Abstract
The neuronal circuits of the cerebellar cortex are essential for motor and sensory learning, associative memory formation, and the vestibular ocular reflex. In children and young adults, tumors of the granule cell, the medulloblastomas, represent 40% of brain tumors. We report the differentiation of E14 ES cells into mature granule neurons by sequential treatment with secreted factors (WNT1, FGF8, and RA) that initiate patterning in the cerebellar region of the neural tube, bone morphogenic proteins (BMP6/7 and GDF7) that induce early granule cell progenitor markers (MATH1, MEIS1, ZIC1), mitogens (SHH, JAG1) that control proliferation and induce additional granule cell markers (Cyclin D2, PAX2/6), and culture in glial-conditioned medium to induce markers of mature granule neurons (GABAα6r), including ZIC2, a unique marker for granule neurons. Differentiated ES cells formed classic “T-shaped” granule cell axons in vitro, and implantation of differentiated Pde1c-Egfp-BAC transgenic ES cells into the external granule cell layer of neonatal mice resulted in the extension of parallel fibers, migration across the molecular layer, incorporation into the internal granule cell layer, and extension of short dendrites, typical of young granule cells forming synaptic connections with afferent mossy fibers. These results underscore the utility of treating ES cells with local, inductive signals that regulate CNS neuronal development in vivo as a strategy for cell replacement therapy of defined neuronal populations.
Keywords: cerebellum, CNS development, granule neuron, stem cell
The cerebellar cortex is a remarkably simple laminar structure, with two principal neurons, the granule cell and the Purkinje cell, and a diverse set of interneurons, which modulate the output of the Purkinje cell to the cerebellar nuclei (1). The cerebellar circuitry coordinates movement and balance and functions in sensory discrimination (2) and cognitive processing (3). Long-term depression of parallel fiber synapses onto Purkinje has been assumed to control the simple vestibular ocular reflex. Recent studies on both simple functions and adaptive control of the cerebellum suggest that multiple plasticity mechanisms may contribute to cerebellum-dependent learning. Multiple plasticity mechanisms are probably important to encode memories over different time scales, to regulate the dynamics of movement, and to allow bidirectional changes in the amplitude of movements (4). Although the role of the cerebellum in learning and memory is becoming more complex, the remarkably simple architectonics of the cerebellum make it an attractive model system for developing cell replacement strategies.
Replacement therapy in the cerebellum, like that in other brain regions, depends on the ability to induce progenitor cells to differentiate into cells with specific cell fates. The pluripotent nature of mouse ES cells was formally demonstrated by their ability to contribute to all tissues of adult mice, including the germ line, after their injection into host blastocysts (5). In addition to their developmental potential in vivo, ES cells display a remarkable capacity to form differentiated cell types in culture. Recently, Sato et al. (6) demonstrated that activation of the canonical Wnt pathway could replace the requirement of mouse embryonic fibroblast-conditioned media in the maintenance of undifferentiated hES cells for short periods of time (5–7 days). In addition, Wichterle et al. (7) demonstrated that treating ES cells with the series of signals that induce specific cell populations during normal, in vivo development induces mouse ES cells to differentiate into spinal progenitor cells and subsequently into motor neurons. In the present study we have tested the signals that induce formation of the cerebellar territory (8–10) from rhombomere 1 (11), signals that dorsalize the neural tube specify (12) granule neurons (13), and mitogens that expand the pool of granule cell progenitors (GCPs) in the neonatal cerebellar cortex (14, 15). To monitor ES cell differentiation we measured expression of genes essential for granule neuron development, unique markers for GCPs and differentiated granule neurons, and markers for differentiating granule neurons. We also monitored the expression-specific markers for cerebellar Purkinje neurons (16) and markers for cerebellar astroglia.
During cerebellar development, granule neuron progenitors arise from the boundary of the mesencephalon and metencephalon in an area known as the rhombic lip. Recent studies reveal that the anterior rhombic lip, once thought to exclusively generate granule neurons of the cerebellar cortex (17), generates precursors of the cerebellar and precerebellar nuclei, which project afferent fibers to the cerebellar cortex or receive efferent fibers from the cerebellum. Progenitors of the Purkinje neuron arise from the ventricular zone of the cerebellar territory and express specific transcription factors (20).
Over the past 5 years, the Gene Expression Neuronal Database (GENSAT) Project has used Egfp-BAC transgenic mice to define patterns of CNS gene expression in the developing and adult mouse brain (28). The GENSAT Project has generated hundreds of Egfp-BAC transgenic lines and identified markers for cerebellar granule cells, including Pde1c for granule cells. Pde1c is expressed by GCPs very early in the program of development, commencing at E10, when the earliest GCP markers, including Math1 (18), Zic1,2 (19), and Meis1 (20), are expressed. Expression continues throughout the lifetime of the animal, making Pde1c a granule neuron marker that is expressed at all stages of life. To provide markers for ES differentiation into granule cells, we generated ES cells from Pde1c-Egfp-BAC transgenic mice. To test whether the local signals and transcription factors that establish the cerebellar primordium and specify neural fates in vivo can be harnessed in vitro to direct the differentiation of mouse ES cells into granule neurons, Purkinje cells, and cerebellar glial cells, we studied the influence of cerebellar “organizer molecules,” dorsal signals and proteins that expand the GCP cell population on ES cell differentiation.
Results
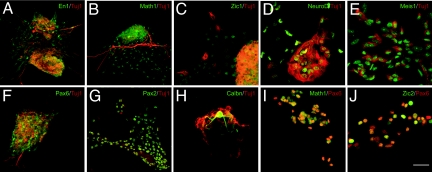
As discussed, inductive signals and transcription factors involved in cerebellar neuron generation have been identified, raising the question of whether these developmental insights can be used to direct stem cells to a cerebellar fate. We show that developmentally relevant signaling factors can induce mouse ES cells to differentiate into midbrain/hindbrain progenitor cells, and subsequently into cerebellar neurons, through a pathway recapitulating that used in vivo. To examine the capacity of ES cells to generate cerebella cells, we cultured mouse ES cells for 2 days to form EBs (1,000 cells) and maintained the EB suspension culture (1–7 days) in medium supplemented with FGF8b and retinoic acid (RA). We observed expression of the neural marker Nestin, the cerebellar marker EN1, PAX2, and NEUROD, but not the GCP markers MATH1 and ZIC1 [supporting information (SI) Fig. 7]. Thus, FGF8b and RA directed ES cells to a midbrain/hindbrain fate and induced expression of the general interneuron marker PAX2.
As the neural tube closes, the expression of Hoxa2 delineates rhombomere 1, which gives rise to the cerebellum (11). The addition of FGF8b and RA to culture medium of EBs induces expression of Otx1, Hoxa2, and Gbx2, suggesting that these two factors are sufficient to induce expression of genes that establish the cerebellar anlagen. Subsequently, soluble factors produced by the roof plate induce dorsal fates (21–23). To examine the influence of dorsalizing factors on ES cells, we treated ES cells with FGFs (FGF4, FGF8B, and bFGF) for 2 days, after which we added FGF8, WNT1, and WNT3a for 3 days. By immunocytochemistry, EBs treated with FGF8, WNT1, and WNT3a contained cells that expressed MATH1 and ZIC1,2, specific markers of cerebellar GCPs (Fig. 1). Thus, treatment of E14 or D3 ES cells with the series of factors that induce the formation of the cerebellar territory directs the differentiation of ES cells toward cerebellar neuronal fates.
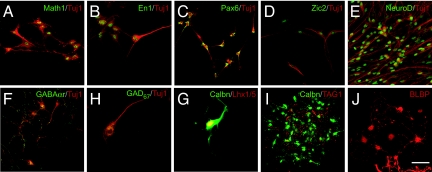
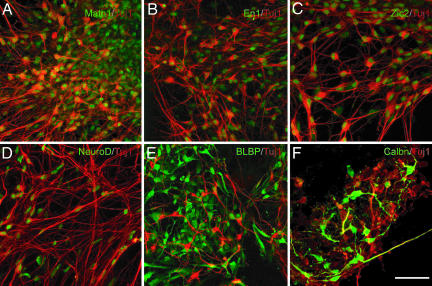
Fig. 1.
Differentiation of ES cells into proliferating EGL cells, Purkinje neurons, and interneurons. (A–D) After 5 days (12 DIV) of differentiation with FGF8b, WNT1 and WNT3a ES cells expressed a marker of the cerebellar territory (green), EN1 (A), and of proliferating EGL cells MATH1 (B), ZIC1 (C), and NEUROD (D). (E–H) After 4 days (16 DIV) of culture with FGF8b, WNT1, WNT3A, and BMP7, GDF7 and BMP6 ES cells expressed the granule neuron markers MEIS1 (E), PAX6 (F), and PAX2 (G) and the Purkinje cell marker CALB1 (H). Differentiated ES cells also expressed neuronal marker class III β-tubulin (TUJ1; A–H) (red). (I and J) ES cells coexpressed PAX6 (red), MATH1, and ZIC2 (green). (Scale bar: 40 μm for A, B, F, and G and 20 μm for C–E and H–J.)
Because prior experiments demonstrated that BMP7, BMP6, and GDF7 are required for the specification of cerebellar granule neurons within the mes/met territory (13), we cultured ES cells in medium supplemented with BMP7, BMP6, and GDF7 for 4 days. Immunocytochemical assays revealed expression of the GCP markers MATH1, MEIS1, PAX6 (24), and PAX2 (25). Although the majority of ES cells expressed GCP markers, a small number of cells expressed CALB1, a marker for cerebellar Purkinje neurons (SI Fig. 8) Double labeling experiments (J) showed that the ES cells expressed multiple markers of GCPs. RT-PCR analysis confirmed these results and demonstrated that the cells did not express markers of spinal cord neurons (26). Thus, treatment of ES cells with dorsalizing signals and with the specific combination of BMPs shown previously to induce GCP fates in vivo appeared to recapitulate the developmental program that generates cerebellar neurons, especially the cerebellar granule cell.
In the developing cerebellar cortex, sonic hedgehog (SHH) (15) and JAG1, the ligand for NOTCH2 (14), induce the remarkable expansion of GCPs. The addition of SHH and JAG1 promoted the rapid proliferation of ES cells and appeared to induce expression of Cyclin D2, which is expressed in neonatal GCPs in the external granule cell layer (EGL) as well as continued expression of specific GCP markers, including MATH1 and PDE1c (SI Fig. 9).
Finally, we examined the role of BDNF, a neurotrophin that promotes GCP differentiation (27). Treatment of proliferating ES cells that expressed GCP markers with BDNF induced expression of the postmitotic GCP markers NEUROD (Fig. 2), the axonal glycoprotein TAG1 (Fig. 3), one of the earliest markers of granule neuron axon extension and the signature protein of the granule cell parallel fiber, and markers of terminal stages of granule cell development. RT-PCR analysis of gene expression in differentiated ES cells confirmed these results (SI Fig. 7 C). Although the vast majority of ES cells expressed GCP markers, some cells expressed Purkinje cell markers (Fig. 3 G–I) and the Bergman cell marker BLBP (Fig. 2J). We did not optimize conditions for the differentiation of ES cells into Purkinje neurons or Bergmann glial cells in the current study.
Fig. 2.
Differentiation of ES cells into cerebellar granule cells. After 16 DIV, ES cells were cultured in N2 Supplement-B and B27 (StemCell Technologies) as well as FGF8b, WNT1, WNT3a, BMP7, GDF7, BMP6, SHH, BDNF, and NT3. The latter factors induced expression of markers of dorsal neurons (green) MATH1 (A), the cerebellar territory EN1 (B), and granule neurons PAX6 (C), ZIC2 (D), NEUROD (E), and GABAα6r (F) as well as markers for Purkinje cell GAD67 (G), LHX1/5 (red, H), and CALB1 (green, H and I) and Bergman glia BLBP (red, J). The postmitotic neuronal markers (red) TUJ1 (A–F) and TAG1 (I) are also shown. (Scale bar: 20 μm for A–H and J and 40 μm for I.)
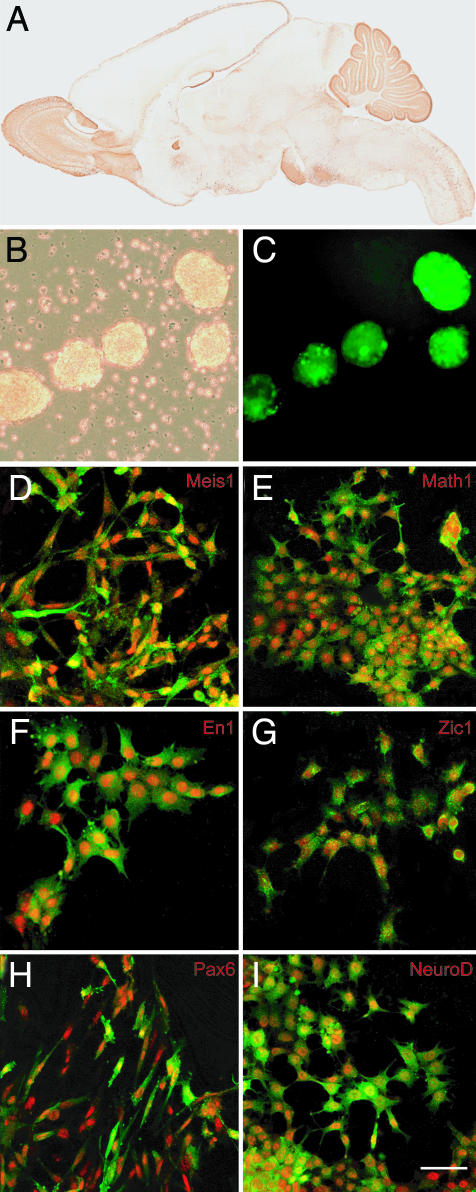
Fig. 3.
Generation of Pde1c-Egfp-BAC ES cells. (A) Sagittal section of P7 Pde1c-Egfp-BAC transgenic mice shows the expression of Pde1c gene in the cerebellar granule cells, cortical Cajal Retzius cells, olfactory bulb, and pontine nucleus. ES cells were derived from F1 Pde1c-BAC 129SvJ heterozygous blastocysts. (B) Bright-field and fluorescence images showing expression of the EGFP marker of Pde1c-Egfp EBs cultured in FGF8b/RA after 4 days (16 DIV). (C) Culturing the ES cells in medium supplemented with FGF8b, WNT1, WNT3a, BMP7, and GDF7 for 4 days (8 DIV) induced expression of the granule neuron markers MEIS1 (red, D) and MATH1 (E), the cerebellar territory marker EN1 (F), and cerebellar granule neuron markers ZIC1 (G), PAX6 (H), and NEUROD (I). (Scale bar: 25 μm for B–I.)
To provide a genetic system where we could monitor expression of a GCP-specific gene at each of the developmental stages we induced in E14 and D3 ES cells, we generated ES cells from the Pde1c-Egfp-BAC transgenic mice (GENSAT Project) (28). In cells of Pde1c-Egfp-BAC transgenic mice, the regulatory elements of the Pde1c gene control expression of the Egfp reporter gene. Moreover, studies on cells of Pde1c-Egfp-BAC transgenic mice show that the expression of EGFP is sufficient to visualize cells that express Pde1c without antibody staining. We therefore generated ES cells from Pde1c-Egfp-BAC transgenic mice (Fig. 3 A–C). ES cells transmitted the Pde1c-Egfp gene through the germ line and, when cultured in the sequence of inducing factors described above, recapitulated the key steps of GCP differentiation. These steps included expression of EN1, which delineates the midbrain/hindbrain territory from which the cerebellum will develop, markers that characterize GCPs, proliferating GCPs, and differentiated granule neurons. Expression of the Egfp reporter gene allowed us to examine the influence of coculturing ES cells with GCPs or purified glial cells and to implant the cells into the native cerebellar cortex, where we could monitor whether they would develop in the complex 3D setting of the brain.
Although ES cells survived in coculture with native cells, many of the wild-type GCP cells aggregated and died when plated at equivalent densities with ES cells. We therefore tested a ranging of cell ratios (differentiated ES cells:native cerebellar cells) and measured plating efficiencies to define the optimal coculture conditions. Plating differentiated ES cells with wild-type GCPs at ratios between 1:103 and 1:104 increased the plating efficiency of both cell types to 90–100%. Three general types of cultures were assayed: ES cells cocultured with the neonatal wild-type GCPs, wild-type neonatal glial cells, or medium conditioned by one of the latter cell types (Fig. 4). After 8 days, Pde1c-Egfp-BAC ES cells cocultured with purified granule cells or astroglial cells expressed GCP markers induced in E14 or D3 ES cells. In contrast, Pde1c-Egfp-BAC ES cells cocultured with nonneuronal feeder cells did not express markers of differentiated cerebellar GCPs. Although differentiated ES cells thrived in cocultures with cerebellar GCPs or glial cells, a larger proportion of ES cells differentiated in coculture with cerebellar astroglial cells. Because coculture experiments raise the possibility that differentiation occurred by the fusion of ES cells with wild-type cells, we examined the influence of medium conditioned by GCPs or purified glial cells. Identical results were obtained with conditioned medium, suggesting that cerebellar GCPs and especially cerebellar glial cells release factors that promote GCP differentiation (SI Fig. 9). The most striking result of culturing Pde1c-Egfp-BAC ES cells in glial-conditioned medium was formation of the signature “T-shaped” axons of the cerebellar granule neurons (Fig. 5). This suggests that Pde1c-Egfp-BAC ES cells expressed genes and signaling pathways required for the development of the polarity of wild-type cerebellar granule neurons (29).
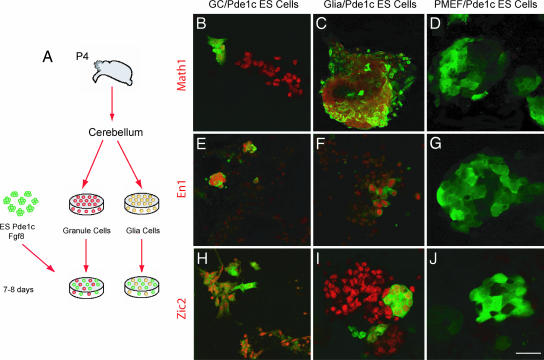
Fig. 4.
Coculture of Pde1c ES cells with medium conditioned by native cerebellar neurons promotes differentiation. (A) Pde1c-Egfp-BAC ES cells were resuspended in serum-free media supplemented with FGF8b for 7 days and cultured in medium conditioned by granule cells (B, E, and H), astroglial cells (C, F, and I), or Pde1c (D, G, and J). ES cells expressed cerebellar granule cells markers (red) MATH1 (B and C), EN1 (E and F), and ZIC2 (H and I). By contrast, ES cells cultured on nonneuronal cells did not express any of the latter markers. (Scale bar: 30 μm for B, C, E, F, H, and I and 20 μm for D, G, and J.)
Fig. 5.
Medium conditioned by native cerebellar cells induces terminal differentiation of markers for cerebellar granule neurons, Purkinje cells, and astroglia. ES cells differentiated into cerebellar neurons in medium supplemented with FGF8 and medium conditioned by purified cerebellar granule neurons or astroglia at two concentrations (0.25 mg/ml or 0.5 mg/ml) (see Materials and Methods). ES cells cultured in the low concentration of granule cell-conditioned medium expressed MATH1 (green, A). (B–E) At the high concentration of granule neuron-conditioned medium, ES cells expressed EN1 (B), a specific marker for the cerebellar territory cells. ES cells cultured in the low concentration of glial cell-conditioned medium expressed the cerebellar specific marker ZIC2 (C), the granule cell marker NEUROD (D), and the glial cell marker BLBP (E). (F) ES cells cultured in the higher concentration of glial-conditioned medium expressed the Purkinje cell marker. The neuronal marker class III β-tubulin (TUJ1) was expressed under all culture conditions (red). (Scale bar: 20 μm.)
To examine whether differentiated Pde1c-Egfp ES cells would integrate into the native cerebellar cortex, migrate along the Bergmann glial fibers into the internal granule cell layer (IGL), and establish connections with mossy fiber afferent axons, we implanted the cells into postnatal day 4 (P4) to P6 cerebellar cortex. As a control, we isolated EGFP+ granule cells from P4 Pde1c-Egfp BAC mice, implanted them into the EGL of P4–P6 neonatal mice, and maintained the animals for 2–9 days before killing them and sectioning to locate EGFP-labeled cells. One to 3 days after implantation, Pde1c-Egfp BAC granule cells migrated from the EGL into the molecular layer (ML) (SI Fig. 10 C and E). These studies suggested that a 3- to 14-day incubation period is suitable for assaying the differentiation of implanted ES cells. Before implantation, ES cells were differentiated and injected (1–2 × 103 cells) into the EGL of neonatal mice (P4–P6). Three days to 2 weeks after implantation Pde1c-Egfp-BAC ES cells migrated across the ML, past the Purkinje cell layer, and into the IGL of the neonatal cerebellum (Fig. 6). Consistent with our previous studies of cerebellar transplantation of embryonic EGL cells (13), numerous EGFP+ ES cells appeared to migrate across the ML on the glial scaffold, transit the Purkinje cell layer, and arrive in the IGL, where native granule cells elaborate dendrites and interact with mossy fibers (Fig. 3 C–F and SI Fig. 10 A and B) and mature granule cells. Implanted EGFP+ neurons in the IGL (9 days after implantation) expressed ZIC2, a specific marker for terminally differentiated granule cells (GABAa6 is expressed in interneurons in other brain regions) (SI Fig. 10 D and F). These findings provide evidence that ES cells can differentiate into cerebellar GCPs that both migrate across the ML and establish connections with mossy fibers via the extension of dendritic arbors in the IGL.
Fig. 6.
Implantation of differentiated ES cells into neonatal cerebellum. (A) Differentiated Pde1c-Egfp-BAC ES cells were implanted into the EGL of P6 mice as described. Three to 9 days after implantation, ES cells expressing the EGFP reporter are present in the EGL, where GCPs are proliferating (top arrow), migrating across the ML along Bergmann glia (middle arrow) and in the IGL, where terminally differentiated granule neurons synapse with mossy fiber afferent axons via the elaboration of dendrites (lower arrow). The polarity and morphological features of differentiated Pde1c-Egfp-BAC ES cells closely resembled those of native granule neurons. (B) Detailed view of postmigratory EGFP+ ES cell extending dendrites (arrow) in the IGL. For orientation, the Purkinje cell layer is labeled with CALB1. (Scale bar: 50 μm for A and 10 μm for B.)
Discussion
Using current principles of cerebellar development, we differentiated E14 ES cells, as well as ES cell lines we derived from Pde1c-Egfp-BAC transgenic mice, to express specific markers of cerebellar granule neurons. Our approach involved treatment of ES cells with “cerebellar organizers” (FGF8 and RA), followed by the addition of local signals that “dorsalize” the cerebellar anlagen (WNTS, BMP7, GDF7, and BMP6), mitogens that expand the cerebellar GCP population (SHH and Jag1), and medium conditioned by cerebellar glial cells. Differentiated ES cells expressed both markers of dorsal neurons (MATH1 and ZIC1) and markers specific for cerebellar granule cell markers (EN1, ZIC2, PAX6, and GABAα6r). In addition, cultured ES cells differentiated into neurons that expressed granule cell markers and the signature T-shaped polarity of cerebellar granule neurons. Upon implantation into neonatal cerebellar cortex, a small number of differentiated ES cells migrated into host neonatal cerebellar cortex to positions normally occupied by mature granule neurons and elaborated dendrites typical of mature cerebellar granule cells forming synaptic contacts with mossy fiber afferents (30). Thus, the protocol developed in the current study supported expression of neurons in the cerebellar territory with molecular and cellular characteristics that are unique to the cerebellar granule neuron.
Our findings demonstrate that ES cells can be differentiated into cells from a particular region of the CNS with high efficiency by a pathway that recapitulates the differentiation of cerebellar neurons in vivo. This study confirms the experimental approach developed by Wichterle et al. (7) and suggests that this experimental strategy can be used systematically to direct the differentiation of specific classes of CNS neurons from ES cells. Our results also underscore the importance of characterizing the normal program of development of various types of CNS neurons. It is especially advantageous to use ES cells derived from mice expressing EGFP in specific CNS cells to test programs of differentiation of various classes of neurons in the brain. The GENSAT Project provides specific markers for the major classes of neurons of brain regions not yet analyzed in as much detail as the spinal cord, hindbrain, and cerebellar cortex (M.E.H. and N. Heintz, unpublished data). Among these are the large-output neurons of cerebral cortex and hippocampus, among other brain regions. Although adult neural “stem cells” generate CNS neurons, they have not yet generated specific output neurons of cortical regions of the brain. The latter will be essential to provide cells suitable for translational studies of cell replacement therapy.
The present results are consistent with a large number of studies on the induction of neural fates in the CNS and of cerebellar development. These studies can be summarized in four steps: the primary neurulation of the ectodermal plate, the establishment of the midbrain/hindbrain domain, formation of the cerebellum from rhombomere 1, and induction of GCP fate by dorsalizing factors (13). The present findings support a key role for FGF8 in the differentiation of cerebellar cells (31) because exposing ES cells to FGF8 induced expression of genes at the mes/met border of the neural tube, where the cerebellum develops. RA was required to induce the expression of En1, a gene required for the formation of the mes/met domain of the neural tube, and to regulate the expression of dorsal neuronal fates within the mes/met domain. The present experiments suggest a key role for RA in the expression of cerebellar neuronal fates.
The expression of Wnt genes is essential for the formation of the midbrain/hindbrain territory (10) and for precerebellar nuclei that project mossy fibers to the cerebellar cortex (32). Our studies on ES cells confirm a key role for WNTs in cerebellar development and in the directed differentiation of ES cells to cerebellar fates, including the two principal classes of cerebellar neurons, granule cells and Purkinje cells. The present experiments indicate that GCP proliferation, induced by SHH and JAG1, expands the ES cell population and induces expression of late GCP markers. Interestingly, recent work indicates that SHH is also a coactivator of RORα, which provides a reciprocal signaling pathway that induces Purkinje cell differentiation in addition to promoting GCP proliferation (33). Our observation that SHH and JAG1 increased the number of Purkinje cells in the ES cells is consistent with a role for SHH in Purkinje cell differentiation.
Culturing differentiated ES cells with the neurotrophin BDNF promoted the survival of ES cells, expression of cellular markers, and elaboration of the polarity of differentiated granule neurons. This result is consistent with the experiments of Segal and colleagues (27), who showed that BDNF promotes the survival of postmitotic EGL cells as they commence differentiation, migrate across the ML, and undergo terminal differentiation in the IGL. These findings suggest that neurotrophins promote late stages of neuronal differentiation in ES cells that, in turn, enable differentiated ES cells to incorporate into appropriate neuronal laminae and/or circuits.
The ability to use soluble factors that allocate cell fates in various brain regions suggests that many other cells can be differentiated from ES cells, when the normal developmental program is known. Thus, defining the steps of normal development in detail optimizes the success of cell replacement therapy in the brain. The present study is consistent with a recent report by Su et al. (34) showing expression of dorsal markers (MATH1, PAX6, and ZIC2) and differentiated interneurons (GABAa6) following a program of induction by WNT3a and BMP4. The study extends their findings by demonstrating an expression of EN1, a marker for the midbrain/hindbrain territory that gives rise to the cerebellar analgen, of unique markers for GCPs (ZIC2), for the incorporation of ES cells into the IGL of the neonatal cerebellum, and the progression of implanted cells through the cardinal steps of granule neuron development: the formation of T-shaped parallel fibers, glial-guided migration of immature neurons across the ML, and the formation of dendritic arbors by postmigratory ES cells in the IGL. These studies suggest that implanted EGFP+ ES cells can form synaptic connections with afferent axons in the position where granule neurons function in the cerebellar circuitry. Neurophysiological studies will be required to establish the connectivity of differentiated ES cells that have been stereotaxically implanted into the neonatal cerebellar cortex. Thus, the present study supports the general idea that inductive signals acting in vivo to pattern the CNS and generate specific classes of cells provide a rational approach to direct ES cell differentiation.
Materials and Methods
The GENSAT Project kindly provided the Pde1c-Egfp-BAC transgenic mouse line. Pde1c-Egfp ES cell lines were derived from Pde1c-Egfp-BAC transgenic mice (28). 129Sv J heterozygous blastocysts were expanded and cryopreserved as described (35). The Rockefeller University Transgenic Facility provided murine E14 ES cells, and we obtained D3 ES cells from American Type Culture Collection (Manassas, VA). We cultured E14, D3, or Pde1c-Egfp-BAC ES cells as cellular reaggregates on tissue culture plates (coated with 0.1% gelatin) in DMEM [supplemented with 15% FBS, 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 2 mM l-glutamine, and penicillin-streptomycin and 1 unit/ml−1 leukemia inhibitory factor (Chemicon)]. In some experiments, we cultured ES cells on a feeder layer of primary embryonic fibroblasts. ES cells were trypsinized (0.25% trypsin-EDTA) for 5 min at 37°C and plated on a Petri dish at a cell density of 2.0–2.5 × 104 cells per square centimeter in DFK5 medium (7). Embryoid bodies formed after 1 day in vivo (DIV). At this point, we supplemented DFK5 medium with FGF8b (100 ng/ml) and all-trans RA (2 μM; Sigma-Aldrich, St. Louis, MO) and cultured the EBs for 4 days.
EBs were plated in BME, supplemented with ITS Supplement-B (StemCell Technologies, Vancouver, BC, Canada), FGF8b and FGF4 (100 ng/ml), and bFGF (20 ng/ml) on glass coverslips or in 60-cm2 TC dishes (Nalge Nunc, Rochester, NY) or Permanox chamber slides (Nalge Nunc) pretreated with fibronectin (5 μg/ml) and laminin (1 μg/ml) for 2 days (7 DIV). We replaced the medium with DMEM, supplemented with FGF8b, WNT1 (50 ng/ml), and WNT3a (50 ng/ml), and cultured the cells for 5 days (12 DIV). (The protocol for Wnt conditioned medium is published in SI Materials and Methods.) To differentiate the ES cells into cerebellar granule cells, we cultured them in BME supplemented with N2 Supplement-B (×100) (StemCell Technologies), B27 (StemCell Technologies), and a BMP [BMP7 (100 ng/ml), GDF7 (100 ng/ml), and BMP6 (20 ng/ml)] (13) for 4 days (16 DIV). To promote proliferation of granule neuron precursors, we cultured the cells in the medium above containing SHH (100 ng/ml), JAG1 (20 ng/ml), NT3 (100 ng/ml), and BDNF (100 ng/ml) for 7–8 days (16–24 DIV). Throughout the period of cell culture (22–23 DIV), we replaced the indicated medium with fresh medium (replacing 50% of the medium) every 2 days. All growth factors (FGF4.8, bFGF, BMP6,7, GDF7, SHH, JAG1, BDNF, and NT3) were purchased from R & D Systems (Minneapolis, MN).
Primary cerebellar granule cells and glial cells purified from P6 C57BL/6J mice (36) were plated on polyd-lysine-coated (0.5 mg/ml; Sigma, St. Louis, MO) 60-cm2 TC dishes or Chamber slides (Nalge Nunc) in neurobasal (NB) medium supplemented with horse serum (10%), FBS (5%), and 2 mM l-glutamine (Gibco-Invitrogen, Carlsbad, CA). In some experiments, we plated a single-cell suspension of Pde1c-Egfp-BAC ES cells (1.5–2 × 103 cells per square cm) on top of a monolayer of purified cerebellar granule cells or glial cells, cultured the cells for 36–72 h, and processed the cultures for immunocytochemistry.
In other experiments, we plated Pde1c-Egfp-BAC ES cells (1.5–2 × 103 cells per square cm) in medium conditioned by either cerebellar granule neurons or cerebellar glial cells. To generate conditioned medium, we harvested medium from granule neurons or glial cells cultured in NB medium supplemented with NB/27 for 3 DIV, concentrated the medium with an Amicon Ultra-4 Centrifugal Filter (Millipore, Bedford, MA), dialyzed the medium against NB, and measured total protein (spent medium concentrate). Conditioned medium was prepared by adding 1.0 mg/ml of the spent medium concentrate to NB basic salt solution (Gibco-Invitrogen) and 2 mM l-glutamine (Gibco-Invitrogen). Cells were cultured in NB medium supplemented with a series of concentrations of conditioned medium ranging from 1:1 to 1:10 (vol/vol) for 1–8 days.
RT-PCR analysis of gene expression profiles of ES cells were as described (14). To analyze the relative level of expression of specific mRNAs, the amount of cDNA was normalized to actin mRNA. We compared neural mRNAs at specific stages of ES cell differentiation to that of undifferentiated ES cells. (Primer sequences, cycle numbers, and annealing temperatures are described in SI Table 1.) Gels were imaged by using a Gel Doc 2000 12-bit CCD camera (Bio-Rad, Athens, OH).
For stereotaxic implantation of differentiated ES cells into neonatal cerebellum, Pde1c-Egfp-BAC ES cells were differentiated as described and implanted in neonatal cerebellar cortex as described previously (13) by using an TransferMan NK2 System (Eppendorf, Westbury, NY) to standardize the coordinates for and volume of the cell injection. The details of the method are provided in SI Materials and Methods. In brief, we injected 1 μl of a cell suspension (1,000 cells) into the EGL and maintained animals with implanted ES cells for 1–15 days, after which animals were deeply anesthetized (Nembutal; Abbot Laboratories) and perfused with 4% paraformaldehyde. We processed the tissue and cut 14-μm cryostat sections, which were immunostained with anti-EGFP antibodies.
Supplementary Material
Acknowledgments
We are grateful to Drs. Thomas M. Jessell and Heinrich Wichterle for helpful discussions and to Drs. Rosalind Segal (Harvard Medical School, Boston, MA), Arthur Kania (College of Physicians and Surgeons of Columbia University, New York, NY), and A. Buchberg (College of Physicians and Surgeons of Columbia University, New York, NY) for generously providing antibodies. We thank Drs. Alex Joyner, Chenwen Yang, Shioaching Gong, and Laurenz Studer for advice on ES cells and Dani Morales for helpful discussions. Traci Jackson provided assistance with mouse husbandry, and Michael Morris helped prepare the manuscript. We are also grateful to our colleagues Dr. Kathryn Zimmerman and Ali Hemmati-Brivanlou for critically reading the manuscript. The GENSAT Project (N. Heintz and M.E.H., Co-Principal Investigators) provided the Pde1c-Egfp-BAC mice used in this study. This work was supported by The Rockefeller University and National Institutes of Health/National Institute of Neurological Disorders and Stroke Grant R0130532-04 (to M.E.H.).
Abbreviations
- EB
embryoid bodies
- EGL
external granule cell layer
- GCP
granule cell progenitor
- Pn
postnatal day n
- ML
molecular layer
- IGL
internal granule cell layer
- DIV
days in vivo
- RA
retinoic acid
- SHH
sonic hedgehog.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610879104/DC1.
References
- 1.Palay SL, Chan-Palay S. The Cerebellar Cortex: Cytology and Organization. New York: Springer; 1974. [Google Scholar]
- 2.Gao JH, Parsons LM, Bower JM, Xiong J, Li J, Fox PT. Science. 1996;272:545–547. doi: 10.1126/science.272.5261.545. [DOI] [PubMed] [Google Scholar]
- 3.Fiez JA. Neuron. 1996;16:13–15. doi: 10.1016/s0896-6273(00)80018-5. [DOI] [PubMed] [Google Scholar]
- 4.Boyden ES, Katoh A, Raymond JL. Annu Rev Neurosci. 2004;27:581–609. doi: 10.1146/annurev.neuro.27.070203.144238. [DOI] [PubMed] [Google Scholar]
- 5.Bradley A, Evans M, Kaufman MH, Robertson E. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 6.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 7.Wichterle H, Lieberam I, Porter JA, Jessell TM. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 8.Joyner AL. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- 9.Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. Development (Cambridge, UK) 1999;126:1189–1200. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- 10.McMahon AP, Bradley A. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 11.Wingate RJ, Hatten ME. Development (Cambridge, UK) 1999;126:4395–4404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- 12.Lee KJ, Dietrich P, Jessell TM. Nature. 2000;403:734–740. doi: 10.1038/35001507. [DOI] [PubMed] [Google Scholar]
- 13.Alder J, Lee K, Jessell T, Hatten ME. Nat Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- 14.Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 15.Wechsler-Reya RJ, Scott MP. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 16.Mason CA, Christakos S, Catalano SM. J Comp Neurol. 1990;297:77–90. doi: 10.1002/cne.902970106. [DOI] [PubMed] [Google Scholar]
- 17.Alder J, Cho NK, Hatten ME. Neuron. 1996;17:389–399. doi: 10.1016/s0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 19.Aruga J. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Morales D, Hatten ME. J Neurosci. 2006;26:12226–12236. doi: 10.1523/JNEUROSCI.3493-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KJ, Jessell TM. Annu Rev Neurosci. 1999;22:261–294. doi: 10.1146/annurev.neuro.22.1.261. [DOI] [PubMed] [Google Scholar]
- 22.Millonig JH, Millen KJ, Hatten ME. Nature. 2000;403:764–769. doi: 10.1038/35001573. [DOI] [PubMed] [Google Scholar]
- 23.Wilson L, Maden M. Dev Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 24.Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Development (Cambridge, UK) 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- 25.Rowitch DH, Kispert A, McMahon AP. Brain Res Dev Brain Res. 1999;117:99–108. doi: 10.1016/s0165-3806(99)00104-2. [DOI] [PubMed] [Google Scholar]
- 26.Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- 27.Borghesani PR, Peyrin JM, Klein R, Rubin J, Carter AR, Schwartz PM, Luster A, Corfas G, Segal RA. Development (Cambridge, UK) 2002;129:1435–1442. doi: 10.1242/dev.129.6.1435. [DOI] [PubMed] [Google Scholar]
- 28.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 29.Solecki DJ, Govek EE, Tomoda T, Hatten ME. Genes Dev. 2006;20:2639–2647. doi: 10.1101/gad.1462506. [DOI] [PubMed] [Google Scholar]
- 30.Mason CA, Morrison ME, Ward MS, Zhang Q, Baird DH. Perspect Dev Neurobiol. 1997;5:69–82. [PubMed] [Google Scholar]
- 31.Sato T, Joyner AL, Nakamura H. Dev Growth Differ. 2004;46:487–494. doi: 10.1111/j.1440-169x.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- 32.Dymecki SM, Tomasiewicz H. Dev Biol. 1998;201:57–65. doi: 10.1006/dbio.1998.8971. [DOI] [PubMed] [Google Scholar]
- 33.Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su HL, Muguruma K, Matsuo-Takasaki M, Kengaku M, Watanabe K, Sasai Y. Dev Biol. 2006;290:287–296. doi: 10.1016/j.ydbio.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Roach ML, McNeish JD. Methods Mol Biol. 2002;185:1–16. doi: 10.1385/1-59259-241-4:1. [DOI] [PubMed] [Google Scholar]
- 36.Hatten ME. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.