local r.m.s.d. is 0.2 Å; Figure 4B and Table I). Similar to the HCV IRES, CSFV IRES domain IIa also adopts a bent conformation (bend angle 60.4±9.1° versus 84.5±9.6° for HCV, Lukavsky et al, 2003), which is introduced by its adenine-rich, asymmetric, internal loop (Figure 4C). The lower stem formed by the Watson–Crick base pairs extends into this internal loop by one non-canonical base pair formed between nucleotides A75 and C122 on the 5′- and 3′-side of the loop, respectively. A75 and C122 stack on top of the C74–G123 Watson–Crick base pair and form a single hydrogen bond between A75–N3 and C122–N4 (average distance is 2.80±0.08 Å) evidenced by a medium NOE between A75–H2 and C122–H5 (Figure 4A). This unusual base pair narrows the ribose C1'–C1' distance to 8.7±0.02 Å, as compared with regular A-form RNA (10.6 Å) and allows for the base moiety of A121 to stack across the strand on top of the base moiety of A75. This defined position of the base of A121 is supported by NOEs between the A121-H2 proton and A75-, G76-ribose protons (Figure 4A). The remaining single-stranded residues, A119 and A120, both form a continuous base stacking below the last G76–C118 Watson–Crick base pair of the upper helix, which orients the bases of A120 and A121 almost at a right angle to each other and introduces a 90° kink in the backbone between A120 and A121 reflected in non-A-form values for torsion backbone angles (Figure 4C and Supplementary Table I). In addition, this architecture places the base moiety of A119 directly above the six-membered ring of the purine moiety of A120, evidenced in a strong upfield shift of the A119-H2 proton resonance to 6.45 p.p.m. (Figure 4A). The upper helix contains a stretch of non-canonical base pairs, such as G78–A116, A79–U115, G82–A113, and a dynamic, looped-out residue C80, which introduces a second, minor bend (bend angle 21.7±3.7°), reminiscent of the HCV IRES (bend angle 46.8±13.2°) (Lukavsky et al, 2003). The structure of CSFV IRES domain IIa displays a preservation of the overall shape previously seen in HCV IRES domain IIa, suggesting their conserved bent conformation might be important for domain II function.
Figure 4.
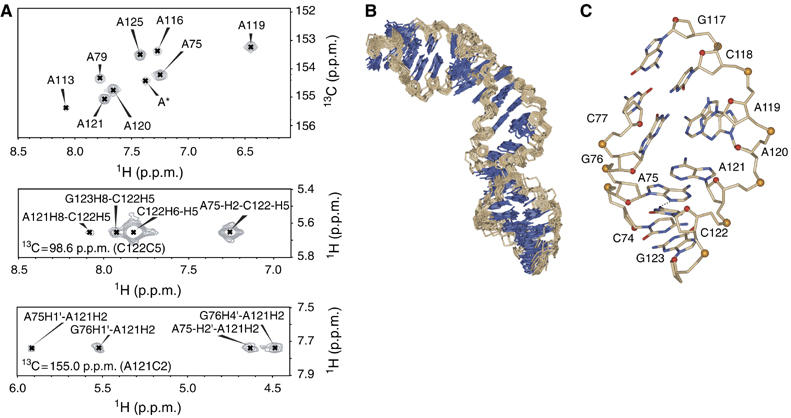
Structure of the CSFV IRES domain IIa. (A) 13C-HSQC (heteronuclear single-quantum coherence) spectra of adenine C2-H2 correlations in CSFV domain IIa and two representative 13C planes from the 3D 13C-edited NOESY-HSQC spectrum showing NOE crosspeaks, which define the structure of the internal loop. Numbering according to Figure 1D. The n+1 residue is labeled A*. (B) Heavy-atom superposition of the final 11 CSFV domain IIa structures calculated with RDCs. Bases are blue and the ribose–phosphate backbone is light brown. (C) Single representative view of the asymmetric internal loop from the minor groove. Numbering according to (A). All nitrogen atoms are blue, oxygen atoms of the bases are red, all carbon atoms of the bases and ribose–phosphate backbone atoms are light brown, except ribose O4′ atoms (red spheres) and phosphorus atoms (yellow spheres). The hydrogen bond between A75-N3 and C122-N4 is indicated as a dashed line.
Table 1.
Structural statistics
| Experimental restraints | |
| NOE-derived distance restraintsa | 606 |
| Hydrogen bonding distance and planarity restraints | 25 |
| Dihedral restraints | 271 |
| RDC-derived angular restraints | 107 |
| R.m.s. deviation from experimental restraintsb | |
| Distance (Å) | 0.017±0.0005 |
| Dihedral (deg) | 0.66±0.02 |
| RDC (Hz)c | 1.31±0.04 |
| R.m.s. deviation from ideal geometriesb | |
| Bonds (Å) | 0.003±0.00003 |
| Angles (deg) | 0.84±0.005 |
| Impropers (deg) | 0.43±0.02 |
| R.m.s. deviation from the average structure (Å)b | |
| All RNA | 0.94 |
| Upper stem (G76–C84 and G111–C118) | 0.68 |
| Lower stem (C70–C74 and G123–G127) | 0.90 |
| Internal loop and adjacent WC base pairs (C74–G76 and C118–G123) | 0.20 |
| aOnly meaningful, non-fixed distance constraints were used. | |
| bAverage and s. d. values obtained from 11 converged structures, which had no distance restraint violation above 0.2 Å and dihedral restraint violation >5°. | |
| cThe axial (Da) and rhombic (R) component of the alignment tensor used in the final structure calculation are Da=−13.09 and R=0.182, respectively. | |
Conserved structural motifs within domain II are required for eIF2 release
To examine which of the conserved structural motifs found in both HCV and CSFV domain II are required for 80S ribosome assembly, we used deletion mutants of HCV and CSFV IRESes, in which the conserved domain II hairpin loop (ΔdIIb), loop E motif (ΔloopE) or loop IIa-bend (ΔdIIa) were deleted individually or all together (ΔdII) (Figure 1B and C). As some of these mutations could also affect formation of binary IRES–40S complexes or simply sterically hinder 60S subunit joining, sucrose density gradient analyses were used to confirm that none of the mutations blocked binary IRES–40S complex formation as well as factorless assembly of IRES–80S particles (Lancaster et al, 2006) (Supplementary Figure 2). The effect of these domain II mutations on 80S ribosome formation was then monitored by sucrose density gradient analyses of the assembly reactions in RRL with 32P-labeled IRES RNAs in the presence of cycloheximide. Deletion of the entire domain II as well as deletion of individual conserved structural elements in both the HCV and CSFV IRES RNAs led to accumulation of 48S complexes and therefore greatly reduced 80S formation as compared with wt IRESes (Figure 5A and B). As domain II promotes eIF5-mediated eIF2 release, we questioned whether all the conserved domain II motifs are essential for this step. The efficiency of eIF2 release from the purified 48S complexes assembled onto wt or domain II mutant (ΔdIIb, ΔloopE, ΔdIIa orΔdII) CSFV and HCV IRES RNAs was examined. Immunoblotting with antibodies against eIF2α, eIF3-p66 and eIF5 was used to confirm that none of the mutations affected the recruitment of eIF2, eIF3 or eIF5 to 48S complexes (Supplementary Figure 3). Incubation of wt CSFV or HCV 48S complexes in the presence of excess GTP and eIF5 led to near-complete dissociation of eIF2 from 48S complexes (Figure 5C and D). Deletion of the entire domain II as well as individual domain II motifs, in contrast, showed a five-fold decrease in the levels of eIF2 release for both HCV and CSFV 48S complexes (Figure 5C and D).
Figure 5.
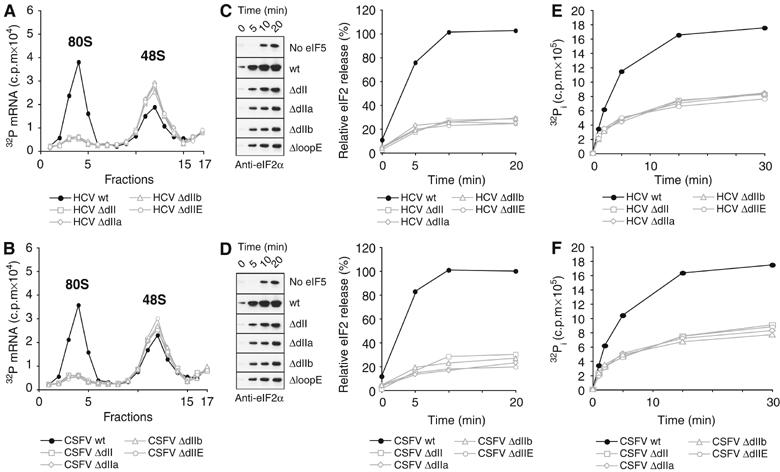
Conserved structural motifs within the HCV and CSFV domain II are required for 80S ribosome formation. (A–F) Specific mutations of HCV and CSFV IRES RNAs (ΔdII, ΔdIIa, ΔIIb and ΔloopE) are depicted in Figure 1B and C. (A) Sucrose density gradient analysis of wt and mutant HCV IRES ribosomal complexes. Assays were performed in RRL pre-incubated with cycloheximide. The positions of ribosomal complexes are indicated above appropriate peaks. The first fractions have been omitted for clarity. (B) Sucrose density gradient analysis of wt and mutant CSFV IRESes in RRL. Assays were performed as for HCV IRESes (A). (C) Analysis of eIF5-mediated eIF2 release from wt and mutant HCV IRES 48S complexes assembled and purified in the absence of recombinant eIF5. Release of eIF2 was induced by the addition of excess GTP and eIF5 and assayed over time by immunoblotting with antibody against eIF2α. A control reaction without the addition of eIF5 was used to quantify background dissociation of eIF2 from 48S complexes (left). Band intensities were measured to quantify the relative amount of eIF2 release from 48S complexes (right) and normalised to the initial amount of eIF2 in untreated 48S complexes (right). Data represent the average of three experiments. (D) Analysis of eIF5-mediated eIF2 release from wt and mutant CSFV IRES 48S complexes. Assays and data analyses were performed as for HCV IRESes (C). (E) Analysis of eIF5-induced GTP hydrolysis in wt and mutant HCV IRES 48S complexes assembled and purified in the absence of recombinant eIF5. 48S complexes were treated with γ32P-GTP and eIF5 and 32P–Pi quantified at different time points reflecting the amount of total GTP hydrolysis. Data represent the average of four experiments. (F) Analysis of eIF5-induced GTP hydrolysis in wt and mutant CSFV IRES 48S complexes. Assays were performed as for HCV IRESes (E).
As dissociation of eIF2/GDP from 48S complexes occurs upon eIF5-mediated GTP hydrolysis (Das and Maitra, 2001), the effect of domain II mutations on GTP hydrolysis was also examined. Purified 48S complexes assembled onto wt and domain II mutant HCV or CSFV IRESes were incubated in the presence of γ32P-GTP and eIF5, and total inorganic 32P–Pi was measured over time. Near complete hydrolysis of eIF2-bound GTP was observed only for the wt HCV and CSFV IRESes, whereas all domain II mutants displayed a 50% reduction in the hydrolysis efficiency over the same time period (Figure 5E and F). These data clearly demonstrate that eIF5-mediated GTP hydrolysis and the subsequent release of eIF2/GDP are both promoted by the conserved structural motifs within HCV and CSFV domain II and that none of the motifs is dispensable.
Discussion
The HCV IRES domain II is essential for IRES function (Rijnbrand et al, 1995), but its precise role remained unclear. It has been suggested that this domain positions the AUG start codon on the 40S subunit through interactions with the HCV ORF, or indirectly by modulating 40S subunit conformation (Kolupaeva et al, 2000; Spahn et al, 2001). On the other hand, a domain III-only IRES, lacking domain II, still forms 48S complexes, but subsequent 80S ribosome assembly is impaired (Ji et al, 2004; Otto and Puglisi, 2004). This defect in 80S formation could therefore be either a consequence of improper 48S assembly or caused by a blockade of events downstream of 48S complex formation. We show that the IRES domain III alone is sufficient for 48S assembly. Our experiments demonstrate that the IRES domain III alone mediates proper base pairing between Met-tRNAiMet and the authentic HCV AUG start codon, and that IRES domain II is not required for AUG start codon selection process (Figure 2A and C). Instead, domain II controls 80S ribosome formation downstream of 48S assembly (Figure 2B). While both wt and ΔdII 48S complexes recruit eIF5 with equal efficiency (Figure 3A), only the wt IRES displays efficient eIF5-induced activation of GTP hydrolysis by eIF2 and subsequent release of eIF2/GDP, whereas both processes are impaired in the ΔdII IRES 48S complex (Figure 3 and 5). These combined data suggest that HCV IRES domain II is required to actively promote both eIF5-mediated GTP hydrolysis and subsequent eIF2/GDP release from 48S complexes.
The IRES domain II-dependent mode of eIF2 release clearly contrasts the canonical initiation, where P-site codon–anticodon base pairing alone commits the 48S complex to eIF2 release and subsequent steps of 80S ribosome assembly (Unbehaun et al, 2004; Algire et al, 2005). In canonical initiation, eIF1 and eIF1A aid to locate the proper AUG start codon during scanning while eIF1 also acts as a negative regulator by inhibiting premature Pi release and thereby dissociation of eIF2/GDP prior to AUG start codon selection (Unbehaun et al, 2004; Algire et al, 2005). Both domain III and eIF1 bind to the 40S platform and their binding is proposed to modulate 40S conformation (Spahn et al, 2001; Lomakin et al, 2003; Maag et al, 2006). It is not yet known whether the structural changes imposed on the 40S platform upon binding of IRES domain III or eIF1 are similar or different, but their effect is similar in that dissociation of eIF2/GDP is blocked. In canonical initiation, the inhibitory effect of eIF1 is relieved upon recognition of the AUG start codon, which weakens the binding affinity of eIF1 to the 40S platform thereby allowing efficient GTP hydrolysis, Pi release and subsequent release of eIF2/GDP (Unbehaun et al, 2004; Algire et al, 2005). The HCV IRES domain III, on the other hand, being a part of the mRNA stays bound to the 40S subunit upon AUG start codon selection and would thereby retain its inhibitory effect on eIF2 release beyond establishment of codon-anticodon base pairing. To counteract this effect a stimulatory element is therefore required to promote both GTP hydrolysis and subsequent eIF2/GDP release, which is the precise role of domain II during IRES-mediated initiation (Figure 6).
Figure 6.
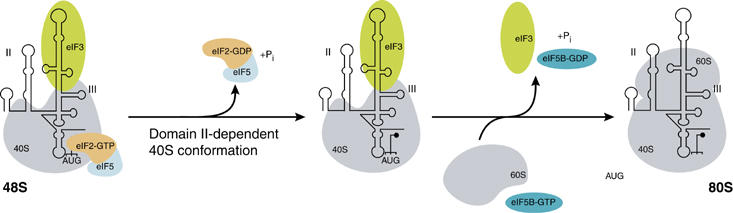
Model of HCV IRES-meditated ribosomal subunit joining. HCV IRES domain II modulates 40S ribosomal subunit conformation to promote eIF5-dependent hydrolysis of eIF2-bound GTP and subsequent release of eIF2/GDP. The release of the remaining factor eIF3 requires eIF5B and GTP hydrolysis and occurs during the joining of the 60S ribosomal subunit to form the 80S ribosome. HCV IRES domains II and III and the AUG start codon are indicated.
Domain II binds to the ribosomal head and platform near the E-site, where it interacts with rpS5 (Fukushi et al, 2001) and causes conformational changes in the 40S subunit (Spahn et al, 2001). It is tempting to speculate that these IRES domain II-dependent structural rearrangements triggered by its interaction with the rpS5, induce a favourable configuration of eIF5 and the ternary complex components (eIF2/GTP/Met-tRNAiMet) on the 40S subunit. This rearrangement might be responsible for the increased rate of hydrolysis of eIF2-bound GTP and also the weakening of the binding affinity of eIF2/GDP to the 40S subunit allowing their subsequent dissociation from the complex. Once eIF2 is released, the remaining factor eIF3 can also be displaced efficiently from the HCV IRES 48S particle. The release of eIF3 requires eIF5B and GTP hydrolysis and occurs during the joining of the 60S subunit in the same mode as in the canonical initiation pathway (Figure 6). The observed, basal levels of eIF2 release from ΔdII 48S complexes might reflect that a domain II-induced 40S conformation can also be adopted without domain II, but it occurs less efficiently or frequently (Figure 3B, ΔdII). These altered dynamic properties of the 40S subunit upon ΔdII IRES binding can also explain the consistently weaker toeprint observed in ΔdII 48S complexes (Pestova et al, 1998b; Otto and Puglisi, 2004) (Figure 2A). The only slight, two-fold increase in GTP hydrolysis in wt IRES 48S complexes, as compared with ΔdII IRES, suggests that whereas eIF2/GDP release is strongly linked to the domain II-dependent 40S conformation, GTP hydrolysis by eIF2 itself depends mainly on the interaction with its GTPase activator, eIF5, within the 48S particle (Unbehaun et al, 2004; Algire et al, 2005).
To better understand how the function of domain II is encoded in a variety of IRES RNAs, we dissected the role of conserved domain II elements found in different HCV-like IRESes. The previously determined solution structure of the HCV IRES domain II showed three characteristic features, an apical hairpin loop, a loop E motif and an internal asymmetric loop, which is responsible for an approximately 90° bend in the structure (Lukavsky et al, 2003). The fact that this bent conformation is retained in the HCV IRES upon binding to the 40S subunit suggested that this preformed conformation might be functionally important (Spahn et al, 2001). HCV-like IRESes within the Flaviviridae family share the same major structural domains and functional similarities with the HCV IRES (Brown et al, 1992; Wang et al, 1995; Rijnbrand et al, 1997; Pestova et al, 1998b; Honda et al, 1999; Pestova and Hellen, 1999). Moreover, the primary sequences of the apical loop and the loop E motif are strictly conserved among HCV, CSFV and BVDV, but their basal domain IIa, containing an asymmetric internal loop, differs significantly. The structure of CSFV domain IIa determined by NMR spectroscopy revealed that this domain also adopts a bent conformation in solution (Figure 4B and C). The shape conservation of the basal domain IIa between HCV and CSFV suggests that the functional role of this asymmetric loop is to place the apical conserved motifs (hairpin loop and loop E) at the interface between 40S ribosomal head and platform, where they interact with rpS5 and other ribosomal proteins and possibly rRNA to modulate 40S subunit conformation. If the bent conformation of domain IIa is eliminated by deleting single-stranded nucleotides within the asymmetric internal loop, levels of eIF2 release and GTP hydrolysis are reduced to 20 and 50%, respectively; the same reduction seen with ΔdII IRESes (Figure 5). Deletion of the other structural motifs also gives rise to the same reduction, suggesting that each conserved structural element is required for correctly positioning IRES domain II within the IRES 48S complex.
The two major RNA domains of HCV and HCV-like IRESes play distinct roles during IRES-mediated initiation. Domain III recruits the 40S subunit, eIF3 and the ternary complex and mediates the assembly of 48S complexes at the correct AUG start codon. Therefore, canonical initiation factors from the eIF4 family, which recruit the 43S particle to the 5′-capped mRNA, as well as eIF1 and eIF1A, which aid scanning and start codon selection, are not required in the IRES-mediated pathway. Domain II, on the other hand, displays a novel function, which lacks a canonical counterpart. By modulating 40S conformation, domain II promotes eIF5-dependent GTP hydrolysis and eIF2/GDP release and commits the complex to 80S ribosome formation. This commitment checkpoint solely depends on AUG start codon recognition in the canonical counterpart. It remains to be seen whether other viral or cellular IRES RNAs with different domain organisation and eIF requirement also encode the distinct functional domains found in the HCV IRES-like family. Affinity-purified IRES 48S complexes used in this study to dissect their eIF composition and release as a function of individual IRES domains could provide a useful tool to answer this open question.
Materials and methods
Preparation of tagged IRES RNAs
Standard PCR and cloning techniques were used to create a DNA construct containing the wt HCV IRES (1–426), the primer sequence for toeprinting and the streptomycin aptamer for purification using overlapping primers as previously described (Locker et al, 2006). DNA template for the wt CSFV IRES (1–452, GenBank database number JO4358) was constructed similarly. DNA templates for the domain II deletion mutants (ΔdII, ΔdIIa, ΔdIIb and ΔloopE) were prepared by PCR using the wt HCV or CSFV plasmids and appropriate primers. PCR products were digested with EcoRI and HindIII and ligated into pUC18 digested with the same enzymes. RNA oligonucleotides were transcribed in vitro from the above plasmids, linearised with EcoRI and purified by size-exclusion chromatography as previously described (Lukavsky and Puglisi, 2004). Radiolabeled IRES RNAs were transcribed as above in the presence of α32P-CTP (3000 mCi/mmol) and purified by denaturing polyacrylamide gel electrophoresis (PAGE). For crosslinking experiments, IRES RNAs were transcribed as above with the addition of 25% 4-thio-UTP and purified using RNeasy Mini kits (Qiagen). An RNA containing 4-thiouridine only within domain II (wt-lig) was prepared by transcribing and purifying individual parts and then by ligation of the parts with T4 RNA ligase as previously described (Kim et al, 2002).
Preparation of factors and ribosomal subunits
Details of the preparation of factors and ribosomal subunits are given in Supplementary Methods.
In vitro assembly and affinity purification of the 48S complexes
The 48S complexes were assembled in RRL onto IRES RNAs in the presence of 2 mM GMPPNP and then isolated using a StreptoTag-based affinity purification method with the streptomycin aptamer as described before (Locker et al, 2006). To test eIF5 recruitment to the 48S complexes, RRL was supplemented with purified, recombinant eIF5 (final concentration 1 μM). The 48S complexes used for eIF release experiments and GTP hydrolysis assay were assembled in the absence of recombinant eIF5 and purified as above, except that the GMPPNP concentration was lowered to 0.2 mM. The 48S complexes used for sucrose density gradient analyses were assembled onto 32P-labeled RNA (specific activity 100 000 c.p.m/pmol mRNA) and purified as described above. Details of the StreptoTag-based affinity purification method are also described in Supplementary Methods.
Analysis of 48S and 80S complexes
Toeprinting analysis was performed in buffer A (20 mM Tris pH 7.6, 100 mM KOAc, 2.5 mM MgCl2 and 2 mM DTT) as described previously (Wilson et al, 2000). Sucrose density gradient centrifugation of ribosomal particles was performed in buffer A containing 10–50% sucrose as previously described (Anthony and Merrick, 1992; Pestova et al, 1996; Wilson et al, 2000).
Analysis of eIF2 and eIF3 release from 48S complexes
For eIF5-induced eIF release experiments, 25 pmoles of purified 48S complexes were incubated for 15 min at 37°C in buffer A with 1.5 mM GTP and 100 pmole of eIF5 in a total volume of 150 μl. Ribosomal complexes were pelleted by centrifugation and the presence of eIF2 and eIF3 in pellets and supernatants was tested by immunoblotting (see below). Samples containing 5 pmoles of 48S complexes assembled onto 32P-labeled IRES RNAs and incubated in 50 μl of buffer A (with 1.5 mM GTP and without or with 20 pmoles of eIF5) for 15 min at 37°C were analysed by sucrose density gradient centrifugation.
For eIF5B-induced eIF release experiments, 25 pmoles of 48S complexes were incubated in buffer A (150 μl) containing various combinations of 1.5 mM GTP, 100 pmoles of eIF5, 100 pmoles of eIF5B and 40 pmoles of 60S subunits for 15 min at 37°C and assayed as above. The 80S ribosome assembly was monitored by sucrose density gradient analysis using samples containing 5 pmoles of 48S complexes assembled onto 32P-labeled IRES RNAs and incubated for 15 min at 37°C in 50 μl of buffer A supplemented with combinations of 1.5 mM GTP, 20 pmoles of eIF5, 20 pmoles of eIF5B and 10 pmoles of 60S subunits.
Immunoblotting
To test eIF composition of 48S complexes, proteins from 2.5 pmoles of purified particles were resolved by electrophoresis on 4–12% NuPAGE gels and detected using antibodies against eIF2α (Abcam, ab5369), eIF3-p66 (PTGlab, 10219–1-AP) or eIF5 (SantaCruz Biotechnology, sc-282), then with appropriate HRP-conjugated secondary antibodies (Abcam) followed by phosphoimager analysis. To test eIF release from 48S complexes, pellets from the centrifugation step were resuspended in gel loading buffer (100 mM Tris pH 6.8, 2 mM DTT, 4% SDS, 0.2% bromophenol blue and 20% glycerol) while the corresponding supernatants were TCA extracted, precipitated and resuspended in the same buffer and analysed as above. For quantitative analysis of eIF2 release, supernatant samples were collected at various time points and analysed for the presence of eIF2 by immunoblotting while a reaction mixture lacking eIF5 served as a control. Band intensities were quantified using a phosphoimager and normalised to the initial eIF2 quantity in the 48S complexes. Nonspecific eIF2 dissociation observed in the control was subtracted to calculate the percentage of eIF2 released.
GTP hydrolysis assays
48S complexes (2.5 pmoles) were incubated in buffer A with 100 μM GTP, 50 μCi of γ32P-GTP and 10 pmoles of eIF5 in a total volume of 200 μl for 15 min at 37°C. A 20 μl volume of aliquots was removed at various time points and assayed for the amount of inorganic 32P–Pi release (Conway and Lipmann, 1964). Data were averaged from four independent experiments.
Crosslinking and Northern blot analysis
Purified 48S complexes (8 pmoles) assembled onto 4-thiouridine-labeled IRES RNAs were irradiated at 360 nm on ice for 30 min using a handheld light source. Total RNA was extracted using the acetic acid/MgCl2 method (Hardy et al, 1969) and separated on a 4% denaturing PAGE with in vitro-transcribed tRNAiMet (Pestova and Hellen, 2001) and tagged HCV IRES RNA as positive and negative controls, respectively. Gels were probed for the presence of tRNAiMet by Northern blotting as described (Locker et al, 2006).
NMR sample preparation
RNA oligonucleotides comprising CSFV IRES domain IIa were prepared by in vitro transcription from linearised plasmid DNA using phage T7 RNA polymerase, followed by gel filtration (Lukavsky and Puglisi, 2004) and equilibration in centrifugal devices against the final buffer (10 mM sodium phosphate buffer pH 6.4, 50 mM NaCl). Unlabeled and 13C,15N-labeled RNA oligonucleotides were prepared as previously described (Lukavsky et al, 2003). NMR samples were prepared in a Shigemi NMR tube (280 μl containing 4% or 100% D2O and 0.25 mM d12-EDTA) at RNA concentrations of 0.8–1.5 mM. Weakly aligned NMR samples were prepared by addition of 8 mg ml−1 of filamentous phage Pf1 (Hansen et al, 1998).
NMR spectroscopy
NMR spectroscopy data were recorded and analysed as described in Supplementary Methods.
Structure calculation and analysis
The structures were calculated and analysed as described in Supplementary Methods.
Accession codes
The coordinates of the CSFV IRES domain IIa have been deposited in the Protein Data Bank under accession number 2HUA.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Data
Acknowledgments
We thank members of the K Nagai and PJ Lukavsky groups for helpful discussions and Y Shibata for helpful discussions and comments on the paper. NL is supported by a career development fellowship from MRC.
References
- Algire MA, Maag D, Lorsch JR (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262 [DOI] [PubMed] [Google Scholar]
- Anthony DD, Merrick WC (1992) Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem 267: 1554–1562 [PubMed] [Google Scholar]
- Brown EA, Zhang H, Ping LH, Lemon SM (1992) Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res 20: 5041–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Maitra U (1991) Function of eukaryotic initiation factor 5 in the formation of an 80S ribosomal polypeptide chain initiation complex. J Biol Chem 266: 14039–14045 [PubMed] [Google Scholar]
- Chaudhuri J, Das K, Maitra U (1994) Purification and characterization of bacterially expressed mammalian translation initiation factor 5 (eIF-5): demonstration that eIF-5 forms a specific complex with eIF-2. Biochemistry 33: 4794–4799 [DOI] [PubMed] [Google Scholar]
- Conway TW, Lipmann F (1964) Characterization of a ribosome-linked guanosine triphosphatase in Escherichia coli extracts. Proc Natl Acad Sci USA 52: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ghosh R, Maitra U (2001) Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J Biol Chem 276: 6720–6726 [DOI] [PubMed] [Google Scholar]
- Das S, Maitra U (2001) Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog Nucleic Acid Res Mol Biol 70: 207–231 [DOI] [PubMed] [Google Scholar]
- Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN (2003) Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett 533: 99–104 [DOI] [PubMed] [Google Scholar]
- Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K (2001) Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem 276: 20824–20826 [DOI] [PubMed] [Google Scholar]
- Godchaux W III, Adamson SD, Herbert E (1967) Effects of cycloheximide on polyribosome function in reticulocytes. J Mol Biol 27: 57–72 [DOI] [PubMed] [Google Scholar]
- Hansen MR, Mueller L, Pardi A (1998) Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol 5: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Hardy SJ, Kurland CG, Voynow P, Mora G (1969) The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry 8: 2897–2905 [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612 [DOI] [PubMed] [Google Scholar]
- Honda M, Beard MR, Ping LH, Lemon SM (1999) A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol 73: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA (2004) Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA 101: 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliampakou KI, Psaridi-Linardaki L, Mavromara P (2002) Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett 511: 79–84 [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR (2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657–704 [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA (2001) Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA (1999) The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol 292: 513–529 [DOI] [PubMed] [Google Scholar]
- Kim I, Lukavsky PJ, Puglisi JD (2002) NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J Am Chem Soc 124: 9338–9339 [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU (2000) An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AM, Jan E, Sarnow P (2006) Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12: 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ (2006) Affinity purification of eukaryotic 48S initiation complexes. RNA 12: 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD (2003) Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Puglisi JD (2001) RNAPack: an integrated NMR approach to RNA structure determination. Methods 25: 316–332 [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Puglisi JD (2004) Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA 10: 889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Puglisi JD (2005) Structure determination of large biological RNAs. Methods Enzymol 394: 399–416 [DOI] [PubMed] [Google Scholar]
- Lytle JR, Wu L, Robertson HD (2002) Domains on the hepatitis C virus internal ribosome entry site for 40 s subunit binding. RNA 8: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Algire MA, Lorsch JR (2006) Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol 356: 724–737 [DOI] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR (2005) A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275 [DOI] [PubMed] [Google Scholar]
- Merrick WC (1979) Evidence that a single GTP is used in the formation of 80S initiation complexes. J Biol Chem 254: 3708–3711 [PubMed] [Google Scholar]
- Merrick WC (2004) Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332: 1–11 [DOI] [PubMed] [Google Scholar]
- Odreman-Macchioli F, Baralle FE, Buratti E (2001) Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem 276: 41648–41655 [DOI] [PubMed] [Google Scholar]
- Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD (2002) Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA 8: 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD (2004) The pathway of HCV IRES-mediated translation initiation. Cell 119: 369–380 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU (1998a) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU (1999) Internal initiation of translation of bovine viral diarrhea virus RNA. Virology 258: 249–256 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU (2001) Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA 7: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU (1998b) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ (1996) Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2: 867–878 [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W (1995) Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett 365: 115–119 [DOI] [PubMed] [Google Scholar]
- Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJ, Bredenbeek PJ (1997) Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol 71: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW (1997) Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89: 831–838 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40 s ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Stade K, Rinke-Appel J, Brimacombe R (1989) Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5′ with respect to the decoding site. Nucleic Acids Res 17: 9889–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Willis AE (2004) Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23: 3200–3207 [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Le SY, Ali N, Siddiqui A (1995) An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1: 526–537 [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P (2000) Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Data


