Abstract
All cells are unavoidably exposed to chemicals that can alkylate DNA to form genotoxic damage. Among the various DNA lesions formed, O6-alkylguanine lesions can be highly cytotoxic, and we recently demonstrated that O6-methylguanine (O6MeG) and O6-chloroethylguanine (O6CEG) specifically initiate apoptosis in hamster cells. Here we show, in both hamster and human cells, that the MutSα branch of the DNA mismatch repair pathway (but not the MutSβ branch) is absolutely required for signaling the initiation of apoptosis in response to O6MeGs and is partially required for signaling apoptosis in response to O6CEGs. Further, O6MeG lesions signal the stabilization of the p53 tumor suppressor, and such signaling is also MutSα-dependent. Despite this, MutSα-dependent apoptosis can be executed in a p53-independent manner. DNA mismatch repair status did not influence the response of cells to other inducers of p53 and apoptosis. Thus, it appears that mismatch repair status, rather than p53 status, is a strong indicator of the susceptibility of cells to alkylation-induced apoptosis. This experimental system will allow dissection of the signal transduction events that couple a specific type of DNA base lesion with the final outcome of apoptotic cell death.
Apoptosis, or programmed cell death, induced by mutagenic carcinogens is vital because it eliminates cells harboring mutagenic DNA damage from the body. If allowed to survive, such cells could contribute to carcinogenesis. In fact, a large fraction of human tumors contain cells that have been potentially compromised in their ability to undergo apoptosis because they are mutated in the p53 tumor suppressor gene (1). Such a deficiency in the apoptotic response may not only accelerate the carcinogenic process but also render the resultant tumor cells resistant to radiation or chemotherapy. However, apoptosis induced by some DNA-damaging agents is independent of p53 function (2–4), and it is therefore important to identify p53-independent pathways involved in signaling such programmed cell death.
Ionizing radiation (IR) is an efficient inducer of apoptosis and an efficient inducer of DNA damage. IR has thus become the prototypic agent for DNA-damage-induced apoptosis, which is generally believed to be p53-dependent (5, 6). However, because IR creates many types of DNA damage, it has been difficult to determine which particular lesions signal apoptosis (7). Moreover, the fact that IR damages cellular components other than DNA further obscures exactly how IR signals apoptosis (8). Indeed, the fact that DNA-damaging agents can activate an apoptotic pathway in enucleated cells challenges the notion that the major signal for apoptosis originates from DNA damage (9). Recently, we and others demonstrated that a specific type of DNA base damage can signal apoptosis (10, 11). It had been shown that simple alkylating agents such as N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and chemotherapeutic alkylating agents such as the chloronitrosoureas efficiently signal apoptosis. But, like IR, alkylating agents damage other cellular components and produce over a dozen different DNA lesions (12). Therefore, it was not possible to determine the precise origin of the signal for apoptosis. However, expression of the O6-methylguanine (O6MeG) DNA methyltransferase protein (MGMT) virtually eliminated apoptosis, demonstrating that O6-alkylguanines are the initiating lesions, because these are the only DNA lesions efficiently repaired by MGMT (13). Here, we explore how O6-alkylguanine DNA lesions signal apoptosis.
In the absence of MGMT-mediated O6MeG repair, the DNA mismatch repair (MMR) pathway has a strong influence on MNNG cytotoxicity. MMR-deficient cells can resist MNNG-induced cytotoxicity, even when O6MeG lesions persist in the genome; hence this phenomenon was termed alkylation tolerance (14). Apparently, the cytotoxicity of O6MeG actually requires a functional MMR pathway, and MMR-deficient cells can repeatedly replicate genomes containing mutagenic O6MeG lesions. Whether such alkylation tolerance contributes to the susceptibility of MMR-deficient individuals to hereditary nonpolyposis colon cancer (HNPCC) and other cancers remains to be determined (15). Here, we explore the mechanism by which MMR renders O6MeG cytotoxic, and thus how MMR-deficient cells achieve alkylation tolerance.
Mammalian MMR occurs via a pathway homologous to, but more complex than, that of Escherichia coli (16). Mismatch-containing heteroduplex DNA is bound by one of two mammalian heterodimeric protein complexes, MutSα (17, 18) or MutSβ (19–21); the complexes consist of MSH2 paired with either MSH6 (for MutSα) or MSH3 (for MutSβ). Each complex has a different substrate range: MutSα binds primarily to single base pair mismatches and single extrahelical bases, whereas MutSβ binds primarily to heteroduplexes with larger stretches of extrahelical bases (20–24). Once bound, MutS heterodimers interact with other MMR proteins to elicit excision of single-stranded DNA containing the mismatch plus neighboring nucleotides, followed by DNA synthesis and ligation (16). It has been shown that alkylation tolerance occurs in MutSα-deficient cells (14, 25) and that MutSα can bind and process O6MeG-containing duplexes in vitro (26, 27). Whether MutSβ binds such duplexes is not known. In this study, we explore the role of both p53 and MMR (initiated by either MutSα or MutSβ) in the induction of apoptosis by alkylating agents.
MATERIALS AND METHODS
Cell Culture and Drug Treatment. The human lymphoblastoid cell lines TK6 (28) and MT1 (29) were gifts from W. G. Thilly (Massachusetts Institute of Technology, Cambridge, MA) and were grown as described (30). TK6-E6–5E (hereafter, TK6-E6) and TK6-E6–20C (hereafter, TK6-E6C) cells (31) were gifts from J. B. Little (Harvard School of Public Health). Chinese hamster DC3F and DC3F/A3 1.5 clone 1 (hereafter A3) cells were gifts from J. L. Hamlin (University of Virginia, Charlottesville, VA) and were cultured as described (32). All media contained 50 units/ml penicillin G (Sigma) and 50 μg/ml streptomycin sulfate (Sigma). TK6-M12 and TK6-P1 cell lines were generated by transfection of TK6 cells with the p500 vector containing MGMT or the vector alone, respectively (10). TK6-M12 cells contain significant levels of MGMT activity, whereas TK6-P1 cells contain undetectable levels (data not shown).
For drug treatments, logarithmically growing cells were used. MNNG (Sigma), in 0.1 M sodium acetate, pH 5.0, was added directly to the culture medium; because of the short half-life of MNNG, the medium was not replaced. Cells were treated with 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU; from J. Johnson, Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute, Bethesda, MD; BCNU stock solutions were in 100% ethanol and the minimum dilution into medium was 1000-fold) and methyl-lexitropsin (from B. Gold, University of Nebraska Medical Center, Omaha; stock solution in DMSO made immediately before use) in serum-free medium for 1 h, then cultured with serum-containing medium. For IR treatment, cells were irradiated in a 60Co ICN GR-9 GammaRad Irradiator with a dose rate of approximately 10 cGy/sec. Survival of TK6 and MT1 cells (33) and DC3F and A3 cells (34) was determined 10 days after treatment, using colony-formation assays.
Measurement of Apoptosis. For microscopic analysis, TK6, MT1, TK6-E6, and TK6-E6C cells were fixed in methanol/acetic acid (3:1) for 10 min and dropped onto slides. DC3F and A3 cells were fixed with methanol vapor for 10 min and methanol liquid for 20 min. All cells were then stained with 10 μg/ml Hoechst 33258 (Sigma) for 10 min. A Nikon Eclipse E800 fluorescence microscope was used to count the percentage of condensed nuclei characteristic of apoptosis from a minimum of 500 cells per sample, as described previously (35).
For fluorescence-activated cell sorting analysis, cells were fixed in 70% ethanol and stained with 40 μg/ml propidium iodide (Sigma) and 10 μg/ml RNase A (Sigma) for 1 h before analysis on either an Ortho 2150 Cytofluorograf (Cyonics/Uniphase) or a Coulter Elite flow cytometer (36).
Western Blot Analysis.
Total cellular extracts were prepared by incubating cells in lysis buffer (20 mM Tris⋅HCl, pH 8.0/137 mM NaCl/10% glycerol/1% Nonidet P-40/10 mM EDTA/0.5 mM PMSF/1 mM benzamidine) for 30 min. Extract protein (30 μg), measured by using the Bradford assay (ref. 37; Bio-Rad), was separated by denaturing polyacrylamide gel electrophoresis, transferred to nitrocellulose, probed with 1:1000 antibody, and visualized by enhanced chemiluminescence (ECL kit; Amersham). The primary antibodies used were α-p53 (clones DO-1 and Pab240) and α-p21 (clone C-19) (Santa Cruz Biotechnology).
Northern Blot Analysis.
RNA was isolated as described (38), and 20 μg was separated on 1% agarose, transferred to a nylon membrane, and hybridized to a 1.1-kb fragment of the mMsh3 cDNA. pGC1587, containing the mMsh3 cDNA, was kindly provided by G. F. Crouse (Emory University, Atlanta) (39). Equal loading was verified by probing with a fragment of the human β-actin cDNA (CLONTECH).
Methyltransferase Assay.
Cell-free extracts were prepared as previously described (40) and stored at −80°C. Protein concentrations were determined by the Bradford assay. Methyltransferase activity in cell extracts was measured as previously described (41). Briefly, extracts were incubated with Micrococcus luteus DNA that had been methylated with [3H]MNU (18 Ci/mmol or 0.9 Ci/mmol, Amersham; 1 Ci = 37 GBq), for 1 h at 37°C. Proteins were precipitated in 1 M perchloric acid at 70°C for 1 h and resuspended in 10 mM NaOH, and the associated 3H was measured by scintillation counting.
Band-Shift Assays.
Protein extracts were prepared as described (42), and the concentration was determined by the Bradford assay. The oligonucleotide 5′-GCTAGCAAGCTTTCGATTCTAGAAATTCGGC-3′ was 32P-labeled at the 5′ end and annealed (43) to the following oligonucleotides: 5′-GCCGAATTTCTAGAATCGGCTTGCTAGC-3′ (TTT loop), 5′-GCCGAATTTCTAGAATCGAGAGCTTGCTAGC-3′ (GT mismatch), or 5′-GCCGAATTTCTAGAATCGAAAGCTTGCTAGC-3′ (non-mismatched) (17). Oligonucleotides were synthesized by Ransom Hill Bioscience (Ramona). Band-shift and competition assays were performed as described (42).
RESULTS
We previously demonstrated that a signal for the initiation of apoptosis emanates from O6MeG DNA lesions (10). DNA duplexes containing O6MeG can be bound and processed in vitro by MutSα (26, 27), and cells deficient in either MSH2 or MSH6 are MNNG-tolerant, particularly in the absence of MGMT (14). Taken together, these data suggested that MMR participates in generating a signal for apoptosis at the site of O6MeG DNA lesions (10). We therefore set out to determine whether MMR is indeed involved in signaling apoptosis in response to MNNG, and to establish the roles of MutSα and MutSβ in initiating such a signal.
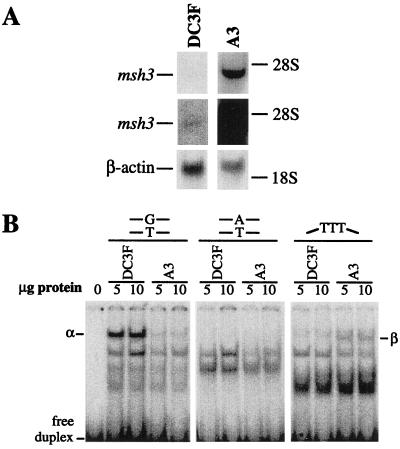
MutSα, but Not MutSβ, Mediates MNNG-Induced Cell Death and Apoptosis. Although MSH2 forms a complex with both MSH6 and MSH3 (to form MutSα and MutSβ, respectively), the ratio of MutSα to MutSβ is normally about 10:1 (24, 44). However, when the MSH3 gene is co-amplified with its neighboring DHFR (dihydrofolate reductase) gene (as it is in several methotrexate-resistant cell lines), the MutSβ complex predominates, and such cells become effectively MutSα-deficient (44, 45). We compared the MNNG sensitivity of two Chinese hamster cell lines that are expected to so differ in their MutSα to MutSβ ratios, namely a MMR-wild-type lung fibroblast cell line, DC3F, and its msh3/dhfr-amplified derivative, DC3F/A3 1.5, clone 1 (hereafter designated A3). A3 contains about 300 copies of msh3/dhfr (46). We confirmed by Northern analysis that msh3 mRNA is indeed overexpressed in A3 cells (Fig. 1A). We also established that MutSα binding activity to a G⋅T mismatch-containing duplex, while present in wild-type DC3F cells, is virtually absent from msh3-amplified A3 cells (Fig. 1B). In contrast, MutSβ binding activity to a duplex containing three unpaired nucleotides was detected only in msh3-amplified A3 cells (Fig. 1B). MutSα or MutSβ binding activity to a perfectly matched duplex was not detected for either cell type. Thus, msh3 amplification resulted in the expected depletion of MutSα, and increase in MutSβ, activity. Note that both DC3F and A3 hamster cell lines have no detectable MGMT activity (data not shown).
Figure 1.
Northern and band-shift analyses of DC3F and A3 Chinese hamster lung fibroblasts. (A) Northern blot of total RNA probed with mMsh3 cDNA probe. The middle panels show an overexposure of the top panels. The positions of the 28S and 18S rRNA bands are shown. The size of the msh3 transcript is in agreement with a previous study (39). The lower panels represent total RNA probed with human β-actin cDNA, to verify equal loading. (B) Band-shift assays in which extract proteins were incubated with labeled DNA oligonucleotide duplexes containing a mismatch (G⋅T), three unpaired nucleotides (TTT), or no mismatch (A⋅T). The reaction mixtures were electrophoresed in 6% polyacrylamide gels; the positions of the DNA duplexes in complex with MutSα (α) and MutSβ (β) are shown, and other unlabeled bands represent nonspecific protein⋅DNA complexes. The specificity of MutSα and MutSβ binding was verified by competition assays using a 40-fold molar excess of unlabeled duplex (data not shown), and results were consistent with a previous study (45).
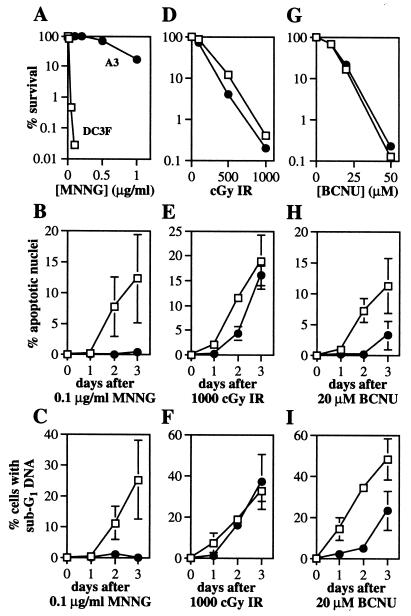
The MutSα-deficient/MutSβ-proficient A3 cells turned out to be remarkably resistant to MNNG-induced cytotoxicity (as measured by colony formation), compared with the parental DC3F cells (Fig. 2A). Moreover, while apoptosis was induced efficiently by MNNG in DC3F cells (as measured by both microscopic and cell sorting analyses), apoptosis was virtually absent in the similarly treated MutSα-deficient A3 cells (Fig. 2 B and C). We infer that MutSα, but not MutSβ, mediates MNNG-induced cell death and apoptosis. The extreme differences in cell killing and apoptotic response to MNNG do not extend to an agent that fails to produce O6MeG DNA lesions, namely IR (Fig. 2 D, E, and F), indicating that A3 and DC3F cells are equally capable of programmed cell death.
Figure 2.
Cell survival (A, D, and G) and apoptosis (B, C, E, F, H, and I) after treatment of DC3F (□) and A3 (●) cells with MNNG (A, B, and C), IR (D, E, and F) and BCNU (G, H, and I). Representative survival curves are shown. Apoptosis was measured by nuclear morphology (B, E, and H) and fluorescence-activated cell sorting analysis (C, F, and I). Each point represents the mean ± SD of at least two independent determinations. Note the different y-axis scales.
MutSα Influences Apoptosis, but Not Cell Killing, Induced by O6-Chloroethylguanine (O6CEG) DNA Lesions. The extreme cytotoxicity of numerous alkylating agents is exploited clinically for cancer chemotherapy, and among these agents the bifunctional chloronitrosoureas are highly favored (47). BCNU, or carmustine, is particularly cytotoxic, in part because BCNU-induced O6CEG lesions undergo a molecular rearrangement to create a replication-blocking DNA interstrand crosslink between the N1 of guanine and the N3 of the opposing cytosine (48). Like methyl groups at the O6 position of guanine, chloroethyl groups at the same position can be repaired by transfer to the MGMT protein, thus preventing formation of the lethal crosslink and averting cell death. We previously demonstrated that MGMT-mediated repair prevents the induction of both cell killing and apoptosis by BCNU (10, 49), and here we test the possibility that O6CEG, like O6MeG, is processed by MutSα to generate the signal for apoptosis.
To our surprise, the influence of MMR on BCNU-induced cell killing versus apoptosis was radically different. The depletion of MutSα had no detectable influence on BCNU cytotoxicity, as measured by colony formation (Fig. 2G). In contrast, MutSα depletion had a marked inhibitory effect on BCNU-induced apoptosis (Fig. 2 H and I). A plausible model to explain this apparent contradiction is presented in Discussion.
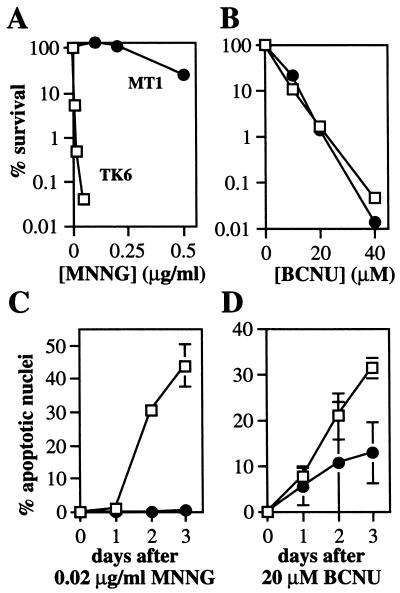
Human MutSα Signals Apoptosis at O6-Alkylguanine Lesions. In a parallel set of experiments, we show that the MutSα complex in human cells also participates in signaling apoptosis in response to O6MeG and O6CEG DNA lesions. Human lymphoblastoid TK6 cells are wild type for MMR, but the TK6 derivative, MT1, is MutSα-deficient due to a mutation in the MSH6 gene (50). Note that neither cell line contains detectable MGMT activity (data not shown). TK6 and MT1 cells were examined for MNNG- and BCNU-induced cell death and apoptosis (Fig. 3). The MutSα deficiency in MT1 cells produced extreme MNNG resistance (as measured by colony-forming ability) but, as with the hamster cells, the MutSα deficiency had no detectable influence on BCNU resistance (Fig. 3A and B). Moreover, MT1 cells were extremely resistant to apoptosis induced by MNNG and, like the MutSα-deficient hamster cells, moderately resistant to apoptosis induced by BCNU (Fig. 3C and D).
Figure 3.
Cell survival (A and B) and apoptosis (C and D) after treatment of human lymphoblastoid TK6 (□) and MT1 (●) cells with MNNG (A and C) and BCNU (B and D), as described in Fig. 2. Apoptosis was quantitated by nuclear morphology.
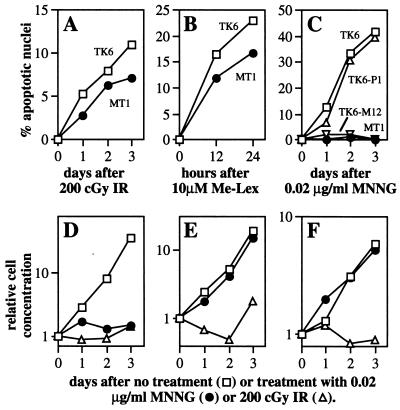
To eliminate the possibility that MT1 cells had simply lost the ability to undergo apoptosis in response to any stimulus, and to be sure that the MutSα-mediated apoptosis observed in these human cells was indeed initiated by O6-alkylguanine, we did the following control experiments. We demonstrated that (i) two agents that do not produce O6-alkylguanine (7, 51) can induce apoptosis in both MT1 and TK6 cells (Fig. 4 A and B); (ii) MGMT-mediated O6-alkylguanine repair eliminates MNNG-induced apoptosis in TK6 cells, indicating that, as well as being MutSα-dependent, such apoptosis is O6-alkylguanine dependent (Fig. 4C); and (iii) cell growth inhibition by IR was independent of MGMT and MutSα status, whereas growth inhibition by MNNG was dependent on both the absence of O6MeG repair and the presence of MutSα (Fig. 4 D–F).
Figure 4.
Apoptosis and cell growth after treatment of human lymphoblastoid cells. Apoptosis was quantitated by nuclear morphology; for A–C, TK6 (□), MT1 (●), TK6 containing control vector, TK6-P1 (▵), TK6 expressing MGMT, TK6-M12 (▿). Me-Lex, methyl-lexitropsin. Cell density was monitored for TK6-P1 (D), TK6-M12 (E), and MT1 (F) after exposure to MNNG and IR.
Role of p53 in MNNG-Induced Apoptosis. Many commonly used Chinese hamster cell lines are defective in p53 function and have been shown to bear a mutation in the p53 DNA-binding domain (52, 53). The DC3F Chinese hamster cells used here displayed defective p53 induction and a diminished G1 arrest in response to IR (data not shown), as would be expected for p53-deficient cells. Moreover, their ability to readily undergo gene amplification implies loss of p53 (54, 55). Therefore, it seems unlikely that the signal for apoptosis initiated by MutSα recognition of O6-alkylguanine DNA lesions is transmitted by the p53 tumor suppressor protein in DC3F hamster cells. However, since TK6 and MT1 human cells express functional p53 protein (56), we set out to examine the possible involvement of p53 in alkylation-induced MMR-dependent apoptosis in human cells.
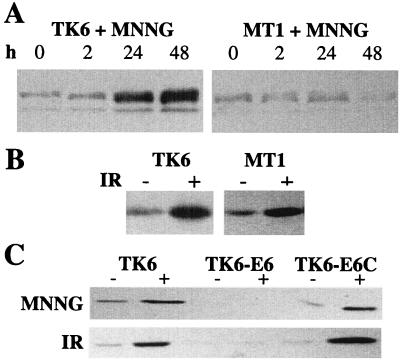
It is well established that exposure of mammalian cells to certain agents that damage DNA results in p53 protein stabilization (probably by means of p53 phosphorylation) (5). Fig. 5A shows that MNNG can induce p53 levels by 10-fold in TK6 cells, but not in MT1 cells. In other words, p53 is induced dramatically by MNNG, but induction depends on a functional MutSα complex. As a control, IR induced p53 equally well in TK6 and MT1 cells (≈5.5-fold), demonstrating that MT1 cells are not simply unresponsive for p53 stabilization (Fig. 5B). Also, IR induced p21 protein levels similarly in TK6 and MT1 cells, suggesting that p53 is functional in both cell lines (data not shown).
Figure 5.
Western blot analysis of p53 expression after 0.02 μg/ml MNNG (A and C) or 1,000 cGy of IR (B). p53 expression was measured 2 or 24 h after IR or MNNG treatment, respectively. Equal amounts of protein (determined by Bradford assay) were loaded onto the gels.
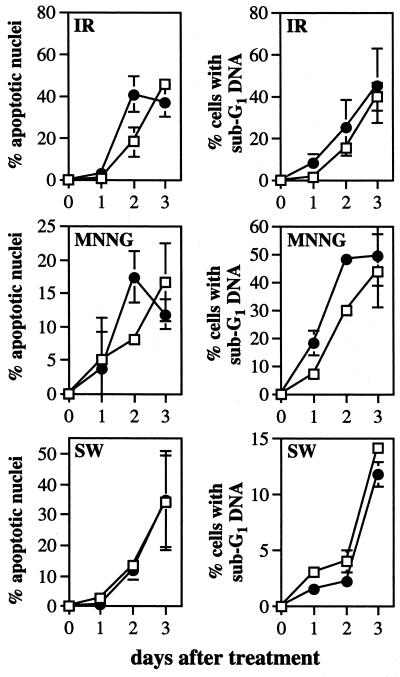
We have established that the induction of apoptosis and p53 by MNNG are each dependent upon functional MutSα in human cells. However, we could not deduce from these results whether the MNNG-induced MutSα-dependent apoptosis was, in fact, p53-dependent. To address this question, we determined whether p53-deficient derivatives of TK6 cells were still susceptible to MNNG-induced apoptosis. A p53 deficiency was achieved by expression of the human papillomavirus 16 E6 gene, whose product targets p53 for rapid degradation (31, 57). As is evident from Fig. 5C, p53 is virtually absent in TK6 cells expressing the E6 gene (TK6-E6), even after MNNG and IR exposure; Fig. 5C also shows that TK6 cells with a control plasmid (TK6-E6C) display normal p53 induction in response to MNNG and IR. Moreover, p21 protein levels were not induced by IR in TK6-E6 cells, demonstrating that p53 function is abrogated in these cells; p21 was induced in TK6-E6C cells (data not shown). It was previously shown that E6 expression delayed, but did not eliminate, the onset of IR-induced apoptosis, indicating that such apoptosis may be only partially p53-dependent; those results are confirmed in Fig. 6 Top). Likewise, we find that MNNG-induced apoptosis may be only partially p53-dependent because E6 expression merely delayed MNNG-induced apoptosis (Fig. 6 Middle). As a control, the induction of apoptosis in response to serum starvation was unaffected by p53 status, as would be expected (Fig. 6 Bottom) (58). Taking the Chinese hamster and human lymphoblastoid cell line results together, we infer that MNNG-induced apoptosis is largely p53-independent.
Figure 6.
Apoptosis in TK6-E6 (□) and TK6-E6C (●) cells after 200 cGy of IR (Top), 0.02 μg/ml MNNG (Middle), and serum withdrawal (Bottom), as described in Fig. 2. Apoptosis was quantitated by nuclear morphology (Left) and cell sorting analysis (Right).
DISCUSSION
The molecular events required for the execution of apoptosis in response to cell surface signals (e.g., tumor necrosis factor α and Fas ligand binding) have been very well characterized (59, 60). In contrast, the molecular events required to initiate apoptosis in response to different types of DNA base damage remain obscure, in part because DNA-damaging agents also damage other cellular components such as RNA, lipids, and proteins. Using sets of isogenic cell lines differing in the repair of a single type of DNA base lesion, we demonstrated that a potent signal for apoptosis emanates from O6-alkyguanine DNA lesions (10, 11). Now we show that for this class of DNA base damage, the first step in the programmed cell death signaling pathway requires the MutSα branch of the DNA MMR pathway, and that such MutSα-dependent signaling of apoptosis is largely p53-independent. These results are consistent with two recent studies showing that MutSα mediates apoptosis induced by the methylating agent temozolomide (61), and that MutSβ is not required for MNNG-induced cytotoxicity (62). However, p53 is required for MNNG-induced apoptosis in the mouse small intestine (63), suggesting that the requirement for p53 likely depends on cell type.
The p53 status of tumor cells has often been taken to indicate whether a tumor will respond well (i.e., be sensitive) to radiation or chemotherapy (64). However, the results presented here demonstrate that for methylating agents, the MMR status of tumor cells may be a more accurate indicator of responsiveness, at least in the absence of adequate O6MeG repair by MGMT. Indeed, the biological response of mammalian cells to O6MeG DNA lesions is likely determined by the balance between MGMT-mediated O6MeG repair and MutSα-mediated signaling of apoptosis. Clearly, if O6MeG lesions are efficiently converted to normal guanine residues by MGMT, O6MeG’s biological consequences are eliminated. However, MGMT levels vary by more than 100-fold between different tissues and between different individuals (40, 65); since MMR proteins are also likely to vary between tissues and individuals, it seems certain that the balance between MGMT and MutSα will be far from constant.
O6MeG lesions that escape MGMT-mediated repair can have one of two distinctly different, mutually exclusive, biological effects, depending upon whether they are processed by the MutSα branch of the MMR pathway. O6MeGs that escape MutSα recognition frequently direct the misincorporation of thymine during replication; should the resulting O6MeG⋅T mispairs also escape MutSα recognition they will produce G⋅C to A⋅T transition mutations in daughter cells (14, 66). Only a small fraction of such mutational events will be lethal, and the vast majority of mutant cells will persist in the population; indeed, some of the mutant cells may acquire a growth advantage. In contrast, MutSα recognition of O6MeG elicits a signal for apoptotic cell death, and such cells vanish from the population. Methylating agents not only are used in the clinic for chemotherapy (e.g., temozolomide, streptozotocin, procarbazine, and dacarbazine) but also are present in our environment, in tobacco smoke, in our diet, and in cells as natural metabolites. Thus, the relative balance between MGMT and MMR, as well as determining the responsiveness of tumor cells to chemotherapy, may affect spontaneous mutation rates and relative sensitivities to environmental alkylating agents between different tissues and different individuals. An extreme example of such an imbalance would occur in MMR-deficient individuals with hereditary nonpolyposis colon cancer; in these individuals, tissues with high MGMT levels would not be at risk, but tissues with low MGMT levels would be highly vulnerable to the accumulation of cells harboring G⋅C to A⋅T transition mutations.
How the balance between MGMT and MMR affects the biological response to O6CEG lesions (induced by chloronitrosoureas such as BCNU) is more complex. The difference stems in part from the fact that O6CEGs that escape both MGMT-mediated repair and MutSα recognition, inevitably undergo a molecular rearrangement to form highly cytotoxic DNA interstrand crosslinks (48). Thus, while our evidence suggests that MutSα-recognition of O6CEG lesions does indeed signal apoptotic cell death, O6CEG lesions are destined to cause cell death even in the absence of MutSα recognition. Our evidence further suggests that DNA interstrand crosslinks induce cell death by necrosis rather than apoptosis. Since MGMT-mediated repair removes an O6CEG lesion that can signal cell death by apoptosis (by means of MutSα), or removes an O6CEG lesion that could form a crosslink to kill cells by necrosis, MGMT status is probably more important than MMR status as an overall indicator of chloronitrosourea sensitivity. However, while MMR status does not seem to influence the ultimate cytotoxicity of O6CEG lesions, it does influence the mechanism by which cells die. Knowing whether an agent will kill cells by necrosis or apoptosis can be important for chemotherapy regimen choices because necrotic cells elicit a strong and undesirable inflammatory response, whereas apoptotic cells do not (67).
The precise mechanism by which MutSα recognition of O6-alkylguanine in the genome signals apoptosis remains to be determined. Whether O6-alkylguanine recognition and signal transduction occurs before or after replication of the lesion (with O6-alkylguanine paired to C or T, respectively) is unclear, and whether excision repair in the vicinity of the lesion is actually required for signaling the initiation of programmed cell death is also unclear. The most commonly cited model for MMR-mediated cell death at O6MeG lesions invokes futile cycling of MMR excision events opposite the lesion to create persistent DNA strand breaks and gaps (14, 29, 68, 69). However, little formal proof for this model is available. Such putative breaks and gaps may indeed signal apoptosis in response to O6-alkylguanines, but our evidence suggests that signaling need not involve p53, the classic signal transduction molecule thought to respond to DNA breaks and gaps (70, 71).
MMR status has been reported to influence the biological response of cells to numerous agents, including several methylating agents, the platinum-containing anticancer drugs cisplatin and carboplatin, the antimetabolite 6-thioguanine, and the topoisomerase II inhibitors etoposide and doxorubicin (72). Whether MutSα versus MutSβ recognition was involved was not always established, and it remains possible that MutSβ can recognize certain DNA lesions. Thus, the model system described here, to examine the molecular events initiated by MutSα recognition (exclusively at O6-alkylguanine DNA lesions), may model the signaling events that occur at a wide variety of different DNA lesion types. It should now be possible to dissect the molecular signaling events involved specifically in DNA-damage-induced apoptosis (as opposed to apoptosis induced by damage at the cell membrane or other cellular locations) because these events should be apparent in MGMT-deficient/MutSα-proficient cells, and absent from both MGMT-proficient and MutSα-deficient cells.
Acknowledgments
We are grateful to Drs. Joyce L. Hamlin, John B. Little, and William G. Thilly for sharing their cell lines, to Dr. Barry Gold for providing methyl-lexitropsin and to Amy Imrich (Harvard School of Public Health) for performing fluorescence-activated cell sorting analysis, which was supported by National Institute on Environmental Health Sciences Center Grant P30 ES00002–36. This work was supported by grants from the National Cancer Institute (R01-CA55042) and the National Institute on Environmental Health Sciences (P01-ES03926). M.J.H. was supported by the Training Program in Environmental Health Sciences T32 ES07155–15. L.D.S. is a Burroughs Wellcome Toxicology Scholar.
ABBREVIATIONS
- O6MeG
O6-methylguanine
- O6CEG
O6-chloroethylguanine
- MGMT
O6MeG DNA methyltransferase
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- BCNU
1,3-bis(2-chloroethyl)-1-nitrosourea
- IR
ionizing radiation
- MMR
mismatch repair
References
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 3.Dou Q P, An B, Will P L. Proc Natl Acad Sci USA. 1995;92:9019–9023. doi: 10.1073/pnas.92.20.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheikh M S, Burns T F, Huang Y, Wu G S, Amundson S, Brooks K S, Fornace A J, Jr, el-Deiry W S. Cancer Res. 1998;58:1593–1598. [PubMed] [Google Scholar]
- 5.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 6.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 7.Ward J F. Prog Nucleic Acids Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 8.Goodhead D T. Int J Rad Biol. 1989;56:623–634. doi: 10.1080/09553008914551841. [DOI] [PubMed] [Google Scholar]
- 9.Devary Y, Rosette C, DiDonato J A, Karin M. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- 10.Meikrantz W, Bergom M A, Memisoglu A, Samson L. Carcinogenesis. 1998;19:369–372. doi: 10.1093/carcin/19.2.369. [DOI] [PubMed] [Google Scholar]
- 11.Tominaga Y, Tsuzuki T, Shiraishi A, Kawate H, Sekiguchi M. Carcinogenesis. 1997;18:889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- 12.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. New York: Plenum; 1983. pp. 55–78. [Google Scholar]
- 13.Zak P, Kleibl K, Laval F. J Biol Chem. 1994;269:730–733. [PubMed] [Google Scholar]
- 14.Karran P, Hampson R. Cancer Surveys. 1996;28:69–85. [PubMed] [Google Scholar]
- 15.Marra G, Boland C R. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 16.Modrich P. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 17.Drummond J T, Li G M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 18.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 19.Habraken Y, Sung P, Prakash L, Prakash S. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 20.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 21.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risinger J I, Umar A, Boyd J, Berchuck A, Kunkel T A, Barrett J C. Nat Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 23.Umar A, Risinger J I, Glaab W E, Tindall K R, Barrett J C, Kunkel T A. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genschel J, Littman S J, Drummond J T, Modrich P. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 25.Umar A, Koi M, Risinger J I, Glaab W E, Tindall K R, Kolodner R D, Boland C R, Barrett J C, Kunkel T A. Cancer Res. 1997;57:3949–3955. [PubMed] [Google Scholar]
- 26.Duckett D R, Drummond J T, Murchie A I, Reardon J T, Sancar A, Lilley D M, Modrich P. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceccotti S, Aquilina G, Macpherson P, Yamada M, Karran P, Bignami M. Curr Biol. 1996;6:1528–1531. doi: 10.1016/s0960-9822(96)00758-0. [DOI] [PubMed] [Google Scholar]
- 28.Skopek T R, Liber H L, Penman B W, Thilly W G. Biochem Biophys Res Commun. 1978;84:411–416. doi: 10.1016/0006-291x(78)90185-7. [DOI] [PubMed] [Google Scholar]
- 29.Goldmacher V S, Cuzick R A, Jr, Thilly W G. J Biol Chem. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 30.Kat A, Thilly W G, Fang W H, Longley M J, Li G M, Modrich P. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, Li C Y, Little J B. Oncogene. 1997;14:1661–1667. doi: 10.1038/sj.onc.1201026. [DOI] [PubMed] [Google Scholar]
- 32.Leu T H, Hamlin J L. Mol Cell Biol. 1992;12:2804–2812. doi: 10.1128/mcb.12.6.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furth E E, Thilly W G, Penman B W, Liber H L, Rand W M. Anal Biochem. 1981;110:1–8. doi: 10.1016/0003-2697(81)90103-2. [DOI] [PubMed] [Google Scholar]
- 34.Risinger J I, Umar A, Barrett J C, Kunkel T A. J Biol Chem. 1995;270:18183–18186. doi: 10.1074/jbc.270.31.18183. [DOI] [PubMed] [Google Scholar]
- 35.Meikrantz W, Gisselbrecht S, Tam S W, Schlegel R. Proc Natl Acad Sci USA. 1994;91:3754–3758. doi: 10.1073/pnas.91.9.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicoletti I, Migliorati G, Pagliacci M C, Grignani F, Riccardi C. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 37.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 39.Wells J, Held P, Illenye S, Heintz N H. Mol Cell Biol. 1996;16:634–647. doi: 10.1128/mcb.16.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moritz T, Mackay W, Glassner B J, Williams D A, Samson L. Cancer Res. 1995;55:2608–2614. [PubMed] [Google Scholar]
- 41.Sassanfar M, Dosanjh M K, Essigmann J M, Samson L. J Biol Chem. 1991;266:2767–2771. [PubMed] [Google Scholar]
- 42.Jiricny J, Hughes M, Corman N, Rudkin B B. Proc Natl Acad Sci USA. 1988;85:8860–8864. doi: 10.1073/pnas.85.23.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiricny J, Wood S G, Martin D, Ubasawa A. Nucleic Acids Res. 1986;14:6579–6590. doi: 10.1093/nar/14.16.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond J T, Genschel J, Wolf E, Modrich P. Proc Natl Acad Sci USA. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Proc Natl Acad Sci USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma C, Leu T H, Hamlin J L. Mol Cell Biol. 1990;10:1338–1346. doi: 10.1128/mcb.10.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ludlum D B. Cancer Invest. 1997;15:588–598. doi: 10.3109/07357909709047601. [DOI] [PubMed] [Google Scholar]
- 48.Ludlum D B. Mutat Res. 1990;233:117–126. doi: 10.1016/0027-5107(90)90156-x. [DOI] [PubMed] [Google Scholar]
- 49.Maze R, Carney J P, Kelley M R, Glassner B J, Williams D A, Samson L. Proc Natl Acad Sci USA. 1996;93:206–210. doi: 10.1073/pnas.93.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papadopoulos N, Nicolaides N C, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K, Kinzler K W, et al. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 51.Engelward B P, Allan J M, Dreslin A J, Kelly J D, Wu M M, Gold B, Samson L D. J Biol Chem. 1998;273:5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 52.Lee H, Larner J M, Hamlin J L. Gene. 1997;184:177–183. doi: 10.1016/s0378-1119(96)00592-6. [DOI] [PubMed] [Google Scholar]
- 53.Hu T, Miller C M, Ridder G M, Aardema M J. Mutat Res. 1999;426:51–62. doi: 10.1016/s0027-5107(99)00077-9. [DOI] [PubMed] [Google Scholar]
- 54.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 55.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 56.Xia F, Wang X, Wang Y H, Tsang N M, Yandell D W, Kelsey K T, Liber H L. Cancer Res. 1995;55:12–15. [PubMed] [Google Scholar]
- 57.Crook T, Tidy J A, Vousden K H. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 58.Vaux D L, Haecker G, Strasser A. Cell. 1994;76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 59.Muzio M. Int J Clin Lab Res. 1998;28:141–147. doi: 10.1007/s005990050035. [DOI] [PubMed] [Google Scholar]
- 60.Singh A, Ni J, Aggarwal B B. J Interferon Cytokine Res. 1998;18:439–450. doi: 10.1089/jir.1998.18.439. [DOI] [PubMed] [Google Scholar]
- 61.D’Atri S, Tentori L, Lacal P M, Graziani G, Pagani E, Benincasa E, Zambruno G, Bonmassar E, Jiricny J. Mol Pharmacol. 1998;54:334–341. doi: 10.1124/mol.54.2.334. [DOI] [PubMed] [Google Scholar]
- 62.Hinz J M, Meuth M. Carcinogenesis. 1999;20:215–220. doi: 10.1093/carcin/20.2.215. [DOI] [PubMed] [Google Scholar]
- 63.Toft N J, Winton D J, Kelly J, Howard L A, Dekker M, te Riele H, Arends M J, Wyllie A H, Margison G P, Clarke A R. Proc Natl Acad Sci USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weller M. Cell Tissue Res. 1998;292:435–445. doi: 10.1007/s004410051072. [DOI] [PubMed] [Google Scholar]
- 65.Gerson S L, Trey J E, Miller K, Berger N A. Carcinogenesis. 1986;7:745–749. doi: 10.1093/carcin/7.5.745. [DOI] [PubMed] [Google Scholar]
- 66.Singh J, Su L, Snow E T. J Biol Chem. 1996;271:28391–28398. doi: 10.1074/jbc.271.45.28391. [DOI] [PubMed] [Google Scholar]
- 67.Kerr J F, Winterford C M, Harmon B V. Cancer. 1994;73:2013–2026. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 68.Kaina B, Ziouta A, Ochs K, Coquerelle T. Mutat Res. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- 69.Galloway S M, Greenwood S K, Hill R B, Bradt C I, Bean C L. Mutat Res. 1995;346:231–245. doi: 10.1016/0165-7992(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 70.Huang L C, Clarkin K C, Wahl G M. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson W G, Kastan M B. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fink D, Aebi S, Howell S B. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]