Abstract
Many genomic loci contain transcription units on both strands, therefore two oppositely oriented transcripts can overlap. Often, one strand codes for a protein, whereas the transcript from the other strand is non-encoding. Such natural antisense transcripts (NATs) can negatively regulate the conjugated sense transcript. NATs are highly prevalent in a wide range of species—for example, around 15% of human protein-encoding genes have an associated NAT. The regulatory mechanisms by which NATs act are diverse, as are the means to control their expression. Here, we review the current understanding of NAT function and its mechanistic basis, which has been gathered from both individual gene cases and genome-wide studies. In parallel, we survey findings about the regulation of NAT transcription. Finally, we hypothesize that the regulation of antisense transcription might be tailored to its mode of action. According to this model, the observed relationship between the expression patterns of NATs and their targets might indicate the regulatory mechanism that is in action.
Keywords: natural antisense transcript (NAT), regulatory RNA, RNA interference, tile array, noise dampening
Introduction
Natural antisense transcripts (NATs) are endogenous RNA molecules containing sequences that are complementary to other transcripts. NATs can be divided into cis-NATs, which are transcribed from opposing DNA strands at the same genomic locus, and trans-NATs, which are transcribed from separate loci. cis-NAT pairs display perfect sequence complementarity (as expected from their genomic overlap), whereas trans-NAT pairs display imperfect complementarity and can therefore target many sense targets to form complex regulation networks (Li et al, 2006).
cis-NATs were first detected in viruses (Barrell et al, 1976), then in prokaryotes (Tomizawa et al, 1981; Wagner & Simons, 1994) and finally in eukaryotes (Knee & Murphy, 1997; Kumar & Carmichael, 1998; Williams & Fried, 1986). The importance of NATs is now apparent, and their widespread prevalence has been reported in many genomes (see next section). cis-NATs can be categorized according to their relative orientation and degree of overlap; head-to-head (5′ to 5′), tail-to-tail (3′ to 3′) or fully overlapping (Fig 1). All genome-wide studies, except one (Katayama et al, 2005), have reported the tail-to-tail orientation to be the most prevalent. Overlapping transcripts might comprise two protein-encoding genes, one protein-encoding and one non-encoding gene, or two non-encoding transcripts.
Figure 1.
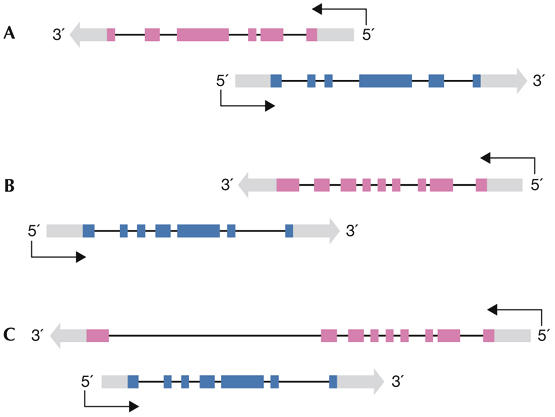
Relative orientation of cis-natural antisense transcript pairs. (A) Headto-head (5′ to 5′) overlap involving 5′-untranslated regions and coding exons. (B) Tail-to-tail (3′ to 3′) overlap. (C) Fully overlapping (one gene included entirely within the region of the other). Coloured boxes represent exons, grey boxes represent untranslated regions.
The conservation of the phenomenon across kingdoms (Wagner & Flardh, 2002) implies that it might constitute a common mechanism for regulating gene expression. NATs have been proposed to regulate the expression of their target genes at several levels including transcription, messenger RNA processing, splicing, stability, cellular transport and translation. They are also linked to monoallelic gene expression through mechanisms that include genomic imprinting, X-inactivation and clonal expression. However, apart from a few experimentally validated cases (Korneev et al, 1999; Kramer et al, 2003; Sleutels et al, 2003), the physiological roles of antisense transcription and the underlying mechanisms are largely unknown.
Here, we summarize recent findings about the extent of the NAT phenomenon, the modes of regulation by NATs, and the transcriptional regulation of NATs and their targets. We focus on the extensively characterized cis-NATs and suggest that a relationship exists between the mechanism of antisense function and the transcriptional regulation of sense and antisense transcripts: some mechanisms of antisense action require co-expression with its target, some require a time delay between the onset of antisense and sense transcription, and other mechanisms manifest themselves in anti-correlated expression patterns of the regulator and its target. This concept could be used to infer the regulatory mechanism of action, given the expression profiles of antisense and their sense targets.
Extent of the NAT phenomenon
Genome-wide natural antisense transcription has been reported in various animal and plant species including humans (Chen et al, 2004; Sun et al, 2006; Yelin et al, 2003), mice (Carninci et al, 2005; Katayama et al, 2005; Kiyosawa et al, 2003; Sun et al, 2006), rats and chickens (Sun et al, 2006), Drosophila (Misra et al, 2002; Sun et al, 2006), nematodes (Sun et al, 2006), rice (Osato et al, 2003), Arabidopsis (Jen et al, 2005; Wang et al, 2005) and yeast (David et al, 2006; Havilio et al, 2005). The estimated extent of the phenomenon, measured in terms of the percentage of transcriptional units involved in an overlap, ranges from 5% to 29% in animals (excluding nematodes, which have a much lower percentage of 0.5%) and from 7% to 9% in plants. Most estimates are based on the alignment of full-length cDNAs and expressed sequence tags (ESTs) to the genome, and the identification of overlapping transcripts on opposite strands. Such a procedure is limited to the detection of cis-NATs, implying that the extent of the NAT phenomenon might be much broader. Some predictions have been experimentally validated using methods such as reverse transcription–PCR (RT-PCR) and microarrays containing strand-specific probes (Chen et al, 2004; Yelin et al, 2003).
In mice, all known full-length cDNAs have been clustered into 43,553 transcriptional units and mapped onto the genome (Katayama et al, 2005). Almost 29% (12,519) of all mapped transcriptional units were found to overlap with a cDNA that mapped to the opposite strand, greatly exceeding any previous prediction.
In humans, comparable numbers of overlapping transcript pairs were reported by Yelin and colleagues (2,667; Yelin et al, 2003) and Chen and colleagues (2,940; Chen et al, 2004). Both groups aligned ESTs and cDNAs to the genome to create transcriptional clusters and experimentally validated some of their predictions. Chen and colleagues reported that nearly 22% (5,880) of their 26,741 transcriptional clusters form sense–antisense pairs, which is the highest number estimated for humans. Furthermore, the data sets published by the two research groups overlap by only 38%, indicating that the total number of human sense–antisense pairs might be even greater.
A genome-wide tile array (Bertone et al, 2004) revealed 10,595 novel human transcripts, 1,187 of which lie antisense to annotated exons. Annotation of the Drosophila genome identified 1,027 sense–antisense pairs, representing 15% of the 13,379 genes (Misra et al, 2002). In plants, annotation of full-length rice cDNAs revealed 687 overlapping cluster pairs, amounting to around 7% of all cDNA clusters (Osato et al, 2003), and similar percentages were obtained in Arabidopsis thaliana (Table 1; Wang et al, 2005). Genome-wide antisense transcription in yeast was recently reported (Havilio et al, 2005) and further supported by the construction of high-density tile arrays of the Saccharomyces cerevisiae genome (David et al, 2006). When grown in rich medium, 85% of the genome was expressed, including hundreds of non-encoding transcripts. Antisense transcripts were identified for up to 1,555 of the genes. These findings confirm that antisense transcription is a highly conserved phenomenon that spans the animal, plant and fungal kingdoms.
Table 1.
Genome-wide natural antisense transcripts in several eukaryotic species
| Species | Transcripts involved in overlap | Total transcripts | Percentage (%) | References |
|---|---|---|---|---|
| Human | 5,880 | 26,741 | 22 | Chen et al, 2004; Sun et al, 2006 |
| Mouse | 12,519 | 43,553 | *29 | Katayama et al, 2005 |
| Rat | 548 | 11,332 | 5 | Sun et al, 2006 |
| Chicken | 356 | 7,390 | 5 | Sun et al, 2006 |
| Drosophila | 2,054 | 13,379 | *15 | Misra et al, 2002; Sun et al, 2006 |
| Rice | 1,374 | 20,477 | 7 | Osato et al, 2003 |
| Arabidopsis | 2,680 | 29,993 | 9 | Wang et al, 2005 |
| Nematode | 76 | 14,406 | 0.5 | Sun et al, 2006 |
| Yeast | 610 | 7,598 | *8 | David et al, 2006 |
*Sun and colleagues recently reported a significantly lower proportion ofonly 12% for mouse (2,212 out of19,100),and a slightly higher proportion of17% for Drosophila (1,814 out of10,542; Sun et al, 2006). Estimations in yeast are based on both poly(A) RNA and total RNA.
Regulating the regulator
Most coding cis-NAT pairs overlap in their untranslated regions (UTRs). Such overlaps often involve alternative polyadenylation, which creates transcript variants that differ in their 3′ termini. Similarly, transcripts with heterogeneous transcription start sites might form head-to-head overlaps. A key question is whether such alternative (3′ or 5′) end processing is intentional—forming regulated transcript overlaps—or does it result from ‘leakage' of the RNA transcription machinery?
To address this question, Dahary and colleagues examined the evolution of cis-NATs (Dahary et al, 2005). They defined a set of consecutive gene pairs in the human genome and identified their orthologous gene pairs in both mouse and Fugu. The human genes were divided into sense–antisense pairs and pairs that were transcribed from the same strand. The authors assumed that if sense–antisense pairs carried a beneficial function, selection would work against their separation in related species. Indeed, 23.3% (55 out of 236) of the human sense–antisense pairs remained consecutive in Fugu, compared with only 13.5% (170 out of 1,250) of the same-strand pairs. Moreover, although the Fugu genome is much more compact than the human genome, the average distance between sense–antisense gene pairs was only slightly greater in humans than in Fugu, whereas same-strand pairs were significantly further apart (up to 11-fold).
If antisense transcription is indeed beneficial, how is it regulated? A recent study (Cawley et al, 2004) mapped the binding sites of three human transcription factors—SP1, c-Myc and p53—to chromosomes 21 and 22 using chromatin immunoprecipitation (ChIP)-on-chip technology (Ren et al, 2000). Surprisingly, 36% of the binding sites mapped within or immediately 3′ to well-characterized protein-encoding genes and were associated with non-encoding RNAs. Moreover, many overlapping sense–antisense transcripts showed correlated expression. Some overlapping transcripts were flanked by binding sites for the same transcription factor, implying that the sense and corresponding antisense transcripts might in fact be co-regulated. Similar results were obtained for the human transcription factor cAMP-response-element-binding protein (Impey et al, 2004).
Chen and colleagues used their comprehensive human antisense database to study the relationship between the expression profiles of sense and corresponding antisense transcripts on a genome-wide scale (Chen et al, 2005a). They found that sense–antisense gene pairs tend to be co-expressed or inversely expressed more frequently than would be expected by chance. Moreover, co-expressed and inversely expressed sense–antisense pairs have striking conservation throughout evolution. Both conservation and coupled sense–antisense expression were more prevalent in tail-to-tail NAT pairs, suggesting that such an orientation is not only the most abundant, but also more likely to have a regulatory function (Sun et al, 2005).
Chen and colleagues also reported that antisense genes, especially those that are evolutionarily conserved, have particularly short introns in humans, mice and Drosophila (Chen et al, 2005b; Sun et al, 2006). The authors suggested that, in the case of the antisense genes, the purpose of short introns is not to allow a high level of expression or spurious expression, as has been shown for other genes (Castillo-Davis et al, 2002; Hurst et al, 1996), but to address the need for a rapid response.
Although the above studies suggest that antisense transcription is tightly regulated and evolutionarily conserved, they should be regarded with some caution. The observed co-expression of cis-NATs might originate from the known tendency of genes in close proximity (even in the same genomic strand) to co-express as a result of local chromatin structure or shared regulatory elements (Cohen et al, 2000; Lercher et al, 2003). It is possible that proximal co-expressed genes that were not selected against seem to be ‘tailored' by evolution to serve a regulatory purpose. A recent study introduced the intriguing concept of “neutral expression” (Yanai et al, 2004). The authors of this study argue that mutations that alter gene expression might not always be sufficiently deleterious to be eliminated by purifying selection, and therefore might be fixated in the population by random drift. According to this idea, the possibility that some NATs represent cases of residual transcription cannot be entirely eliminated.
Coupling NAT mechanisms of action to their regulation
The observation of both negative and positive correlations of sense–antisense levels suggests that their mechanisms of action might be diverse. Indeed, well-documented NAT examples point to four mechanisms (Lavorgna et al, 2004): transcriptional interference, RNA masking, double-stranded RNA (dsRNA)-dependent mechanisms and chromatin remodelling (see below). Each mechanism requires different associations between sense and antisense expression patterns; some mechanisms require the concomitant presence of sense and antisense transcripts, whereas others impose their mutual exclusion (Fig 2). We propose that the regulation of sense and antisense transcription is coupled to serve the different regulatory mechanisms. Furthermore, because the type of coupling is characteristic of the regulatory mechanism, we suggest that the relationship between sense and antisense transcription profiles can hint at the mechanism at work as well as the ultimate biological outcome.
Figure 2.
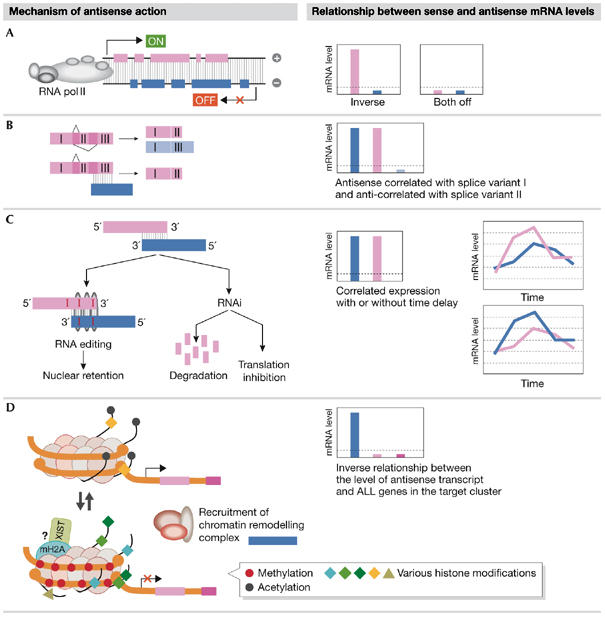
The main mechanisms by which natural antisense transcripts regulate gene expression. Each mechanism is accompanied by what it requires from, or imposes on, the relationship between the levels of sense and antisense transcripts. (A) Transcriptional interference. Two bulky RNA polymerase II complexes on opposite DNA strands might collide with and stall one another. The interference occurs mostly in the elongation step, resulting in either transcription arrest or transcription in one direction (sense or antisense) only. Such a mechanism might occur in cases in which inverse expression is observed. (B) RNA masking. A specific case is shown in which the antisense masks a splice site on the sense pre-mRNA sequence. This prevents a given splice variant from being formed and shifts the balance towards splice variants that do not require splicing of the masked region. Such a mechanism could be observed by correlated expression of the antisense and favoured splice variant and an inverse relationship with the repressed variant. (C) Double-stranded RNA-dependent mechanisms such as RNA editing and RNA interference require the simultaneous presence of sense and antisense transcripts for duplex formation, and might therefore account for the observed co-expression of numerous sense–antisense pairs. A delay in expression of sense compared with antisense (or vice versa) is also possible as long as there is a period in which both transcripts are present (see Fig 3). (D) Chromatin remodelling. Transcription of non-encoding antisense transcripts is involved in monoallelic gene expression, including genomic imprinting, X-inactivation and clonal expression of lymphocyte genes. In these processes, antisense transcripts have been suggested to silence the expression of nearby gene clusters by chromatin remodelling, most likely through the recruitment of histone-modifying enzymes. If such mechanisms are in action, an inverse expression profile of the antisense compared with all genes in the silenced cluster would be expected.
To illustrate this point, we predict two biological outcomes that might result from a delayed initiation of transcription between the sense and antisense transcripts (or vice versa). First, if the sense gene is initially transcribed up to a certain level, then antisense transcription begins and subsequently promotes sense degradation, then the anticipated outcome is a delayed shutdown of the sense gene. Second, if antisense transcription precedes sense transcription, the biological outcome might be the dampening of stochastic fluctuations (noise) in the level of the sense transcripts; the antisense level sets a threshold and only sense transcripts that exceed it are effectively expressed (Fig 3). Noise dampening was shown to be obtained by another type of regulatory RNA, microRNAs (Hornstein & Shomron, 2006), and we hypothesize that antisense transcripts might fulfill the same function. Differences in transcription activation times might be encoded by differential affinities of the sense and antisense promoters to a shared transcription factor, assuming that such a regulator is an activator and that it accumulates with time.
Figure 3.
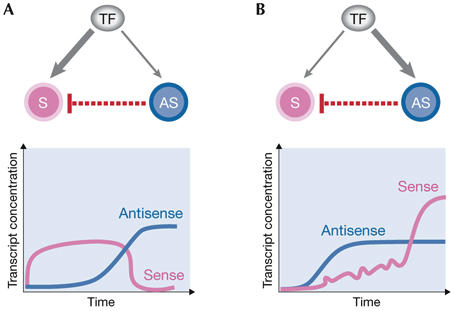
Differences in activation times of the sense compared with the antisense transcript. Such differences might be easily encoded in differential affinities to a shared transcription factor, assuming that this transcription factor is an activator and that it accumulates with time. (A) A higher affinity of the transcription factor to the sense transcript might result in a delayed shutdown, whereby the transcription factor initially activates transcription of the sense messenger RNA up to a certain level and only then is triggered by antisense transcription. The delayed antisense transcription prevents the sense transcript from exceeding the level it has reached when antisense transcription is switched on. (B) A higher affinity of the transcription factor to the antisense transcript. In this case, antisense transcription precedes sense transcription and acts as a buffer for the sense transcript. When the transcription factor accumulates, transcription of sense mRNA begins, but only sense transcripts exceeding the threshold set by the antisense level can be effectively translated. This generates a step-like function in the concentration of the sense transcript. Fluctuations in the amount of sense transcript below the threshold are dampened.
Sense and antisense transcripts might be regulated not only at the transcriptional level, but also at the level of mRNA stability. Therefore, differences in mRNA half-lives of the two transcripts might also be predictive of antisense function. A well-characterized example is the hok–sok system of the R1 plasmid (Gerdes et al, 1990), in which differences in mRNA stability result in delayed activation of the sense-encoded protein. The R1 plasmid encodes a host killing protein (hok) and its antisense suppressor sok; however, killing of host cells occurs when the plasmid is no longer present. This is accomplished by combining a stable and initially inert toxin with an unstable inhibitor. Hok mRNA accumulates in a stable and inert state and is slowly processed into a transcriptionally active form. In plasmid-containing cells, the active hok is targeted by sok, resulting in translation inhibition and message degradation. Plasmid-free cells show rapid loss of the unstable sok, therefore on activation of hok, its suppressor is no longer present and the host cell is killed.
Below we describe four proposed mechanisms of NAT action and accompany each with the resulting relationship between sense and antisense transcript levels.
Transcriptional interference. The presence of an overlapping transcriptional unit might stall sense transcription owing to the collision of two bulky RNA polymerase II complexes on opposite strands. This is most apparent in the transcription elongation step as has been shown for the yeast gene pair GAL10 and GAL7 (Table 2; Prescott & Proudfoot, 2002). Competitive transcriptional interference could be the underlying mechanism when anti-correlated expression levels of sense and antisense are observed. Such interference might alternatively result in the shutdown of both transcripts (Fig 2A).
Table 2.
Well-characterized examples of antisense regulatory function
| Mechanism | Example | Reference |
|---|---|---|
| Transcriptional interference | The yeast GAL10 and GAL7 genes, organized naturally in tandem, were rearranged in a convergent orientation. Transcription initiation was carried out normally but, as soon as the two transcripts began to overlap, elongation stalled and mRNA levels were severely reduced. | Prescott & Proudfoot, 2002 |
| RNA masking | Expression of RevErb, overlapping erbA (encoding α-thyroid hormone receptor) strongly correlates with an increase in the ratio of splice variants erbAα1/erbAα2. RevErb overlaps only with erbAα2 and is thought to block its splicing by masking splicing regulatory cis-elements. Artificial antisense RNAs complementary to the Erbα2-specific exon were shown to efficiently and specifically block ErbAα2 splicing in vitro. | Hastings et al, 1997; Munroe & Lazar, 1991 |
| dsRNA-dependent mechanisms | Salt tolerance in Arabidopsis is regulated by two siRNAs produced from a pair of tail-to-tail overlapping protein-encoding genes: P5CDH (a stress-related gene) and SRO5 (of unknown function). When both transcripts are present, an RNA duplex is formed and two types of siRNA are produced: 24 nucleotides (nt) and 21 nt. The 24-nt siRNA causes initial cleavage of the P5CDH transcript, which is followed by the generation of 21-nt siRNA and further cleavage of the P5CDH transcript. The expression of SRO5 is induced by salt and this induction is required to initiate siRNA formation. | Borsani et al, 2005 |
| dsRNA-dependent mechanisms | Response to iron deficiency in Cyanobacteria is mediated through the formation of an RNA duplex. Cyanobacteria responds to iron deficiency by expressing IsiA (iron stress-induced protein A), which forms a giant ring structure around photosystem I. IsiA is regulated by its cis-encoded antisense IsrR (iron stress-repressed RNA). Artificial overexpression of IsrR under iron stress causes a strongly diminished number of IsiA–photosystem supercomplexes, whereas IsrR depletion results in premature expression of IsiA. The mRNAs IsrR and isiA form a perfect duplex and undergo coupled degradation. | Duhring et al, 2006 |
| dsRNA-dependent mechanisms | Silencing of Drosophila melanogaster Stellate repeats in testis is essential for male fertility and involves dsRNA. Stellate silencing is mediated by the homologous Su (Ste) tandem repeats. Both strands of the repressor repeats are transcribed producing a sense–antisense duplex. This duplex is further cleaved into short 25–27-nucleotide fragments, which confer specific silencing of the Stellate repeats. | Aravin et al, 2001 |
| Effect on methylation | The head-to-head overlapping gene pair Sphk1/Khps1 have been shown to undergo antisense-induced methylation. | Imamura et al, 2004 |
RNA masking. Sense–antisense duplex formation might mask cis-elements residing in either of the transcripts and hinder processes that require protein–RNA interactions such as splicing, mRNA transport, polyadenylation, translation and degradation. The best-characterized example of this mechanism is the antisense transcript for the thyroid hormone receptor gene erbAα, which shifts the balance between two splice variants through the masking of a splice site (Table 2; Hastings et al, 1997). Such a mechanism would result in a correlated expression level of the antisense and the regulated transcript (Fig 2B).
dsRNA-dependent mechanisms and RNA interference. There is accumulating evidence that antisense transcripts might function through the activation of dsRNA-dependent mechanisms such as RNA editing and RNA interference (RNAi). Such mechanisms require the simultaneous existence of sense and antisense transcripts for duplex formation, and might therefore account for the observed co-expression of numerous sense–antisense pairs (Fig 2C; Chen et al, 2005a).
RNA editing involves the deamination of adenosines to inosines in dsRNA (Bass, 2002) and is thought to constitute part of the nuclear defence strategy against dsRNA. Hyper-editing of long, perfect RNA duplexes can result in their nuclear retention (Zhang & Carmichael, 2001) or cytoplasmic degradation (Scadden & Smith, 2001). However, it was recently shown that the level of RNA editing in sense–antisense overlapping areas—apart from the Alu regions within them—is negligible in both humans and mouse (Neeman et al, 2005). Regulation through RNA editing is therefore not likely to be one of the main mechanisms for antisense action.
RNAi is another component of the defence against dsRNA. RNAi involves cleavage of the dsRNA by the enzyme Dicer into 21–23-nucleotide duplexes. These duplexes are further separated into single strands and become part of the RNA-induced silencing complex (RISC). The RISC eventually either degrades cognate mRNAs with great specificity or represses their translation (Meister & Tuschl, 2004; Mello & Conte, 2004). Several precedents suggest that sense–antisense transcription can induce gene silencing through an RNAi-dependent mechanism. For example, salt tolerance in Arabidopsis is regulated by two small interfering RNAs (siRNAs) produced from a pair of tail-to-tail overlapping protein-encoding genes: P5CDH (a stress-related gene) and SRO5. Salt induces SRO5 transcription. When both genes are transcribed, an RNA duplex is formed and siRNAs are produced that ultimately cleave the P5CDH transcript (Table 2; Borsani et al, 2005). The same mechanism could apply to other eukaryotic cis-NAT pairs. In fact, 11 Arabidopsis siRNAs have been mapped to complementary regions of overlapping transcripts, suggesting that these overlapping transcripts might feed into the RNAi machinery (Wang et al, 2005). Other processes shown to involve dsRNA are the response to iron deficiency in cyanobacteria (Duhring et al, 2006) and the maintenance of male fertility in Drosophila (Table 2; Aravin et al, 2001). So far, however, there has been no evidence for mammalian antisense transcripts acting through duplex formation.
Antisense involvement in methylation and monoallelic expression. dsRNA can induce the methylation and silencing of corresponding genes. For example, thalassaemia—a form of anaemia—is caused by antisense-induced DNA methylation (and silencing) of the human haemoglobin 2 gene (Tufarelli et al, 2003).
Monoallelic expression includes X-chromosome inactivation, genomic imprinting and allelic exclusion in B and T lymphocytes. X-chromosome inactivation is a mechanism that balances the expression of X-chromosome-encoded genes in mammalian females. The silencing of one of the X chromosomes is mediated through a large non-encoding RNA (Xist), which recruits a histone-modifying protein complex. Xist is repressed by its antisense Tsix, thus the X chromosome expressing the antisense remains active (Ogawa & Lee, 2002). Imprinted genes are genes for which only one allele—maternal or paternal—is actively transcribed. There are about 100 known human and mouse imprinted genes; they are clustered in the genome and often have both DNA methylation and non-encoding antisense transcripts. Several studies have indicated that imprinting is not mediated through the formation of a sense–antisense RNA duplex (Sleutels et al, 2003; Thakur et al, 2004), but rather through the modification of chromatin structure or methylation patterns in the vicinity of the imprinted allele. In lymphocytes, immunoglobulins and T-cell receptors undergo clonal selection through which one allele is silenced while the other undergoes recombination. Extensive antisense transcription occurs before and during recombination and is believed to function by inducing an open chromatin structure that is accessible to recombination. In all these cases, non-encoding antisense transcription affects an entire gene cluster, rather than merely the overlapping sense transcript, and exerts its effect by chromatin remodelling, probably through the recruitment of histone-modifying enzymes. We therefore predict an inverse expression profile for the antisense and all the genes in the silenced cluster (Fig 2D).
Despite these well-characterized cases, it is not clear which of these mechanisms might apply to a wider set of antisense RNAs.
Discussion
There has been a revolution in our understanding of the regulatory role of non-encoding RNAs in recent years. Genome-wide technologies reveal that a significant proportion of all genomes is transcribed, and might thus fulfill regulatory functions (Carninci, 2006). The possibility that transcribed RNAs represent leakage of the transcription machinery exists, but evidence for a selected process is convincing. In this review, we have discussed one type of non-encoding RNA and suggest that its transcriptional, and post-transcriptional, regulation is tailored to its various regulatory roles.

Michal Lapidot

Yitzhak Pilpel
Acknowledgments
Y.P. is an incumbent of the Rothstein Career Development Chair in Genetic Diseases and a recipient of a Young Investigator award from the European Molecular Biology Organization. M.L. is a Fellow of the Horowitz Foundation for Complexity Sciences. We thank the Ben May Charitable Trust for grant support.
References
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Barrell BG, Air GM, Hutchison CA 3rd (1976) Overlapping genes in bacteriophage φX174. Nature 264: 34–41 [DOI] [PubMed] [Google Scholar]
- Bass BL (2002) RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P et al. (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P (2006) Tagging mammalian transcription complexity. Trends Genet 22: 501–510 [DOI] [PubMed] [Google Scholar]
- Carninci P et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA (2002) Selection for short introns in highly expressed genes. Nat Genet 31: 415–418 [DOI] [PubMed] [Google Scholar]
- Cawley S et al. (2004) Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD (2004) Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res 32: 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD (2005) Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense–antisense transcripts. Trends Genet 21: 326–329 [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD (2005) Human antisense genes have unusually short introns: evidence for selection for rapid transcription. Trends Genet 21: 203–207 [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM (2000) A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet 26: 183–186 [DOI] [PubMed] [Google Scholar]
- Dahary D, Elroy-Stein O, Sorek R (2005) Naturally occurring antisense: transcriptional leakage or real overlap? Genome Res 15: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhring U, Axmann IM, Hess WR, Wilde A (2006) An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA 103: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Thisted T, Martinussen J (1990) Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol Microbiol 4: 1807–1818 [DOI] [PubMed] [Google Scholar]
- Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH (1997) Expression of the thyroid hormone receptor gene, erbAα, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res 25: 4296–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havilio M, Levanon EY, Lerman G, Kupiec M, Eisenberg E (2005) Evidence for abundant transcription of non-coding regions in the Saccharomyces cerevisiae genome. BMC Genomics 6: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N (2006) Canalization of development by microRNAs. Nat Genet 38 (Suppl 1): S20–S24 [DOI] [PubMed] [Google Scholar]
- Hurst LD, McVean G, Moore T (1996) Imprinted genes have few and small introns. Nat Genet 12: 234–237 [DOI] [PubMed] [Google Scholar]
- Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K (2004) Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun 322: 593–600 [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH (2004) Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6: R51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y (2003) Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res 13: 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee R, Murphy PR (1997) Regulation of gene expression by natural antisense RNA transcripts. Neurochem Int 31: 379–392 [DOI] [PubMed] [Google Scholar]
- Korneev SA, Park JH, O'Shea M (1999) Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci 19: 7711–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK (2003) Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421: 948–952 [DOI] [PubMed] [Google Scholar]
- Kumar M, Carmichael GG (1998) Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev 62: 1415–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G (2004) In search of antisense. Trends Biochem Sci 29: 88–94 [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD (2003) Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res 13: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Qin L, Guo ZM, Liu L, Xu H, Hao P, Su J, Shi Y, He WZ, Li YX (2006) In silico discovery of human natural antisense transcripts. BMC Bioinformatics 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D Jr (2004) Revealing the world of RNA interference. Nature 431: 338–342 [DOI] [PubMed] [Google Scholar]
- Misra S et al. (2002) Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol 3: RESEARCH0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe SH, Lazar MA (1991) Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem 266: 22083–22086 [PubMed] [Google Scholar]
- Neeman Y, Dahary D, Levanon EY, Sorek R, Eisenberg E (2005) Is there any sense in antisense editing? Trends Genet 21: 544–547 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT (2002) Antisense regulation in X inactivation and autosomal imprinting. Cytogenet Genome Res 99: 59–65 [DOI] [PubMed] [Google Scholar]
- Osato N et al. (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ (2002) Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA 99: 8796–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B et al. (2000) Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Scadden AD, Smith CW (2001) Specific cleavage of hyper-edited dsRNAs. EMBO J 20: 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Tjon G, Ludwig T, Barlow DP (2003) Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J 22: 3696–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Hurst LD, Carmichael GG, Chen J (2005) Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res 33: 5533–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Hurst LD, Carmichael GG, Chen J (2006) Evidence for variation in abundance of antisense transcripts between multicellular animals but no relationship between antisense transcription and organismic complexity. Genome Res 16: 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Gondor A, Grange T, Ohlsson R, Kanduri C (2004) An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol 24: 7855–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J, Itoh T, Selzer G, Som T (1981) Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci USA 78: 1421–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34: 157–165 [DOI] [PubMed] [Google Scholar]
- Wagner EG, Flardh K (2002) Antisense RNAs everywhere? Trends Genet 18: 223–226 [DOI] [PubMed] [Google Scholar]
- Wagner EG, Simons RW (1994) Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 48: 713–742 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Fried M (1986) A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3′ ends. Nature 322: 275–279 [DOI] [PubMed] [Google Scholar]
- Yanai I, Graur D, Ophir R (2004) Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. Omics 8: 15–24 [DOI] [PubMed] [Google Scholar]
- Yelin R et al. (2003) Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379–386 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG (2001) The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475 [DOI] [PubMed] [Google Scholar]


