Abstract
The maintenance of ionic homeostasis in response to changes in the environment is essential for all living cells. Although there are still many important questions concerning the role of the major monovalent cation K+, cytoplasmic K+ in bacteria is required for diverse processes. Here, we show that enzyme IIANtr (EIIANtr) of the nitrogen-metabolic phosphotransferase system interacts with and regulates the Escherichia coli K+ transporter TrkA. Previously we reported that an E. coli K-12 mutant in the ptsN gene encoding EIIANtr was extremely sensitive to growth inhibition by leucine or leucine-containing peptides (LCPs). This sensitivity was due to the requirement of the dephosphorylated form of EIIANtr for the derepression of ilvBN expression. Whereas the ptsN mutant is extremely sensitive to LCPs, a ptsN trkA double mutant is as resistant as WT. Furthermore, the sensitivity of the ptsN mutant to LCPs decreases as the K+ level in culture media is lowered. We demonstrate that dephosphorylated EIIANtr, but not its phosphorylated form, forms a tight complex with TrkA that inhibits the accumulation of high intracellular concentrations of K+. High cellular K+ levels in a ptsN mutant promote the sensitivity of E. coli K-12 to leucine or LCPs by inhibiting both the expression of ilvBN and the activity of its gene products. Here, we delineate the similarity of regulatory mechanisms for the paralogous carbon and nitrogen phosphotransferase systems. Dephosphorylated EIIAGlc regulates a variety of transport systems for carbon sources, whereas dephosphorylated EIIANtr regulates the transport system for K+, which has global effects related to nitrogen metabolism.
Keywords: leucine toxicity, nitrogen-metabolic phosphotransferase system (PTS), potassium transporter TrkA, protein–protein interaction, signal transduction
The well defined phosphotransferase system (PTS) is composed of two general cytoplasmic proteins, enzyme I (EI) and histidine phosphocarrier protein (HPr), and some sugar-specific components collectively known as enzymes II (1). The carbohydrate PTS, especially for glucose uptake, occupies a central position in bacterial physiology as a result of the identification of multiple regulatory functions superimposed on the transport functions, such as regulation of chemotaxis by EI (2); regulation of glycogen breakdown by HPr (3, 4); regulation of the global repressor Mlc by the membrane-bound glucose transporter EIICBGlc (5–7); and regulation of carbohydrate transport and metabolism (1, 8, 9), the metabolic flux between fermentation and respiration (10), and adenylyl cyclase activity (11) by EIIAGlc. These regulatory functions of the carbohydrate PTS depend on the phosphorylation state of the involved components, which have been shown to increase in the absence and decrease in the presence of a PTS sugar substrate.
The nitrogen-metabolic PTS consists of enzyme INtr (EINtr, an EI paralog encoded by ptsP), NPr (an HPr paralog encoded by ptsO), and enzyme IIANtr (EIIANtr, an EIIAMtl paralog encoded by ptsN) (12–14). The genes encoding NPr and EIIANtr are located on the same operon as rpoN, which led to the proposal that this pathway is involved in nitrogen metabolism. Because phosphoryl transfer to a specific substrate has not yet been demonstrated for the nitrogen PTS, it has been suggested that the role of this pathway is in regulation (12). In contrast to the extensive body of data dealing with regulation mechanisms associated with the carbohydrate PTS, substantially less is known about regulatory mechanisms connected with the nitrogen-metabolic PTS.
Recently, we reported that dephosphorylated EIIANtr is required for derepression of the ilvBN operon, which encodes acetohydroxy acid synthase I (AHAS I), the enzyme catalyzing the first step common to the biosynthesis of the branched-chain amino acids (15). Because the derepression effect was only observed under specific growth conditions, we proposed that dephosphorylated EIIANtr binds to some undefined regulator for that operon. Here, we used ligand fishing to isolate a protein factor interacting with EIIANtr that regulates ilvBN expression and leucine sensitivity in Escherichia coli. The binding partner was identified as TrkA, a K+ transporter. We demonstrate that K+ serves as an important signal controlling both the transcription and activity of AHAS I.
Results
Specific Interaction Between the Dephosphorylated Form of EIIANtr and the K+ Transporter TrkA.
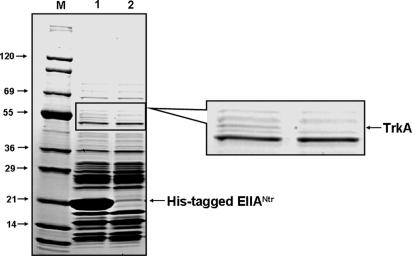
We previously proposed that an additional unknown factor was involved in the requirement of dephosphorylated EIIANtr for the derepression of the ilvBN operon. To search for a factor that interacts with dephosphorylated EIIANtr, we used ligand fishing. A crude extract from E. coli MG1655 was mixed with EIIANtr or a His6-tagged form of EIIANtr (His–EIIANtr) and subjected to pull-down assays by using a metal-affinity resin. We found a protein band (apparent molecular mass of ≈50 kDa) specifically eluting in a fraction containing His–EIIANtr (Fig. 1). Peptide mapping of the protein band indicated that it corresponds to the K+ transporter TrkA, a cytoplasmic component of the low-affinity K+ transport system, Trk (16).
Fig. 1.
Specific interaction of TrkA with EIIANtr. Ligand fishing to search for protein factor(s) interacting with EIIANtr. Crude extract prepared from MG1655 grown in 500 ml of LB to A600 of 2.0 was mixed with 2 mg of either purified His–EIIANtr (lane 1) or EIIANtr (lane 2), and these mixtures were passed through metal-affinity chromatography columns containing 500 μl of TALON resin. After washing the columns with 20 mM Hepes buffer (pH 7.5) containing 200 mM NaCl three times (2 ml), proteins bound to the resin were eluted with the same buffer (500 μl) containing 200 mM imidazole, and the eluates were analyzed by SDS/PAGE followed by staining with Coomassie brilliant blue (Bio-Rad, Hercules, CA). In-gel digestion followed by MALDI-TOF analysis revealed that the protein band bound specifically to His–EIIANtr corresponded to TrkA (marked with an arrow). Tefco (Tokyo, Japan) Wide Range protein standards were used as molecular mass markers (lane M).
To explore whether formation of a TrkA–EIIANtr complex occurs in vivo, TrkA with or without N-terminal His6 residues was expressed in cells lacking the chromosomal trkA gene. Crude extracts prepared from these cells were subjected to a pull-down assay by using an affinity resin. His6-tagged TrkA (His–TrkA) was bound to a Co2+-affinity column [see supporting information (SI) Fig. 7A, lane 4], and proteins eluted with 200 mM imidazole were analyzed by SDS/PAGE. EIIANtr was coeluted with His–TrkA bound to the beads, whereas no EIIANtr was recovered from the elution fraction when crude extract prepared from cells expressing unmodified TrkA was used (SI Fig. 7B). The tight in vivo interaction between EIIANtr and TrkA was also confirmed by using the plasmid pCRduet coexpressing His–EIIANtr and TrkA. When the crude extract prepared from cells overexpressing the two proteins was loaded onto a TALON metal-affinity column, TrkA copurified with His–EIIANtr (see SI Fig. 8). These results support the contention that the tight interaction between TrkA and EIIANtr also occurs in vivo.
To measure the binding constant between TrkA and EIIANtr, the technique of surface plasmon resonance was used (4). EIIANtr was immobilized on a CM5 sensor chip, and TrkA was allowed to flow over the surface. Because the similarity of the sequence identity between EIIAGlc and EIIANtr is only ≈13%, immobilized EIIAGlc was used as a control. When purified TrkA was exposed to immobilized EIIAGlc, no interaction was detected (Fig. 2A, sensorgram b). In contrast, TrkA bound to immobilized EIIANtr (Fig. 2A, sensorgram a). The dissociation constant (Kd) for the interaction between EIIANtr and TrkA was determined to be ≈8.4 × 10−7 M.
Fig. 2.
Analysis of the interaction between EIIANtr and TrkA by surface plasmon resonance. (A) Specific interaction of TrkA with EIIANtr. Purified EIIANtr and EIIAGlc were immobilized on the carboxymethylated dextran surface of a CM5 sensor chip. Based on the assumption that 1,000 resonance units (RUs) corresponds to a surface concentration of 1 ng/mm2, the proteins EIIAGlc and EIIANtr were immobilized to surface concentrations of 1.5 and 2.0 ng/mm2, respectively. TrkA (5 μg/ml) was allowed to flow over the EIIANtr (sensorgram a) and EIIAGlc (sensorgram b) surfaces for 12 min each. The dissociation constant Kd for the interaction between EIIANtr and TrkA was determined by using BIAevaluation 2.1 software (BIAcore, Uppsala, Sweden) to be ≈8.4 × 10−7 M. (B) Phosphorylation state-dependent interaction between TrkA and EIIANtr. TrkA (5 μg/ml) was allowed to flow over the EIIANtr surface for 5 min in each sensorgram. The phosphorylated and dephosphorylated EIIANtr surfaces were generated by reversible phosphoryl transfer reactions between NPr and EIIANtr, as described in Materials and Methods. Sensorgram a shows TrkA binding to the immobilized EIIANtr surface without any treatment. In sensorgram b, TrkA was injected after the immobilized EIIANtr surface had been phosphorylated by exposing it to a mixture of EINtr and NPr in the presence of phosphoenolpyruvate (PEP), then flushing with running buffer to remove PEP and other PTS proteins. In sensorgram c, dephosphorylated NPr was allowed to flow over the phosphorylated EIIANtr surface generated in sensorgram b to dephosphorylate the surface before TrkA was injected.
Because the interaction with target proteins and regulatory functions of the PTS depend on the phosphorylation state of the involved proteins (5), the effect of phosphorylation of EIIANtr on its affinity for TrkA was checked (Fig. 2B). The phosphorylated and dephosphorylated EIIANtr surfaces were generated by reversible phosphoryl transfer reactions between NPr and EIIANtr. When purified TrkA was exposed to immobilized EIIANtr, a high-affinity interaction was detected (Fig. 2B, sensorgram a). After immobilized EIIANtr was phosphorylated by allowing a mixture of EINtr and NPr in the presence of phosphoenolpyruvate (PEP) to flow over the surface and subsequently washing with running buffer, an interaction with TrkA was hardly detectable (Fig. 2B, sensorgram b). The same EIIANtr surface recovered TrkA binding activity after dephosphorylation when we allowed dephosphorylated NPr to flow through the cell and then flushed it with running buffer (Fig. 2B, sensorgram c). These results provide direct evidence for a phosphorylation state-dependent interaction between TrkA and EIIANtr.
Effect of K+ Concentration and TrkA on Growth and Sensitivity to Ala–Leu of a ptsN Mutant.
Based on the phosphorylation state-dependent interaction between the K+ transporter TrkA and EIIANtr, we envisioned that EIIANtr would regulate the activity of TrkA and therefore the intracellular level of K+. To verify this thesis, we tested the effect of medium K+ concentration on the specific growth rate of the exponentially growing cells of a ptsN mutant and WT. It was previously shown that growth of the ptsN mutant was significantly slower than WT in W salts medium (containing 93 mM K+ ion) (13), and the same growth phenotype was observed in M9 minimal medium (containing 22 mM K+), although growth of the ptsN mutant was comparable with that of WT in LB and M9 minimal medium supplemented with 1% casamino acids (data not shown). As shown in Table 1, the growth rate of the ptsN mutant was markedly slower than that of WT in media containing 20–120 mM K+. As the K+ concentration in M9 minimal medium decreased, however, the growth rate of the ptsN mutant increased. Interestingly, the ptsN mutant exhibited a growth rate similar to that of its parental strain, MG1655, in K1 and K0.1 media (n in Kn corresponds to the millimolar concentration of K+ in the M9 minimal medium). The same effects were observed in M9 minimal medium in which glucose was replaced with lactose or glycerol (data not shown).
Table 1.
Effect of K+ concentration on the growth rate of the ptsN mutant (CR301) in minimal medium
| K+ concentration, mM | Growth rate, hr−1 | |
|---|---|---|
| Wild type | ptsN mutant | |
| 0.1 | 0.78 ± 0.035 | 0.82 ± 0.055 |
| 1.0 | 0.79 ± 0.021 | 0.78 ± 0.042 |
| 5.0 | 0.81 ± 0.014 | 0.47 ± 0.037 |
| 10 | 0.82 ± 0.028 | 0.30 ± 0.071 |
| 20 | 0.83 ± 0.016 | 0.15 ± 0.011 |
| 40 | 0.82 ± 0.010 | 0.08 ± 0.033 |
| 60 | 0.82 ± 0.031 | 0.10 ± 0.004 |
| 120 | 0.81 ± 0.034 | 0.06 ± 0.012 |
Cells grown overnight in LB medium were harvested, washed with K0 medium, and suspended in Kn medium as described in Materials and Methods. The growth rate at 37°C of the cells was monitored by measuring the OD600.
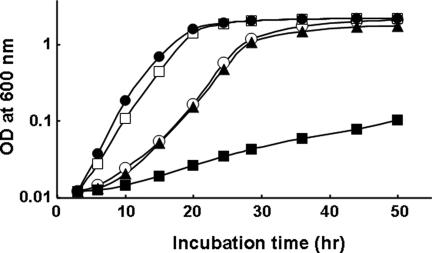
We previously showed that a ptsN mutant was hypersensitive to growth inhibition by the dipeptide Ala–Leu in normal medium (14). From results showing that dephosphorylated EIIANtr interacts with TrkA, it might be predicted that TrkA in ptsN mutant cells would exist in the free form, whereas TrkA would exist in a complex with EIIANtr in ptsO and ptsP mutants because dephosphorylated EIIANtr would accumulate in these mutants. If the nitrogen PTS is epistatic to TrkA in the regulation of sensitivity to LCPs, then the sensitivity of a trkA mutant to Ala–Leu should be independent of mutations in the nitrogen PTS. To test this idea, we constructed three double mutants by deleting the trkA gene in the ptsP, ptsO, and ptsN backgrounds. Although the ptsN mutant was extremely sensitive to LCPs, cells of the ptsN trkA double-mutant strain exhibited normal growth rates, comparable with that of MG1655 in the medium supplemented with 0.5 mM Ala–Leu dipeptide (Fig. 3). Noteworthily, the disruption of trkA in ptsP and ptsO strains neutralized resistance to Ala–Leu to the level seen in MG1655 (data not shown). These results implicate TrkA as the regulator operating downstream of EIIANtr with respect to sensitivity to LCPs.
Fig. 3.
The toxic effect of the Ala–Leu dipeptide on the ptsN mutant strain is neutralized by disruption of the trkA gene. Cells grown overnight in LB were harvested, washed, and suspended in M9 minimal medium containing 0.5% glucose and 0.5 mM Ala–Leu dipeptide. MG1655, Open circles; ptsP mutant CR101, open squares; ptsO mutant CR201, filled circles; ptsN mutant CR301, filled squares; and ptsN trkA double mutant CR302, filled triangles. Strains CR102 (the ptsP trkA double mutant) and CR202 (the ptsO trkA double mutant) showed growth curves similar to that of CR302.
EIIANtr-Mediated Change in AHAS Expression Is Dependent on TrkA.
It was previously reported that the sensitivity of the ptsN mutant to LCPs is due to the low level of AHAS expression, leading to isoleucine pseudoauxotrophy by leucine (15). Thus, we examined the effect of a trkA deletion on AHAS activity (Fig. 4A). AHAS activity in the ptsN mutant cells was significantly increased in the K0 medium compared with that in cells grown in the K20 medium. Furthermore, the introduction of a trkA deletion mutation into the ptsN mutant strain restored AHAS activity to the level of the WT strain, indicating that expression of AHAS is regulated by TrkA. The AHAS level in the trkA mutant was insensitive to the level of K+ in the medium regardless of the presence of the ptsN mutation. This result is consistent with the data in Fig. 3 showing that the trkA mutant was insensitive to Ala–Leu regardless of the presence of the ptsN mutation, further indicating the dependence of sensitivity to LCPs on the intracellular AHAS level.
Fig. 4.
EIIANtr-mediated change in AHAS expression depends on TrkA. (A) AHAS activity assays. Cells of strains MG1655, CR301 (ptsN), CR302 (ptsN TrkA), and EB101 (trkA) grown in LB were harvested, washed, and resuspended in K0 minimal medium containing 0.5% glucose (filled bars) and K20 minimal medium containing 0.5% glucose (open bars). Subsequently, cells were allowed to grow to midlogarithmic phase, and the specific activities of AHAS were measured as described in Materials and Methods. (B) Measurement of the ilvBN transcript by RT-PCR, which was carried out as described previously (15). (C) Primer extension analysis of the ilvBN transcript. RNA was prepared from the indicated cells grown to midlogarithmic phase in M9 minimal medium containing 0.5% glucose by using an RNeasy mini kit (Qiagen), and primer extension assays were carried out as described previously (5).
It was previously shown that the dephosphorylated form of EIIANtr is required for derepression of the ilvBN operon, although ilvIH expression is not affected by a ptsN deletion (15). Therefore, the effect of a trkA deletion as well as the effect of the K+ level in the medium on ilvBN expression were examined. First, the mRNA level of the ilvBN operon was measured by RT-PCR in MG1655, the ptsN mutant CR301 (15), and a ptsN trkA double mutant grown in M9 minimal medium (Fig. 4B). As reported previously (15), the ilvBN transcript level in the ptsN mutant grown in the typical M9 minimal medium containing 22 mM K+ was much lower than that in WT. However, ilvBN expression in the ptsN mutant grown in M9 minimal medium without K+ (K0 medium) was derepressed to the level in WT cells. Significantly, the ilvBN transcript level in the ptsN trkA double mutant was similar to that in WT. To confirm the effect of the trkA deletion on the expression of ilvBN, we also carried out primer extension analysis of the ilvBN transcript. As shown in Fig. 4C, the ptsN deletion caused a significant decrease in ilvBN transcription. Although introduction of the ptsP mutation did not significantly change the ilvBN transcript level in the ptsN mutant, introduction of the trkA mutation recovered ilvBN expression in the ptsN mutant to the level in WT cells. The obvious conclusion is that PtsN-dependent ilvBN expression is mediated by TrkA.
Dephosphorylated EIIANtr Inhibits TrkA-Mediated K+ Uptake.
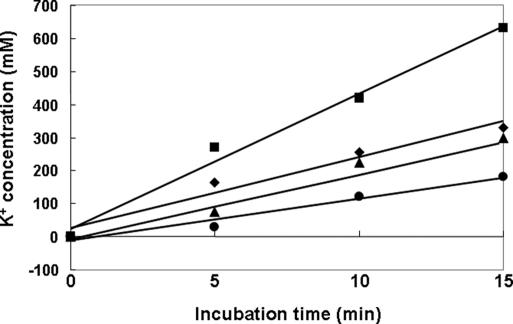
Taken together, our results suggest that TrkA-dependent K+ homeostasis is altered in strains lacking the ptsN gene. It seems clear that low (or no) expression of AHAS in the ptsN mutant was due to the increased level of intracellular K+ resulting from the increased level of the free form of TrkA. Therefore, the rate of K+ accumulation was measured in strains lacking ptsN, ptsO, or a ptsN mutant harboring an expression vector for the H73A mutant form of EIIANtr and compared with that in the WT cells. When cells were shifted from K0 to K20 medium, the K+ uptake rate of the ptsN mutant was higher, whereas that of the ptsO and ptsN cells harboring pCR3(H73A) was lower than those in MG1655 (Fig. 5). These data suggest that dephosphorylated EIIANtr inhibits TrkA-mediated K+ uptake and indicate that an increase of the K+ level in the ptsN mutant associated with the presence of free TrkA negatively affects derepression of AHAS activity.
Fig. 5.
Analysis of intracellular accumulation of K+. Strains MG1655, CR201, and CR301 grown in K0 medium were harvested, washed, and resuspended in K20 minimal medium containing 0.5% glucose. Cells were harvested by filtration at the indicated time points and processed for analysis of K+ content by employing inductively coupled plasma/optical emission spectrometry (ICP-OES) as described in Materials and Methods. MG1655, diamonds; CR201 (ptsO), triangles; CR301 (ptsN), squares; and CR301/pCR3(H73A), circles.
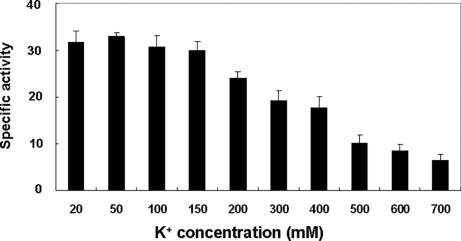
Cytoplasmic K+ concentrations are maintained at relatively constant levels of 300–500 mM in E. coli cells growing in standard media by the activities of several K+ uptake and K+ efflux systems (17, 18). Thus, we tested whether AHAS activity is affected by changes in the concentration of K+. AHAS activity was inhibited by K+ when its concentration was >200 mM in a concentration-dependent manner (Fig. 6). Thus, we concluded that K+ inhibits both AHAS expression and activity.
Fig. 6.
Effect of K+ on AHAS activity. MG1655 cells were grown to midlogarithmic phase in M9 minimal medium containing 0.5% glucose at 37°C. The harvested cell pellet was washed and resuspended in 50 mM Tris·HCl buffer (pH 7.5) containing 20 mM FAD. This suspension was passed through a French pressure cell at 10,000 psi. The lysate was centrifuged at 100,000 × g for 90 min, and the supernatant was used as crude AHAS. The specific activity of AHAS was measured as previously described (15). To check the direct effect of K+ on AHAS activity, KCl was added to the reaction mixture to the indicated concentrations.
Discussion
The maintenance of ionic homeostasis in response to environmental changes is vital to all living cells. K is the major monovalent cellular cation in bacteria such as E. coli. Cytoplasmic K+ is required for processes as diverse as the activation of cytoplasmic enzymes, maintenance of cell turgor, homeostasis of cytoplasmic pH, and adaptation of cells to osmotic conditions (19, 20). K+ concentrations are maintained at relatively constant levels (300–500 mM) in E. coli by the activities of K+ uptake and efflux systems (17, 18). The high-affinity K+ uptake ATPase Kdp (Km for K+ uptake of ≈2 μM) accumulates K+ at low external K+ concentrations, whereas its expression is repressed in K+-replete cells (21). At normal cell turgor, E. coli takes up K+ via two constitutively expressed systems, Trk (Km for K+ uptake of 0.9–1.5 mM) and Kup (Km for K+ uptake of ≈0.37 mM) (22). Trk has an ≈100-fold higher Vmax than Kup has, implying that Trk constitutes the major K+ uptake system under normal growth conditions. Further, Trk, but not Kup, has relatively high specificity for K+ (19, 22).
It has been suggested that complex mechanisms regulate K+ homeostasis in response to a stimulus perceived by cells (19). Recently, it was reported that a protein kinase regulates the K+ transporter Akt1 in Arabidopsis thaliana (23). From the results presented here, it is evident that dephosphorylated EIIANtr interacts with TrkA, resulting in the inhibition of Trk-mediated K+ uptake. The abnormally high level of K+ accumulated by the free form of TrkA in a ptsN mutant inhibits both AHAS expression and activity. This explains the extreme sensitivity of a ptsN mutant and the resistance of a trkA mutant to LCPs. It was previously reported that a mutation in the kefA gene encoding a K+ efflux system made E. coli K-12 cells sensitive to K+ in a synthetic medium (24, 25), indicating that accumulation of high intracellular K+ inhibits the growth of these cells. These results are consistent with our data showing that a ptsN mutant was also sensitive to high medium concentrations of K+ (Table 1). These data, in addition to those showing that sensitivity of the ptsN mutant to LCPs was neutralized in K+-depleted medium, confirm that the slower growth of the ptsN mutant compared with WT is due to higher intracellular K+ concentration.
We studied the relationship of intracellular K+ concentration to AHAS expression. It was recently reported that K salts have a direct inhibitory effect on RNA polymerase at ribosomal promoters (26). It was shown that K+ can act independently of macromolecular repressors or activators by virtue of its ability to directly inhibit RNA polymerase binding to ribosomal promoters. Therefore, K+ may have a similar inhibitory effect on RNA polymerase at the ilvBN promoter or K+ may act through an as yet unidentified regulatory component.
The biological importance of regulation of AHAS expression and activity by the EIIANtr–TrkA complex is worthy of discussion. As pointed out previously (15), cells typically make more proteins involved in amino acid synthesis when they are exposed to an environment depleted of amino acids. Previous studies (27–32) led us to conclude that leucine, one of the branched-chain amino acids whose synthesis is catalyzed by the enzyme AHAS, acts as a second messenger that monitors the environmental amino acid level. Although the three branched-chain amino acids constitute the most abundant protein building blocks in E. coli (27), their synthetic pathways are more complex than those for other amino acids. The regulatory importance of the three branched-chain amino acids is also supported by the existence of three AHAS isozymes with different properties in E. coli. Further, leucine acts as a signaling molecule regulating the leucine-responsive regulatory protein, Lrp (28), a global regulator involved in modulating a variety of metabolic functions including the catabolism and anabolism of amino acids as well as pili synthesis (29). In general, Lrp-related operons functioning in biosynthetic pathways are stimulated by Lrp, whereas those involved in catabolic pathways are repressed. Furthermore, Lrp, via its potent regulation of gltBDF and control of intracellular glutamine concentration, plays a significant role in the regulation of nitrogen assimilation genes (30). The high degree of conservation of Lrp in prokaryotes suggests that it has an important role in all enterobacteria (31, 32), implying that leucine's role as a second messenger is widespread in bacteria.
It is interesting that the paralogous PTSs in E. coli exhibit paralogous regulatory activities: the nitrogen-metabolic PTS inhibits the K+ transporter TrkA by a physical interaction with dephosphorylated EIIANtr, whereas dephosphorylated EIIAGlc of the carbohydrate PTS inhibits several non-PTS permeases via physical interactions (1). Although the phosphorylation state of PTS proteins is known to be controlled by the availability of PTS sugars, the mechanism regulating the phosphorylation state of the nitrogen-metabolic PTS proteins is not understood. The presence of a NifA protein-like putative sensory domain in EINtr suggests that this domain might sense the availability of a ligand that controls the phosphorylation state of the nitrogen-metabolic PTS. Although the search for such a ligand has not yet been successful, changes in nutritional environment seem to signal the nitrogen-metabolic PTS to regulate TrkA activity and intracellular K+ concentrations. K+-dependent changes in AHAS activity then regulate intracellular leucine levels. Subsequently, leucine regulates the expression of numerous genes involved in amino acid metabolism through Lrp. It remains to be established how many additional bacterial pathways are regulated by fluctuations in cellular K+ levels.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions.
The bacterial strains and plasmids used in this study are listed in SI Table 2 and their constructions are described in SI Materials and Methods. Bacteria were cultured as described previously (15, 33).
Purification of Overexpressed Proteins.
Purification of EIIANtr and EIIANtr (H73A) was accomplished as described previously (15). EINtr, NPr, and TrkA were overexpressed in GI698 cells harboring pCR1, pCR2, and pEB1 and purified after procedures previously developed for overexpression and purification of EI (33), HPr (4), and FrsA (10), respectively. His–EIIANtr was purified by using TALON metal-affinity resin (Clontech, Mountain View, CA) according to the manufacturer's instructions, and bound His–EIIANtr was eluted with binding buffer containing 200 mM imidazole. The fractions containing His–EIIANtr were pooled and concentrated in a Macrosep centrifugal concentrator (3K, Pall, East Hills, NY). To obtain homogeneous His–EIIANtr (>98% pure) and to remove imidazole, the concentrated pool was chromatographed on a HiLoad 16/60 Superdex 75 prep grade column (Amersham Biosciences, Little Chalfont, England) equilibrated with buffer A [20 mM Hepes–KOH (pH 8.0), containing 200 mM NaCl].
Ligand Fishing to Search for Proteins Interacting with His6–Tagged EIIANtr.
E. coli MG1655 cells grown overnight in 500 ml of LB were harvested and washed with and resuspended in 30 ml of buffer A in the presence of 100 μg/ml PMSF. Cells were disrupted by passing twice through a French pressure cell at 10,000 psi (1 psi = 6.89 kPa) and then by centrifuging at 100,000 × g for 60 min at 4°C. The supernatant was mixed with 2 mg of EIIANtr or His–EIIANtr, and this mixture was incubated with 500 μl of TALON metal-affinity resin. After incubation at room temperature for 10 min, the mixture was loaded onto a Poly-Prep chromatography column (8 × 40 mm) (Bio-Rad, Hercules, CA). The column was washed with 1 ml of buffer A three times, and the proteins bound to the column were eluted with buffer A containing 200 mM imidazole. Aliquots of the eluted protein sample (10 μl each) were analyzed by SDS/PAGE and then were stained with Coomassie brilliant blue R. The protein band specifically bound to His–EIIANtr was excised from the gel, and in-gel digestion and peptide mapping of tryptic digests were carried out as described previously (34).
Surface Plasmon Resonance Spectroscopy.
Real-time interaction of TrkA with EIIANtr or EIIAGlc was monitored via surface plasmon resonance detection by using a BIAcore 3000 system (BIAcore, Uppsala, Sweden) with some modifications as described previously (4, 5, 10). EIIANtr and EIIAGlc were separately immobilized onto the carboxymethylated dextran surface of a CM5 sensor chip. EIIAGlc and EIIANtr (100 μl, 10 μg/ml) in coupling buffer (10 mM sodium acetate, pH 5.0) were allowed to flow over the sensor chip at 5 μl/min to couple the proteins to the matrix by a N-hydroxysuccinimide/1-ethyl-3′-(3-dimethylaminopropyl)carbodiimide reaction (80 μl of mix). Based on the assumption that 1,000 resonance units (RUs) corresponds to a surface concentration of 1 ng/mm2, EIIAGlc and EIIANtr were immobilized to surface concentrations of 1.5 and 2.0 ng/mm2, respectively. The standard running buffer was 10 mM Hepes (pH 7.2), 150 mM NaCl, 10 mM KCl, 1 mM MgCl2, and 0.5 mM EDTA, and all reagents were introduced at a flow rate of 10 μl/min. The sensor surface was regenerated between assays by using the standard running buffer at a flow rate of 100 μl/min for 10 min to remove bound analytes.
To phosphorylate the immobilized EIIANtr, a mixture (50 μl) of phosphoenolpyruvate (PEP) (0.5 mM), EINtr (20 μg/ml), and NPr (10 μg/ml) in standard running buffer was allowed to flow into the flow cell for 10 min. To remove the phosphoryl group from the immobilized phosphorylated EIIANtr, NPr (0.25 mg/ml in standard running buffer) was allowed to flow into the flow cell for 15 min. The sensor surface was regenerated between injections by running standard buffer at a flow rate of 100 μl/min for 10 min to remove bound analytes.
Effect of K+ Concentration on Growth and Sensitivity to Ala–Leu of the ptsN Mutant.
The influence of K+ concentration in the medium on growth rate was tested on the Kn minimal medium (pH 7.0), where n is the K+ concentration in micromolars (24, 35). K120 medium consisted of 48 mM K2HPO4, 24 mM KH2PO4, 8 mM (NH4)2SO4, 0.4 mM MgSO4, 6 μM FeSO4, 1 mM sodium citrate, 1 mg/ml thiamine hydrochloride, and 0.2% glucose. K0 medium was similar, with equimolar amounts of Na salts replacing the potassium phosphate. Kn medium was prepared by mixing suitable proportions of K0 and K115. Fresh cells grown overnight in LB were harvested and washed with K0 medium. After resuspension in Kn medium, specific growth rates of these cells at 37°C were determined by measuring the OD at 600 nm. Growth was also checked in the presence of 0.5 mM Ala–Leu dipeptide in K0 medium where indicated.
Transcript Analysis.
RT-PCR was carried out as described previously (15). Primer extension reactions were carried out as described previously (16). Cells were grown to A600 of 0.5 in M9 minimal medium containing 0.5% glucose, and total E. coli RNA was purified by using an RNeasy mini kit (Qiagen, Valencia, CA) and resuspended in sterile distilled water. Total cell RNA (30 μg) was mixed with 0.2 pmol of the γ-32P-end-labeled primer ilvBPex, 5′-TTG CCA TGC TCC AGT CCT TTT CTT CTG GGC-3′. The mixture was heated to 60°C and then allowed to cool to room temperature over a period of 1 h. After annealing, 700 μM dNTPs, 10 mM MgCl2, 5 mM DTT, 20 mM Tris·HCl (pH 8.3), and 100 units of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) were added to a final volume of 100 μl. After the mixture was incubated at 40°C for 70 min, 2 μl of 0.5 M EDTA was added to the reaction mixture, and the mixture then was incubated at 37°C for 30 min. DNA was precipitated and resolved on 5% polyacrylamide gel with 8 M urea and visualized by autoradiography.
Measurement of Intracellular K+ Concentration.
Intracellular K+ concentration was measured according to the procedure of McLaggan et al. (24) with some modifications. Cells cultured in LB were harvested by centrifugation, washed, and incubated at 37°C in K0 medium. After incubation for 1 h, KCl was added to 20 mM, and the accumulation of K+ was determined. Samples (10 ml each) taken at the indicated time points were rapidly filtered on 47-mm membrane filters [pore size of 0.8 μm; Whatman (Brentford, U.K.) polycarbonate]. The samples were washed dropwise with 1–2 ml of K0 medium (37°C) made slightly hypertonic by the addition of 50 mM NaCl. After the filters were dried, 10 ml of distilled water was added, and the mixtures were heated at 90°C for 10 min. The released K+ was measured by using an inductively coupled plasma/optical emission spectrometer (ICP-OES) (Shimadzu, Kyoto, Japan).
Supplementary Material
Acknowledgments
This work was supported by 21C Frontier Microbial Genomics and Applications Center Program Grant MG05-0202-6-0 of the Republic of Korea. C.-R.L., S.-H.C., and M.-J.Y. were supported by BK21 Research Fellowships from the Korean Ministry of Education and Human Resources Development. A.P. was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute of the National Institutes of Health.
Abbreviations
- PTS
phosphotransferase system
- AHAS
acetohydroxy acid synthase
- LCPs
leucine-containing peptides
- PEP
phosphoenolpyruvate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609897104/DC1.
References
- 1.Postma PW, Lengeler JW, Jacobson GR. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lux R, Jahreis K, Bettenbrock K, Parkinson JS, Lengeler JW. Proc Natl Acad Sci USA. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seok Y-J, Koo B-M, Sondej M, Peterkofsky A. J Mol Microbiol Biotechnol. 2001;3:385–393. [PubMed] [Google Scholar]
- 4.Seok Y-J, Sondej M, Badawi P, Lewis MS, Briggs MC, Jaffe H, Peterkofsky A. J Biol Chem. 1997;272:26511–26521. doi: 10.1074/jbc.272.42.26511. [DOI] [PubMed] [Google Scholar]
- 5.Nam T-W, Cho S-H, Shin D, Kim J-H, Jeong J-Y, Lee J-H, Roe J-H, Peterkofsky A, Kang S-O, Ryu S, Seok YJ. EMBO J. 2001;20:491–498. doi: 10.1093/emboj/20.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S-J, Boos W, Bouche JP, Plumbridge J. EMBO J. 2000;19:5353–5361. doi: 10.1093/emboj/19.20.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka Y, Kimata K, Aiba H. EMBO J. 2000;19:5344–5352. doi: 10.1093/emboj/19.20.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seok Y-J, Sun J, Kaback HR, Peterkofsky A. Proc Natl Acad Sci USA. 1997;94:13515–13519. doi: 10.1073/pnas.94.25.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurley JH, Faber HR, Worthylake D, Meadow ND, Roseman S, Pettigrew DW, Remington SJ. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 10.Koo B-M, Yoon M-J, Lee C-R, Nam T-W, Choe Y-J, Jaffe H, Peterkofsky A, Seok Y-J. J Biol Chem. 2004;279:31613–31621. doi: 10.1074/jbc.M405048200. [DOI] [PubMed] [Google Scholar]
- 11.Park Y-H, Lee BR, Seok Y-J, Peterkofsky A. J Biol Chem. 2006;281:6448–6454. doi: 10.1074/jbc.M512672200. [DOI] [PubMed] [Google Scholar]
- 12.Reizer J, Reizer A, Merrick MJ, Plunkett G, III, Rose DJ, Saier MH., Jr Gene. 1996;181:103–108. doi: 10.1016/s0378-1119(96)00481-7. [DOI] [PubMed] [Google Scholar]
- 13.Powell BS, Court DL, Inada T, Nakamura Y, Michotey V, Cui X, Reizer A, Saier MH, Reizer J., Jr J Biol Chem. 1995;270:4822–4839. doi: 10.1074/jbc.270.9.4822. [DOI] [PubMed] [Google Scholar]
- 14.Peterkofsky A, Wang G, Seok Y-J. Arch Biochem Biophys. 2006;453:101–107. doi: 10.1016/j.abb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Lee C-R, Koo B-M, Cho S-H, Kim Y-J, Yoon M-J, Peterkofsky A, Seok Y-J. Mol Microbiol. 2005;58:334–344. doi: 10.1111/j.1365-2958.2005.04834.x. [DOI] [PubMed] [Google Scholar]
- 16.Dosch DC, Helmer GL, Sutton SH, Salvacion FF, Epstein W. J Bacteriol. 1991;173:687–696. doi: 10.1128/jb.173.2.687-696.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roe AJ, McLaggan D, O'Byrne CP, Booth IR. Mol Microbiol. 2000;35:1235–1243. doi: 10.1046/j.1365-2958.2000.01793.x. [DOI] [PubMed] [Google Scholar]
- 18.Bossemeyer D, Borchard A, Dosch DC, Helmer GC, Epstein W, Booth IR, Bakker EP. J Biol Chem. 1989;264:16403–16410. [PubMed] [Google Scholar]
- 19.Epstein W. Prog Nucleic Acid Res Mol Biol. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 20.Suelter CH. Science. 1970;168:789–795. doi: 10.1126/science.168.3933.789. [DOI] [PubMed] [Google Scholar]
- 21.Rhoads DB, Waters FB, Epstein W. J Gen Physiol. 1976;67:325–341. doi: 10.1085/jgp.67.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bossemeyer D, Schlosser A, Bakker EP. J Bacteriol. 1989;171:2219–2221. doi: 10.1128/jb.171.4.2219-2221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 24.McLaggan D, Jones MA, Gouesbet G, Levina N, Lindey S, Epstein W, Booth IR. Mol Microbiol. 2002;43:521–536. doi: 10.1046/j.1365-2958.2002.02764.x. [DOI] [PubMed] [Google Scholar]
- 25.Cui C, Adler J. J Membr Biol. 1996;150:143–152. doi: 10.1007/s002329900039. [DOI] [PubMed] [Google Scholar]
- 26.Gralla JD, Vargas DR. EMBO J. 2006;25:1515–1521. doi: 10.1038/sj.emboj.7601041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reitzer L. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 28.Willins DA, Ryan CW, Platko JV, Calvo JM. J Biol Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]
- 29.Calvo JM, Matthews RG. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borst DW, Blumenthal RM, Matthews RG. J Bacteriol. 1996;178:6904–6912. doi: 10.1128/jb.178.23.6904-6912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman EB, Lin R. Annu Rev Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 32.Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. Mol Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 33.Seok Y-J, Lee BR, Zhu P-P, Peterkofsky A. Proc Natl Acad Sci USA. 1996;93:347–351. doi: 10.1073/pnas.93.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong J-Y, Kim Y-J, Cho N, Shin D, Nam T-W, Ryu S, Seok Y-J. J Biol Chem. 2004;279:38513–38518. doi: 10.1074/jbc.M406667200. [DOI] [PubMed] [Google Scholar]
- 35.Epstein W, Kim BS. J Bacteriol. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.