Abstract
The chromosomal passenger complex (CPC) of Aurora-B, Borealin, INCENP (inner centromere protein) and Survivin coordinates essential chromosomal and cytoskeletal events during mitosis. Here, we show that the nuclear export receptor Crm1 is crucially involved in tethering the CPC to the centromere by interacting with a leucine-rich nuclear export signal (NES), evolutionarily conserved in all mammalian Survivin proteins. We show that inhibition of the Survivin–Crm1 interaction by treatment with leptomycin B or by RNA-interference-mediated Crm1 depletion prevents centromeric targeting of Survivin. The genetic inactivation of the Survivin–Crm1 interaction by mutation of the NES affects the correct localization and function of Survivin and the CPC during mitosis. By contrast, CPC assembly does not seem to require the Survivin–Crm1 interaction. Our report shows the functional significance of the Survivin–Crm1 interface and provides a novel link between the mitotic effector Crm1 and the CPC.
Keywords: cell cycle, leptomycin B, nucleocytoplasmic transport, trans-dominance
Introduction
Regulated development and cellular homeostasis relies on proper mitosis. It has emerged that the chromosomal passenger proteins Aurora-B, Borealin, inner centromere protein (INCENP) and Survivin form a conserved complex and are key regulators of mitotic events (Vagnarelli & Earnshaw, 2004). These proteins show a typical chromosomal passenger localization pattern during mitosis—at the centromere in prometaphase, on the central spindle during anaphase and at the midbody during cytokinesis (Vagnarelli & Earnshaw, 2004). Interference with each of the subunits of the chromosomal passenger complex (CPC) results in severe mitotic defects and apoptosis (Vagnarelli & Earnshaw, 2004).
Although the low molecular weight would theoretically allow Survivin to access intracellular compartments by passive diffusion, regulated subcellular localization has been suggested for CPC proteins and for other cell-cycle regulators (Rodriguez et al, 2002, 2006; Xu & Massague, 2004). Active nucleocytoplasmic transport takes place through the nuclear pore complex and is regulated by specific signals binding to transport receptors (Weis, 2003). The best-characterized nuclear export signals (NESs) are leucine rich, interact with the export receptor chromosome region maintenance 1 (Crm1) and depend on the small GTPase Ran, which controls the Crm1–substrate interaction (Weis, 2003). The guanine-nucleotide-exchange factor RCC1 (regulator of chromosome condensation) facilitates Ran binding to Crm1, whereas RanBP1, the main regulator of Ran, promotes Crm1 dissociation from Ran.
Besides the CPC, cellular components that regulate nucleocytoplasmic transport in interphase cells are crucially involved in controlling mitotic processes. These are accomplished by the Ran–GTPase system in conjunction with specific receptors of the importin-β family (Dasso, 2005). Recently, Crm1 has been identified as an essential mitotic effector in mammalian cells (Arnaoutov et al, 2005; Wang et al, 2005).
At present, the molecular mechanisms and requirements for centromere targeting and function of the chromosomal passengers, as well as for the Ran–GTPase system, are under intense investigation (Earnshaw, 2005). Here, we provide evidence that an evolutionarily conserved NES in Survivin is crucial for the biological activity of Survivin and show that inactivation of the Survivin–Crm1 interaction affects the correct localization and function of CPC proteins.
Results And Discussion
Leptomycin B affects CPC subunit localization
To provide a molecular link between the CPC and Crm1, we first examined whether the centromeric localization of CPC proteins was impaired on application of the Crm1-specific inhibitor leptomycin B (LMB). We observed that Survivin, Aurora-B, INCENP and Borealin no longer efficiently localized to the centromere on LMB treatment (Fig 1A–D). Similar results were found for Borealin–green fluorescent protein (GFP), and it was found that LMB also affected the centromeric and midbody localization of Survivin–GFP (Fig 1E; supplementary Fig S1A–D online).
Figure 1.
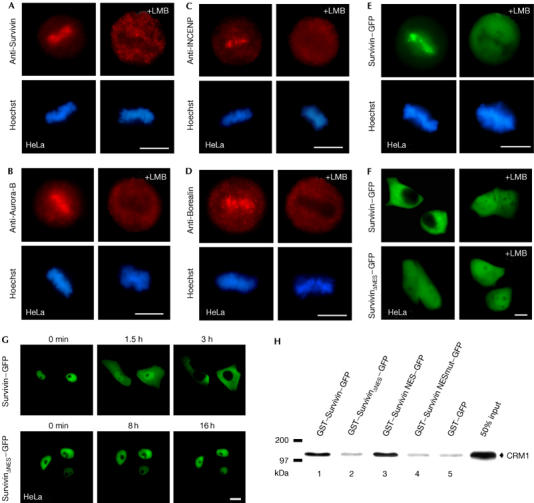
Survivin contains a leptomycin-B-sensitive nuclear export signal interacting with Crm1. (A–E) The centromeric localization of (A) Survivin, (B) Aurora-B, (C) INCENP, (D) Borealin and (E) Survivin–GFP is affected by treatment with leptomycin B (LMB) for 6 h. Endogenous chromosomal passenger complex (CPC) proteins were detected by immunostaining (red). DNA was stained with Hoechst. (F) Survivin–GFP localizes to the cytoplasm and accumulates in the nucleus on LMB treatment, whereas SurvivinΔNES–GFP is equally distributed between the nucleus and the cytoplasm and does not respond to LMB. (G) Nuclear-injected GST–Survivin–GFP is efficiently exported, in contrast with export-deficient GST–SurvivinΔNES–GFP. GFP fusions were detected by fluorescence microscopy. Scale bars, 10 μm. (H) Survivin binds to Crm1 in vitro. 35S-labelled Crm1 protein was incubated with 4 μg of the indicated GST–GFP fusion proteins prebound to glutathione Sepharose beads in the presence of Ran–GTP. Binding of Crm1 to the NES-containing substrates is abolished by mutating the NES. GST–GFP served to control for non-specific binding. GFP, green fluorescent protein; GST, glutathione S-transferase; ΔNES, mutant nuclear export signal; NES, nuclear export signal.
Survivin contains a Crm1 dependent NES
The localization of ectopically expressed Aurora-B–GFP, INCENP or Borealin–GFP was not affected by LMB in interphase cells (not shown), whereas Survivin–GFP accumulated in the nucleus on LMB treatment, as reported previously (Rodriguez et al, 2002, 2006; Fig 1F). As LMB impairs numerous cellular pathways, the effects of LMB need to be verified genetically and biochemically. Deletion mutagenesis (data not shown), inspection of the NES consensus sequence and microinjection experiments identified the Survivin amino acids 89–98 (89VKKQFEELTL98) as the NES (SurvNES). Nuclear-injected full-length glutathione S-transferase (GST)–Survivin–GFP or GST–SurvNES–GFP were quantitatively exported (Fig 1G; supplementary Fig S1E online). By contrast, GST–SurvivinΔNES–GFP and GST–SurvNESmut–GFP, in which two essential leucine residues of the NES were mutated (89VKKQFEELTL98 → 89VKKQFEEATA98; mutated residues are underlined), were not exported (Fig 1G; supplementary Fig S1E online). In contrast with other experimental systems used to identify NESs (Rodriguez et al, 2002, 2006), microinjection of GST–GFP fusions allows the quantification of transport independent of drug treatment or passive diffusion (Heger et al, 2001). Inactivation of the NES in the context of the full-length protein (SurvivinΔNES–GFP) resulted in a protein that no longer localizes to the cytoplasm but is equally distributed between the nucleus and the cytoplasm, and does not respond to LMB (Fig 1F). These results unequivocally define the evolutionarily conserved NES in all mammalian Survivin proteins (supplementary Fig S1F online). The direct NES-mediated Survivin–Crm1 interaction was verified by in vitro interaction studies (Fig 1H). Recombinant GST–Survivin–GFP bound to Crm1 in the presence of Ran–GTP, in contrast with inactive GST–SurvivinΔNES–GFP or GST–GFP alone. No efficient binding was detectable without Ran–GTP or in the presence of LMB (not shown).
Survivin–Crm1 interaction is needed to localize CPC
We observed that in contrast with Survivin–GFP, SurvivinΔNES–GFP failed to localize correctly during mitosis (Fig 2A; supplementary Fig S2A online) and did not localize with Crm1 at the centromere (Fig 2B). The presence of centromeres in cells expressing SurvivinΔNES–GFP was verified by staining with a CREST antiserum (Fig 2C), confirming that impairment of centromeric localization is not due to loss of the centromere structure. A similar localization of NES-deficient Survivin was observed in cell lines on RNA interference (RNAi)-mediated ablation of endogenous Survivin (supplementary Fig S1G online).
Figure 2.
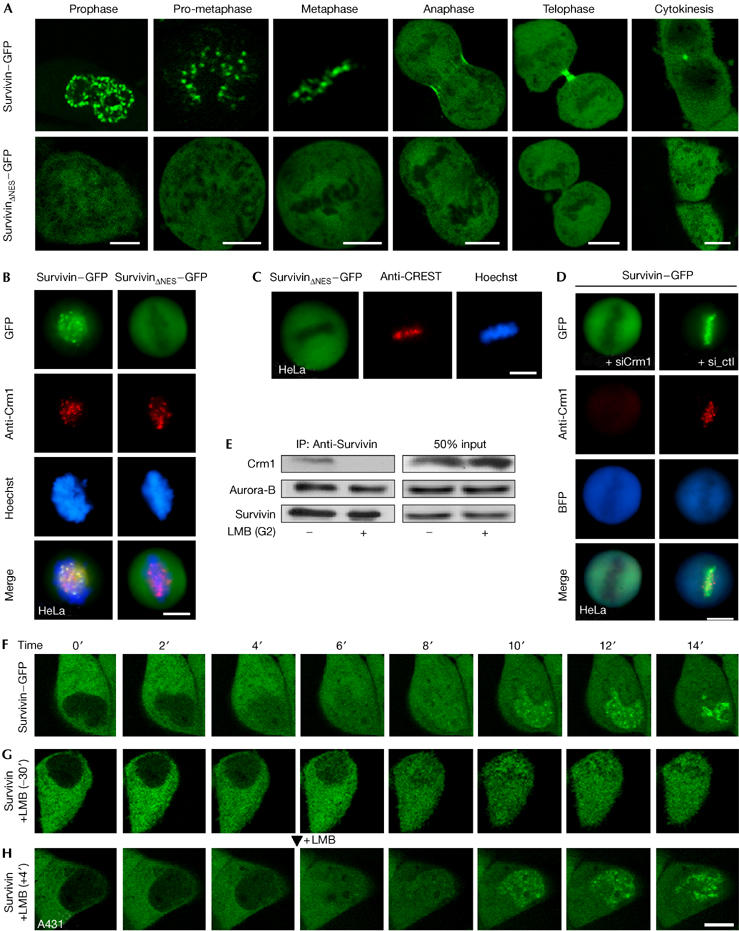
The Survivin–Crm1 interaction is required to tether Survivin to the centromere. (A) The localization of Survivin–GFP and SurvivinΔNES–GFP during mitosis was followed in live A431 cells. (B) Survivin–GFP but not SurvivinΔNES–GFP localizes with Crm1 at the centromere. (C) Staining of centromeres in cells expressing SurvivinΔNES–GFP using a CREST antiserum. (D) RNA-interference-mediated ablation of Crm1 impairs targeting of Survivin–GFP to the centromere. Cells were transfected with a Crm1 or a control (ctl) short interfering RNA together with a BFP expression plasmid as the transfection control. Crm1 was detected by immunostaining (red). (E) Survivin–Crm1 complex formation is abolished by leptomycin B (LMB). Immunoprecipitation of mitotic HeLa cell extracts was performed using Survivin antibody. Crm1, Survivin and Aurora-B were detected by immunoblot. (F–H) Time-lapse imaging of Survivin–GFP during G2/M transition. (G) Pretreatment of late G2 cells with LMB interfered with the (F) centromeric localization of Survivin–GFP, whereas (H) assembled chromosomal passenger complexes at the centromeres were not affected. The arrowhead indicates addition of LMB. Scale bars, 10 μm. GFP, green fluorescent protein; ΔNES, mutant nuclear export signal.
Similar to LMB, RNAi-mediated depletion of Crm1 also impaired targeting of Survivin–GFP or Aurora-B to the centromere (Fig 2D; supplementary Fig S1H online; data not shown). Further evidence that Crm1 and Survivin are physically associated early in mitosis is illustrated in Fig 2E. Crm1 could be recovered in a complex with endogenous Survivin–Aurora-B from mitotic cells, whereas complex formation was abolished on pretreatment with LMB. Equally, the Crm1–Survivin–GFP complex could be precipitated from Survivin–GFP-expressing cells but not from SurvivinΔNES–GFP- or GFP-expressing cells (not shown).
To understand whether CPC assembly or CPC targeting to the centromere requires Crm1, we examined the localization of CPC proteins during the transition from late G2 to prophase in the absence and presence of LMB. At late G2, the localization of Survivin–GFP gradually changed from a predominantly cytoplasmic to a cytoplasmic–nuclear distribution, with a subsequent accumulation at the centromeres in early prophase (Fig 2F; supplementary Fig S2E online). Aurora-B–GFP and Borealin–GFP accumulated at the centromeres with similar kinetics (supplementary Fig S2C–E online). A similar change in localization, except for the centromeric accumulation, was observed for a RevNES–GFP fusion (Rev, for regulator of expression of virion proteins; supplementary Fig S2B online), indicating that the export competence of the cell is gradually lost during breakdown of the nuclear envelope and no active nuclear import of Survivin seems to be involved. By contrast, 30 min pretreatment of late G2 cells with LMB interfered with the centromeric localization of Survivin–GFP (Fig 2G) and Aurora-B–GFP (not shown), whereas the already assembled CPCs at the centromeres were not affected (Fig 2H). By contrast, Ran binding protein 2 (RanBP2) was lost from the centromeres under these conditions (data not shown). As expected, SurvivinΔNES–GFP was not detectable at the centromeres during G2/M transition (supplementary Fig S2A,E online).
Also, Crm1 could not be recovered in a complex with Survivin–GFP from metaphase-enriched cells (supplementary Fig S2F online). Thus, Crm1 seems to be transiently involved in the transport of the CPC to the centromere, rather than in CPC anchoring, consistent with the results from the study of Rodriguez et al (2006). As such, the CPC behaves differently compared with factors such as RanBP2 (Arnaoutov et al, 2005; Rodriguez et al, 2006). This is further supported by showing that Crm1 and Survivin only partly colocalize at the centromere (supplementary Fig S1J online). Other reports indicate that Survivin localizes to the inner centromere, whereas Crm1 has been detected at the outer centromere (Arnaoutov et al, 2005) or even at the centrosomes (Wang et al, 2005), underscoring the dynamic nature of Crm1 during cell-cycle regulation. However, the molecular details of the localization of Crm1 are not yet understood. The NES–Crm1 interaction in nucleophosmin seems to be regulated by phosphorylation (Wang et al, 2005), whereas no phosphorylation of the NES sequence was reported for Survivin (Nousiainen et al, 2006). Also, Survivin mutants, in which the Thr 97 residue in the NES was changed to either alanine (mimicking non-phosphorylation) or aspartic acid (mimicking phosphorylation), still localized to the centromeres (not shown).
NES-deficient Survivin binds to CPC components
We further controlled that the effects of NES-deficient Survivin on CPC targeting and activity are mediated by preventing the Survivin–Crm1 interaction rather than by affecting the ability of Survivin to dimerize or to interact with CPC proteins. First, we immunoprecipitated CPCs from mitotic cells expressing Survivin–GFP, SurvivinΔNES–GFP or GFP. CPCs containing Survivin–GFP or SurvivinΔNES–GFP were recovered with similar efficiencies, whereas the non-CPC kinetochore protein centromere protein A (CENP-A) was not precipitated (Fig 3A). Similarly, immunoprecipitation of the crucial Survivin binding partner Borealin (Gassmann et al, 2004) recovered both Survivin–GFP and SurvivinΔNES–GFP (supplementary Fig S2G online). By precipitating CPCs containing Survivin and also SurvivinΔNES with similar efficiencies from G2-enriched transfectants (Fig 3B), we could show that the inhibitory effects of SurvivinΔNES are not caused by impaired CPC assembly late at the G2/M transition. G2 enrichment was controlled by staining for targeting protein for Xklp2 (TPX2) and Cyclin B1 expression, and no CPC proteins could be precipitated using an antibody against actin, which served as the control (not shown).
Figure 3.
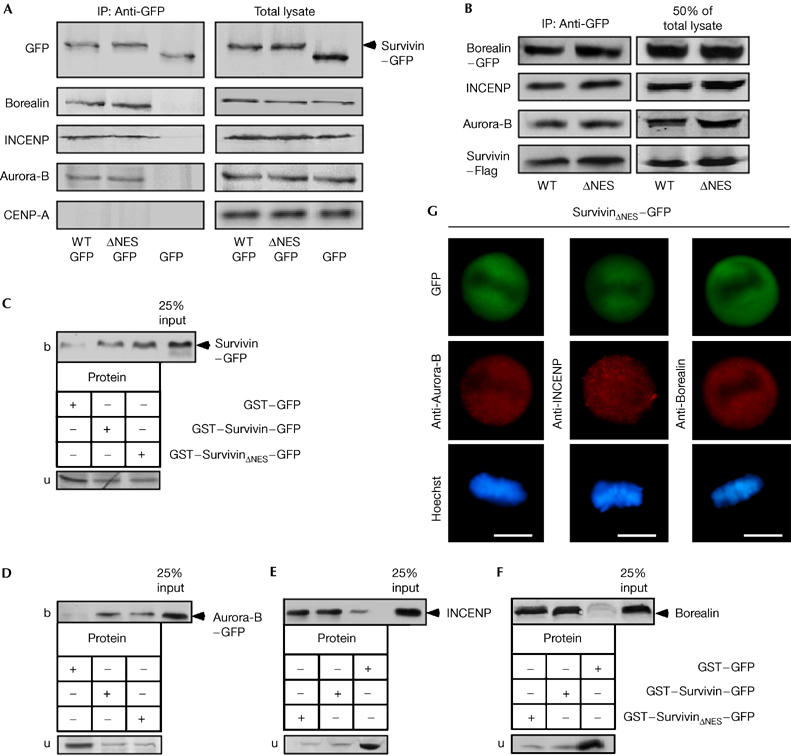
Survivin and SurvivinΔNES interact with subunits of the chromosomal passenger complex in vivo and in vitro. (A) Immunoprecipitations (IPs) of mitotic HeLa cell extracts were carried out with green fluorescent protein (GFP) antibody, and immunoblots were probed with the indicated antibodies. (B) IPs of chromosomal passenger complexes (CPCs) from G2 cells expressing the indicated proteins. CPCs were recovered using immobilized GFP antibody, and precipitated proteins were identified by immunoblot. (C–F) Survivin and SurvivinΔNES bind to CPC proteins in vitro. In vitro-translated, labelled (C) Survivin–GFP, (D) Aurora-B–GFP, (E) INCENP or (F) Borealin was incubated with 4 μg of recombinant GST–Survivin–GFP or GST–SurvivinΔNES–GFP prebound to glutathione Sepharose beads. GST–GFP controlled non-specific binding. Bound (b) and unbound (u) fractions were separated by SDS–polyacrylamide gel electrophoresis, and the proteins were visualized by phosphorimaging. Input: Total amount of in vitro-translated protein added to the immobilized GST–GFP proteins. (G) Expression of SurvivinΔNES–GFP interferes with the centromeric localization of CPC subunits. In SurvivinΔNES–GFP-expressing HeLa transfectants, Aurora-B, INCENP and Borealin were detected by immunostaining (red). DNA was stained with Hoechst. Scale bars, 10 μm. GST, glutathione S-transferase; LMB, leptomycin B; ΔNES, mutant nuclear export signal; WT, wild type.
Second, in vitro interaction assays were used to verify Survivin dimerization and binding of immobilized GST–Survivin–GFP or GST–SurvivinΔNES–GFP to the respective in vitro-translated CPC proteins (Fig 3C–F).
In addition, on overexpression of SurvivinΔNES–GFP, Aurora-B, which colocalized with Survivin–GFP at the centromeres (supplementary Fig S1A online), was not detectable at the centromeres but was found colocalized with SurvivinΔNES–GFP (Fig 3G). Similar results were observed for INCENP and Borealin (Fig 3G). Expression of GFP alone showed no effect on the localization of chromosomal passenger (CP) proteins (not shown). Thus, the genetic inactivation of the NES does not seem to affect significantly the ability of Survivin to dimerize or to interact with other CPC subunits. However, we cannot exclude that the mutations might affect other unknown functions of Survivin.
Survivin–Crm1 interaction is required for CPC function
Various reports show several mitotic defects on interference with Survivin by RNAi or by the expression of trans-dominant Survivin mutants, resulting in multinucleate cells (Vagnarelli & Earnshaw, 2004). Short interfering RNA (siRNA)-resistant Survivin–GFP efficiently counteracted the formation of multinucleate cells on RNAi-mediated ablation of endogenous Survivin, whereas siRNA-resistant SurvivinΔNES–GFP was unable to rescue full mitosis (Fig 4A,B). The dose-dependent increase in multinucleated cells (supplementary Fig S2H,J online), the inhibition of cell proliferation (Fig 4C) and the defects in cell-cycle progression (Fig 4D–F) induced by SurvivinΔNES–GFP characterize NES-deficient Survivin as a trans-dominant protein, and underscore the biological relevance of the Survivin–Crm1 axis.
Figure 4.
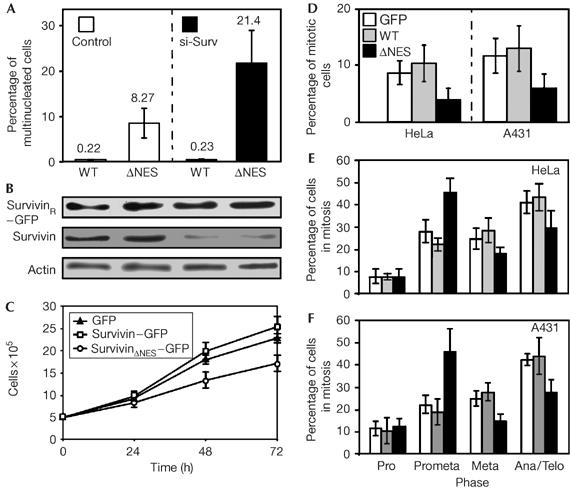
The Survivin–Crm1 interaction is important for correct mitotic progression. (A) HeLa cells stably expressing short interfering RNA (siRNA)-resistant Survivin–GFP fusions (SurvivinR-WT–GFP or SurvivinR-ΔNES–GFP) were transfected with Survivin siRNA (si-Surv) or a control siRNA together with an RFP expression plasmid. The number of cells with two or more nuclei was examined in 200 GFP- and RFP-double-positive cells, and the percentages of multinucleated cells were determined (mean±s.d., n=3). (B) Depletion of endogenous Survivin and similar expression levels were controlled by immunoblot using Survivin antibody. Actin was used as the loading control. (C) Proliferation of A431 cell lines expressing the indicated GFP fusions (mean±s.d., n=2). SurvivinΔNES–GFP-expressing cells show reduced proliferation and (D) less mitotic cells. Mitotic stages were analysed in (E) HeLa or (F) A431 cell lines expressing the indicated GFP proteins (mean±s.d., n=3). At least 80 GFP-positive cells were evaluated for each group. SurvivinΔNES–GFP expression increased the number of cells in pro-metaphase and decreased the number of metaphase and anaphase/telophase cells. GFP, green fluorescent protein; ΔNES, mutant nuclear export signal; WT, wild type.
Conclusion
This study provides evidence for a conserved molecular link between the tethering of the CPC to the centromere and the Crm1–Ran–GTP axis (supplementary Fig S3 online). Borealin forms a complex with Survivin, which can bind to Aurora-B kinase and is incorporated into the CP-holocomplex by interacting with INCENP (Klein et al, 2006). The NES in Survivin mediates the recruitment of Crm1–Ran–GTP, which seems to be involved in guiding the CPC to the centromeres in early prophase by an unknown mechanism. This process might be catalysed by the activity of the guanine nucleotide-exchange factor RCC1 or of TD60 (Vagnarelli & Earnshaw, 2004). Hydrolysis of Ran–GTP, by factors such as RanBPs/Ran–GAP1 (GTPase activating protein 1), might facilitate the release of Crm1. On reassembly of the nuclear envelope, Crm1 functioning as an export receptor might further regulate Survivin function in post-mitotic cells.
Methods
Plasmids. Eukaryotic and bacterial expression constructs for GFP- or FLAG-tagged and untagged versions of Survivin wild type (WT) and mutants were constructed by PCR and by cloning into the vectors pc3 (Invitrogen, Karlsruhe, Germany), pc3–GFP/BFP (blue fluorescent protein) or pGEX–GFP, as described previously (Knauer et al, 2005a; for details, see supplementary information online). Plasmid pc3AuroraB and pc3AuroraB_GFP were constructed by PCR. Plasmids pc3Crm1–HA, pc3-RevNES–GFP and pDsRed have been described previously (Knauer et al, 2005b). Plasmids encoding INCENP (pBSK_hINCENP; Honda et al, 2003), Aurora-B kinase (pGEXAurora-B; Wheatley et al, 2004) and Borealin–GFP (pEGFPN1-Borealin; Gassmann et al, 2004) were reported.
Antibodies and reagents. The following antibodies were used: anti-Survivin, anti-CENP-A, anti-Cyclin B1, anti-TPX2 (Novus Biologicals, Littleton, CO, USA); anti-Aurora-B, anti-β-actin, anti-γ-tubulin, anti-Flag (Sigma-Aldrich, Munich, Germany); anti-Crm1 (Santa Cruz Biotechnology, Heidelberg, Germany); anti-GFP (BD Biosciences, San Jose, CA, USA); and CREST antiserum (Europe Bioproducts Ltd, Cambridge, UK). Antibodies to INCENP and Borealin were from W. Earnshaw. Antibodies to RanBP2 were from F. Melchior. Appropriate Cy-3-conjugated secondary antibodies were used (Dianova, Hamburg, Germany). Reagents were from Sigma, unless stated otherwise.
Cells, transfection and microinjection. Cell lines were maintained and prepared for microinjection or transfected, as described previously (Knauer et al, 2005b). Microinjection and purification of recombinant GST–GFP fusions were performed as described by Knauer et al (2005b). Cells were treated with 10 nM LMB.
Flow cytometry analysis and sorting. Cytometry of GFP-expressing cells was performed as described by Stauber et al (1998).
Immunofluorescence and imaging of cells. Immunofluorescence, observation of cells, quantification, image analysis and presentation were performed as described previously (Stauber et al, 1998; Knauer et al, 2005b). For Crm1 staining, cells were extracted with 0.005% digitonin before fixation. DNA was visualized with Hoechst 33258 or TO-PRO®-3 iodide (Invitrogen).
Immunoprecipitation and immunoblotting. Immunoprecipitation was carried out as described previously (Knauer et al, 2005a). For lysate preparation of mitotically enriched cells, HeLa cells were synchronized at G1/S by treatment for 24 h with 2.5 mM thymidine. Cells were then released for 12 h into medium containing 50 ng/ml nocodazole in the absence or presence of 5 nM LMB. Mitotic cells were collected by shake-off and lysed in immunoprecipitation buffer. For lysate preparation of G2-enriched cells, A431 cells were synchronized with 2.5 mM thymidine and released for 10 h before lysis. Survivin, Borealin or GFP fusion proteins were immunoprecipitated with 3 μg of Survivin, Borealin or GFP antibodies precoupled to protein G-Sepharose (Amersham Pharmacia Biotech, Buckinghamshire, UK). Immunoprecipitated products were detected by immunoblot (Knauer et al, 2005a).
In vitro protein binding assay. Coupled transcription and translation was performed using plasmids pc3Crm1–HA, pc3Survivin–GFP, pBSK_hINCENP, pc3Borealin and pc3AuroraB_GFP, as described previously (Knauer et al, 2005a). Crm1 pull-down assays with recombinant GST–GFP substrates bound to glutathione Sepharose beads and recombinant RanQ69L were carried out as described earlier (Knauer et al, 2005a). For details, see supplementary information online.
RNA interference. RNAi was performed using double-stranded siRNA (Eurogenetec, Searing, Belgium). The targeted regions are as follows: Crm1, 5′-TGTGGTGAATTGCTTATAC-3′; Survivin, 5′-CTGGACAGAGAAAGAGCCA-3′ (residues mutated in siRNA-resistant Survivin are underlined). Cells were treated in parallel with a non-specific control siRNA duplex: 5′-GGTGTGCTGTTTGGAGGTC-3′. siRNA duplexes (each 50 nM) were transfected together with 0.2 μg of red fluorescent protein or BFP expression plasmids, and cells were analysed after 48 h.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by grants from BMBF NGFN (Nationales Genomforschungsnetz) initiative (N1KR-S31T24) and Deutsche Krebshilfe (FKZ:102362).
References
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M (2005) Crm1 is a mitotic effector of Ran–GTP in somatic cells. Nat Cell Biol 7: 626–632 [DOI] [PubMed] [Google Scholar]
- Dasso M (2005) Cell biology: new cog for a familiar machine. Nature 435: 899–900 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC (2005) Cell biology. Keeping Survivin nimble at centromeres in mitosis. Science 310: 1443–1444 [DOI] [PubMed] [Google Scholar]
- Gassmann R, Carvalho A, Henzing AJ, Ruchaud S, Hudson DF, Honda R, Nigg EA, Gerloff DL, Earnshaw WC (2004) Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol 166: 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Lohmaier J, Schneider G, Schweimer K, Stauber RH (2001) Qualitative highly divergent nuclear export signals can regulate export by the competition for transport cofactors in vivo. Traffic 2: 544–555 [DOI] [PubMed] [Google Scholar]
- Honda R, Korner R, Nigg EA (2003) Exploring the functional interactions between Aurora B, INCENP, and Survivin in mitosis. Mol Biol Cell 14: 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein UR, Nigg EA, Gruneberg U (2006) Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of borealin, Survivin, and the N-terminal domain of INCENP. Mol Biol Cell 17: 2547–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer SK, Carra G, Stauber RH (2005a) Nuclear export is evolutionarily conserved in CVC paired-like homeobox proteins and influences protein stability, transcriptional activation, and extracellular secretion. Mol Cell Biol 25: 2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer SK, Moodt S, Berg T, Liebel U, Pepperkok R, Stauber RH (2005b) Translocation biosensors to study signal specific nucleo-cytoplasmic transport, protease activity & protein interactions. Traffic 6: 594–606 [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R (2006) Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA 103: 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Span SW, Ferreira CG, Kruyt FA, Giaccone G (2002) CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein Survivin. Exp Cell Res 275: 44–53 [DOI] [PubMed] [Google Scholar]
- Rodriguez JA, Lens SM, Span SW, Vader G, Medema RH, Kruyt FA, Giaccone G (2006) Subcellular localization and nucleocytoplasmic transport of the chromosomal passenger proteins before nuclear envelope breakdown. Oncogene 25: 4867–4879 [DOI] [PubMed] [Google Scholar]
- Stauber RH, Horie K, Carney P, Hudson EA, Tarasova NI, Gaitanaris GA, Pavlakis GN (1998) Development and applications of enhanced green fluorescent protein mutants. BioTechniques 24: 462–471 [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Earnshaw WC (2004) Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113: 211–222 [DOI] [PubMed] [Google Scholar]
- Wang W, Budhu A, Forgues M, Wang XW (2005) Temporal and spatial control of nucleophosmin by the Ran–Crm1 complex in centrosome duplication. Nat Cell Biol 7: 823–830 [DOI] [PubMed] [Google Scholar]
- Weis K (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441–451 [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Henzing AJ, Dodson H, Khaled W, Earnshaw WC (2004) Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J Biol Chem 279: 5655–5660 [DOI] [PubMed] [Google Scholar]
- Xu L, Massague J (2004) Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol 5: 209–219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


