Abstract
Oral biofilms are multispecies communities, and in their nascent stages of development, numerous bacterial species engage in interspecies interactions. Better insight into the spatial relationship between different species and how species diversity increases over time can guide our understanding of the role of interspecies interactions in the development of the biofilms. Quantum dots (QD) are semiconductor nanocrystals and have emerged as a promising tool for labeling and detection of bacteria. We sought to apply QD-based primary immunofluorescence for labeling of bacterial cells with in vitro and in vivo biofilms and to compare this approach with the fluorophore-based primary immunofluorescence approach we have used previously. To investigate QD-based primary immunofluorescence as the means to detect distinct targets with single-cell resolution, we conjugated polyclonal and monoclonal antibodies to the QD surface. We also conducted simultaneous QD conjugate-based and fluorophore conjugate-based immunofluorescence and showed that these conjugates were complementary tools in immunofluorescence applications. Planktonic and biofilm cells were labeled effectively by considering two factors: the final nanomolar concentration of QD conjugate and the amount of antibody conjugated to the QD, which we define as the degree of labeling. These advances in the application of QD-based immunofluorescence for the study of biofilms in vitro and in vivo will help to define bacterial community architecture and to facilitate investigations of interactions between bacterial species in these communities.
Quantum dots (QD) are semiconductor nanocrystals which have been coupled to biomolecules, such as transferrin (4, 18), immunoglobulin G (IgG) (4), biotin (3), streptavidin (23, 42), avidin (11, 16), nucleic acids (8, 38, 39), peptides (2), serotonin (31), adenine(17), adenine monophosphate (17), and wheat germ agglutinin (17, 18). These conjugates are luminescent probes that bind with specificity and sensitivity to a variety of targets (IgG, antigens, glycoproteins, nucleic acid sequences, and receptors). Unlike traditional fluorophore fluorescence, QD luminescence is photostable and size tunable, with narrow, symmetric emission spectra and broad continuous excitation, allowing excitation of multiple QD with a single wavelength (3, 4, 12, 18, 42). These properties make QD very attractive luminescent labels for biological applications.
In the last decade, significant advances have been made with eukaryotic applications of QD conjugates; however, bacteriological applications are few. The first use of QD for bacterial labeling was reported by Kloepfer et al. in 2003 (18). QD have since been used for labeling, detection, and quantification of Mycobacterium bovis bacillus Calmette-Guérin (25), Escherichia coli O157:H7 (33), and Salmonella enterica serovar Typhimurium (40) and recently for the simultaneous detection of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium (41). The limit and speed for QD-based detection of pathogens were recently expanded with a phage-based assay that achieved detection of 100 bacteria in 1-ml samples in less than an hour (9). These QD-based detection approaches have high specificity and orders of magnitude higher sensitivity than that achievable with traditional fluorophores (15). None of the microbiological applications thus far has addressed the use of QD in biofilms.
Biofilms, defined as bacterial communities growing at interfaces, are a natural mode of growth for numerous bacteria (6) and are characteristic of the oral microflora (27). More than 700 bacterial phylotypes have been identified from oral biofilms (1). Tooth surfaces are colonized in a repeatable and sequential manner in that pioneer species are followed by secondary colonizers (20, 30). We recently characterized, by molecular methods, the microbial diversity of early dental biofilms (7) developed by using a retrievable enamel chip model (29). In the present study, our emphasis is to apply QD-based primary immunolabeling to achieve high single-cell resolution and to study spatiotemporal relationships between community members in these oral biofilm communities.
QD-streptavidin conjugates and QD-F(ab′)2 fragments are commercially available, and many applications use QD-based secondary immunofluorescence. This indirect approach, while more sensitive, limits the number of unique targets that can be recognized simultaneously, because the primary antibodies must be generated in different animal sources. Conjugating the primary antibody directly on the QD surface eliminates this limitation and permits simultaneous application of multiple QD-antibody conjugates.
The three aims in our study were as follows: (i) to compare primary immunofluorescence using QD-antibody conjugates with primary immunofluorescence using typical antibody fluorophore conjugates, such as Alexa Fluor conjugates; (ii) to apply QD-based primary immunofluorescence in in vitro biofilms; and (iii) to use multiple QD conjugates simultaneously in the study of in vivo oral biofilms formed with the retrievable enamel chip model (29). We show that planktonic cultures of oral bacteria can be labeled with QD-conjugated primary antibodies to achieve single-cell resolution, as can in vitro biofilms. For labeling of in vivo oral biofilms, we applied simultaneously QD-based primary immunofluorescence for Veillonella spp. and QD-based primary immunofluorescence of receptor polysaccharide (RPS)-bearing Streptococcus spp. and achieved single-cell resolution that defined bacterial community architecture.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial cultures were maintained short-term at 37°C in a Shel Lab Bactron IV anaerobic chamber (Sheldon Manufacturing, Inc., Cornelius, OR) with an atmosphere of N2:CO2:H2 (90:5:5 ratio). Human oral isolates Streptococcus gordonii DL1 (5) and Streptococcus mutans UA159 (ATCC 700610) were grown under static conditions in brain heart infusion broth (BHI) (Difco, Detroit, MI), and the oral clinical isolate Veillonella spp. strain R1 (26) was grown under static conditions in veillonella broth (Difco).
Biofilms.
Biofilms were formed in flow cells as previously described (28). Briefly, starter cultures were grown anaerobically to mid-exponential phase, and cells were harvested by centrifugation. Cells were washed and resuspended in 25% sterile human saliva (28) or in 28-fold-diluted BHI to an A600 of 0.05. The biofilm growth medium was unsupplemented 25% human saliva except in experiments using S. mutans, for which the medium was 28-fold-diluted BHI with 3 mM sucrose. The flow rate of the medium was 0.1 ml/min unless otherwise indicated. In vivo biofilms were developed by using the retrievable enamel chip model described previously (29). Briefly, enamel chips (x = 2 mm; y = 2 mm; z = 1 mm) were cut from extracted human third molars, ultrasonically cleaned, sterilized with ethylene oxide, fixed with dental wax in bilateral mandibular stents, and worn by a volunteer for 6 h or 8 h.
Antibodies.
Five antibodies were used in this study: anti-S. gordonii DL1 (anti-DL1), preimmune IgG, anti-S. oralis RPS (anti-RPS), anti-Veillonella sp strain R1 (antiveillonella), and SWLA1.2. Anti-DL1 is a purified (ImmunoPure immobilized protein A; PIERCE Biotechnology, Rockford, IL) polyclonal IgG raised against whole cells of S. gordonii DL1: it recognizes the cell surface components of many Streptococcus spp. (27), including S. mutans UA159. Anti-RPS is an affinity-purified antibody against the 1Gn streptococcal receptor polysaccharides (RPS) of S. oralis 34: it recognizes the RPS that are an integral cell wall component of certain Streptococcus spp. (27). Antiveillonella is a purified (ImmunoPure immobilized protein A) polyclonal IgG raised against whole cells of clinical isolate Veillonella sp. strain R1: it recognizes the cell surface components of many Veillonella spp. (26). SWLA1.2 is a monoclonal IgG against S. mutans (32).
Conjugation of antibodies to fluorophores.
The five IgGs (anti-DL1 and its preimmune IgG, anti-RPS, antiveillonella, and SWLA1.2) were fluorescently labeled using Alexa Fluor protein labeling kits (Invitrogen, Carlsbad, CA), following the manufacturer's directions.
The QDot antibody conjugation kit and the Qdot innovator's tool kit are commercial products (Invitrogen) with which proteins can be coupled directly to a QD surface. The former requires reduction of the disulfide bonds, and the latter does not (Qdot antibody conjugation kit; Quantum Dot Invitrogen Nanocrystal Technologies). We used both methods to link primary antibodies to the QD surface for immunofluorescence labeling. Anti-DL1, anti-RPS, and SWLA1.2 were conjugated to QD655 by using the QDot antibody conjugation kit, which was also used to conjugate preimmune IgG to QD525. Antiveillonella and SWLA1.2 were conjugated to QD605 with the Qdot innovator's tool kit. The commercial secondary antibody conjugate Qdot 655 goat F(ab′)2 antirabbit IgG (heavy plus light chains) (Invitrogen) was also used.
For conjugation of primary antibodies to QD655 using the QDot antibody conjugation kit, we modified the manufacturer's protocol to allow calculation of the degree of labeling (DOL) of each conjugate. After conjugation and quenching of the reaction, the QD-antibody reaction mix was applied to a 300-kDa Vivaspin 500-μl centrifugal filter unit (Sartorius Corporation, Edgewood, NY) and centrifuged at 2,000 rpm in 10-min steps until the final volume was 25 μl. The eluate containing the unconjugated IgG was concentrated with 50-kDa Vivaspin 500 μl centrifugal filter units (Sartorius Corporation, Edgewood, NY), and the protein concentration was measured (absorbance at 280 nm) with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). The concentrated QD-antibody conjugate (retentate from 300-kDa spin filter) was purified using size exclusion chromatography according to the manufacturer's recommendation. The DOL was then calculated as moles IgG added to the conjugation reaction minus moles unconjugated IgG (the eluate from the 300-kDa filter) divided by moles QD in conjugate (the retentate from the 300-kDa filter). In our experience, a DOL of 3 to 5 was typical for primary antibody-QD conjugates.
Staining of biofilms and microscopy.
Staining of the biofilm varied according to the target organism. General nucleic acid stains used were SYTO9 or 4′,6′-diamidino-2-phenylindole (Invitrogen) and were applied according to the manufacturer's recommendations. Primary antibody-Alexa Fluor conjugates were applied at a 5-μg protein/ml final concentration for 20 min in phosphate-buffered saline containing 10 mg bovine serum albumin/ml (PBS-BSA). The primary antibody-QD conjugates were used at concentrations that reflect the DOL and the final protein concentration. The typical treatment was with 5 μg protein/ml × DOL for 20 min. For secondary antibody-QD conjugate applications, the biofilms were labeled with primary antibodies for 20 min and the unbound antibodies were washed once with PBS-BSA. The secondary F(ab′)2 fragment-QD conjugates were applied at 20 nM (based on QD nanocrystal extinction coefficient) in PBS-BSA for 20 min. Extracellular polysaccharide was stained by incubating biofilms for 20 min with calcofluor (10 mg/ml in 10 mM sodium phosphate, pH 7.5; Sigma, St. Louis, MO) (35). Labeled planktonically grown cells were examined by epifluorescence microscopy using a 100× oil-immersion lens and a 515 to 560 excitation/590 long pass filter. Images were recorded with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics, Bridgewater, NJ). Biofilms were imaged by confocal microscopy using a Leica TCS/SP2 confocal microscope (Leica Microsystems, Exton, PA).
RESULTS AND DISCUSSION
Labeling of planktonic cells.
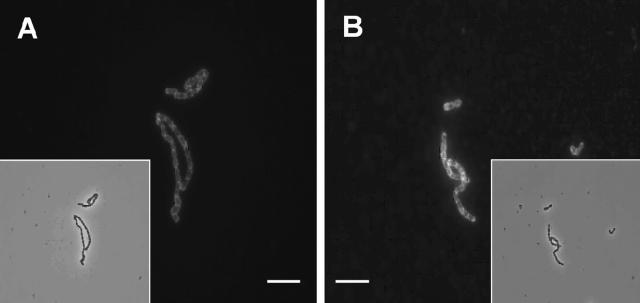
Initially, we compared labeling of planktonic bacteria using the QD-antibody conjugates with labeling using the primary Alexa Fluor conjugates that we have used previously. S. gordonii DL1 cells were grown overnight in liquid culture, washed twice with PBS, resuspended in PBS-BSA containing labeled primary antibody (5 μg/ml) for 20 min, washed once in PBS, and examined by using epifluorescence microscopy (Fig. 1). No difference in the abilities of anti-DL1-QD655 and anti-DL1-Alexa Fluor 546 conjugates to define the cell surface of S. gordonii DL1 was seen. The “halo” surrounding the cells indicated that both types of conjugates yielded excellent single-cell resolution.
FIG. 1.
Planktonically grown S. gordonii DL1 cells labeled with the antibody conjugate anti-DL1-Alexa Fluor 546 (A) or anti-DL1-QD655 (B), showing comparable labeling with the two antibody conjugates as revealed by epifluorescence microscopy. Inserts are transmitted light images; bar, 5 μm.
Brightness and photostability of QD compared with that of organic fluorophores.
QD are reported to be significantly brighter and more photostable than traditional fluorophores. Comparisons include fluorescein isothiocyanate (FITC) and QD605 (22), Alexa Fluor 488 and QD608 (37), Alexa Fluor 488 and QD590 (34), and FITC and QD525 (15). Luminescence with QD has been reported to be 100 times brighter and 10 times more sensitive than that of traditional fluorophores (15) and also to be 420-fold more resistant to photobleaching (34).
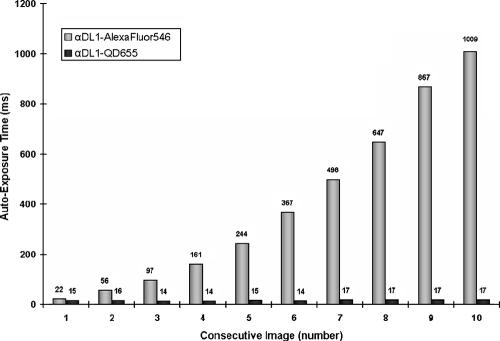
We compared brightness and photostability of Alexa Fluor 546 and QD655 conjugates under constant excitation. Aliquots of a suspension of S. gordonii DL1 were labeled with the QD conjugate or with the Alexa Fluor conjugate. Consecutive images of wet-mounted cells were collected using the camera's autoexposure mode (Fig. 2). The brightness of a sample is reflected by the autoexposure time in milliseconds, i.e., brighter objects have shorter autoexposure times. Bleaching is reflected by the increase of autoexposure time under constant excitation (Fig. 2). Under these conditions, cells labeled with QD conjugates have an initial fluorescence intensity similar to that of traditional fluorophore conjugates, but QD conjugate-labeled bacteria remain brighter longer. By the third exposure, autoexposure time had increased fourfold for the Alexa Fluor conjugate with no change in exposure time for the QD conjugate. Even after acquiring 10 images under constant fluorescence excitation, the exposure time for the QD conjugate had changed little, whereas that for the Alexa Fluor conjugate had increased 45-fold. One application for photostable QD conjugates is in micromanipulation of fluorescent cells, where extended time is required to locate and retrieve the target.
FIG. 2.
S. gordonii DL1 cells labeled with anti-DL1-QD655 conjugates are brighter and more photostable than those labeled with anti-DL1-Alexa Fluor 546. x axis, number of consecutive images under constant fluorescence excitation; y axis, autoexposure time in milliseconds.
QD conjugations to monoclonal and polyclonal antibodies.
Specific surface-located antigens on bacterial cells are recognized by monoclonal antibodies (MAb), and these antigens are present on only a limited number of bacterial species: therefore, MAbs are important for monitoring and quantifying commensal and pathogenic bacteria (13, 14, 32) and for in situ detection of bacterial species in oral biofilms (14). Many QD-based applications with MAbs employ a secondary immunofluorescence approach where initial application of biotinylated MAb is followed by a secondary application of QD-streptavidin conjugates (23), but simultaneous detection of multiple targets is best achieved by using labeled primary antibodies (14).
We sought to conjugate MAbs directly to the QD surface and label cells with the QD-MAb conjugates. SWLA1.2 is a species-specific MAb against S. mutans (14, 32). SWLA1.2 was conjugated to QD655 with the QDot antibody conjugation Kit, and a conjugate with a DOL of 3 (three IgG molecules per QD) was obtained; however, no labeling of S. mutans UA159 planktonic cells was observed with this preparation (data not shown). This absence of reactivity was not due to the lack of conjugation of antibody to QD but might be attributable to the reduction step in the conjugation process. Conjugation of the same MAb to QD605 using the Qdot innovator's tool kit without reduction of the antibody resulted in a conjugate that labeled S. mutans UA159 planktonic cells with good cell surface resolution (data not shown). Accordingly, the sensitivity to reduction for some MAbs prior to direct conjugation to the QD surface can be an important factor for successful application of these QD-MAb conjugates. As described below, sensitivity to reduction has not affected our conjugations using polyclonal antibodies.
DOL and single-cell resolution.
Optimal cellular resolution in immunofluorescence is achieved by proper adjustment of the antibody protein concentration. Typical fluorophore-antibody conjugates have multiple fluorophore molecules conjugated to each IgG. In contrast, QD conjugates typically have multiple antibody molecules conjugated per QD and are therefore multivalent conjugates for which higher protein concentrations are required to achieve optimal cellular resolution. Other investigators also recognized the importance of the protein-QD conjugate concentration; Goldman et al. (10) showed that an increase in the MAb-QD concentration accompanied increased sensitivity and reached saturation at a 120 nM QD concentration. Hahn et al. (15) carried out a thorough investigation on the effects of protein concentration on single-cell resolution. Although the DOL for commercially available QD-streptavidin conjugates ranges from 5 to 10 (Qdot streptavidin conjugates; Quantum Dot Invitrogen Nanocrystal Technologies), experimental evidence (15, 41) indicates that for optimal labeling, the protein concentration of the multivalent QD conjugate must be higher than that used for univalent labels, such as FITC conjugates.
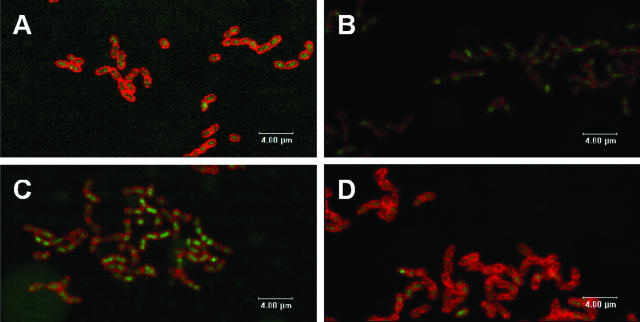
To investigate experimentally the principles of DOL and the concentration (nM) of conjugated QD, we compared the cell surface resolution obtained by using univalent Alexa Fluor-conjugated antibody with that achieved by using multivalent QD-conjugated antibody (DOL = 3) (Fig. 3). The initial application of multivalent QD conjugates in biofilms (Fig. 3B) at the same protein concentration used for univalent Alexa Fluor conjugates (Fig. 3A) resulted in poor definition of the cell surface (compare Fig. 3A and B). By increasing the nanomolar concentration of the QD conjugates (and hence the μg protein/ml), cell surface resolution (Fig. 3C and D) comparable to that observed with Alexa Fluor conjugates was achieved (compare Fig. 3D and A). Thus, the two factors, DOL and nanomolar concentration of QD conjugates, set the optimal concentration of QD conjugate required for optimal immunofluorescence labeling of biofilm cells. We have found that the typical DOL is 3 to 5 and the typical final concentration of QD conjugate for labeling cells is 30 nM.
FIG. 3.
Confocal scanning laser microscopic analysis showing effect of DOL and nanomolar concentration of QD conjugates in obtaining single-cell resolution of S. gordonii DL1 biofilms. Biofilms labeled with the general nucleic acid stain SYTO9 (green) or antibody conjugates (red): A, anti-DL1-Alexa Fluor 546 at 5 μg protein/ml; B, anti-DL1-QD655 at 5 μg protein/ml and 10 nM QD concentration; C, anti-DL1-QD655 at 10 μg protein/ml and 20 nM QD concentration; D, anti-DL1-QD655 at 15 μg protein/ml and 30 nM QD concentration with DOL of 3. Images are single optical sections.
Cell surface resolution with QD primary conjugates.
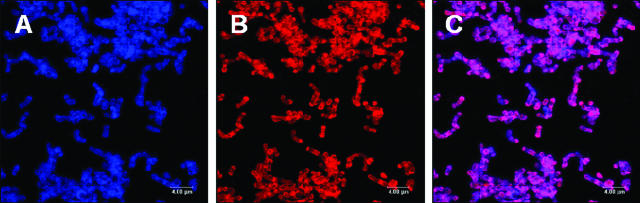
QD-antibody conjugates are larger in size than Alexa Fluor-antibody conjugates, and we hypothesized that each conjugate could label the same cell surface. To compare directly the single-cell resolution achievable by primary immunofluorescence using QD-antibody conjugates or Alexa Fluor-antibody conjugates, a population of cells was labeled with both conjugates (Fig. 4). We labeled S. gordonii DL1 4-h biofilms with anti-DL1-QD655 conjugate (Fig. 4A), and subsequently the same biofilm was labeled with anti-DL1-Alexa Fluor 546 (Fig. 4B). The cell surface resolution in the overlay image (Fig. 4C) reveals that the cells have equivalent resolutions with each conjugate. Neither conjugate interferes with the other in labeling the cells. Thus, QD-antibody conjugates and Alexa Fluor-antibody conjugates can be used simultaneously to label biofilm cells.
FIG. 4.
Confocal scanning laser microscopic images of 4-h S. gordonii DL1 biofilm showing simultaneous and equivalent efficiency of labeling of cells with both anti-DL1-QD655 conjugate (15 μg protein/ml with DOL of 3) and anti-DL1-Alexa Fluor 546 conjugate (5 μg protein/ml). Maximum projection image of the QD655 channel (A, blue), Alexa Fluor 546 channel (B, red), and overlay (C, purple [red plus blue]) showing absence of interference of labeling by conjugates. The biofilm was first labeled with anti-DL1-QD655 conjugate and then with anti-DL1-Alexa Fluor 546 conjugate; bar, 4 μm.
Nonspecific binding.
Nonspecific binding of QD to bacterial cell surfaces has been reported previously (15, 18). Such binding could result from inherent properties of the naked QD surface or from the antibody-conjugated QD where the antibody specificity has been lowered by the conjugation procedure.
We investigated nonspecific binding of commercial QD655 (activated, quenched, and purified) and preimmune IgG conjugated to QD525 in S. gordonii DL1 biofilms. Neither the activated, quenched, and purified QD655 nor the preimmune IgG-QD525 conjugates showed binding in S. gordonii DL1 biofilms (data not shown). Thus, nonspecific binding of QD or of nonimmune antibody-QD conjugates need not be a problem for biofilm work.
Penetration of extracellular polysaccharide by antibody-QD conjugates.
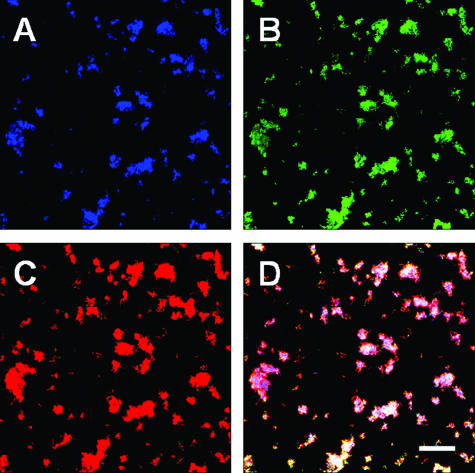
QD-antibody conjugates are much larger than typical fluorophore-antibody conjugates and may be less accessible to cells within biofilms; therefore, they may not label cells as well as the smaller fluorophore-antibody conjugates. Tortuosity was proposed as the most likely explanation for retarded diffusion of fluorescent probes in biofilms (36). Thick biofilms contain extracellular spaces connected as a contorted tunnel through which fluorescent probes must travel to label cells within the biofilm. The presence of extracellular polysaccharides (EPS) increases tortuosity and slows the diffusion of the fluorescent probe. To investigate the effect of the extracellular polysaccharide on the penetration of antibody-QD conjugates, we used S. mutans UA159, which is known to produce abundant EPS when grown in the presence of sucrose. Biofilms of S. mutans UA159 were grown for 18 h, and the EPS in the biofilms was visualized with calcofluor (Fig. 5A). The S. mutans UA159 biofilm cells were labeled with the general nucleic acid stain Syto9 (Fig. 5B) and MAb SWLA1.2-QD605 conjugate (Fig. 5C). Inspection of the three panels and comparison with the overlay panel (Fig. 5D) clearly show that primary MAb-QD-based immunofluorescence is not affected by the presence of EPS in these biofilms. Identical results using polyclonal anti-DL1-QD655 conjugates, which cross-react with S. mutans UA159, were obtained (data not shown). Thus, the large size of QD conjugates (10 to 20 nm) is not a limiting factor in labeling cells in these EPS-containing biofilms (average thickness is 15 μm).
FIG. 5.
Penetration of EPS by MAb SWLA1.2-QD605 conjugate in biofilms grown for 18 h with 28-fold-diluted BHI containing 3 mM sucrose in a flow of 0.2 ml/min. Confocal scanning laser microscopic analysis of biofilms stained with calcofluor (A), general nucleic acid stain SYTO9 (B), or MAb SWLA1.2-QD605 conjugate (C) and overlay image of all channels (D). All images are maximum projection; bar, 4 μm.
Application of secondary immunofluorescence in biofilms.
None of the many QD-based secondary-immunofluorescence studies have investigated biofilm-grown bacteria. We compared primary and secondary immunofluorescence for labeling of 18-h S. gordonii DL1 biofilms by examining pixel intensity. Anti-DL1-QD655 was used for primary immunofluorescence, and anti-DL1 followed by QD655 goat F(ab′)2 antirabbit IgG conjugate was used for secondary immunofluorescence. Consistent with Alexa Fluor-based secondary application, the biofilms labeled by secondary QD-based immunofluorescence had much higher pixel intensities (data not shown). These data indicate that both QD-based secondary- and primary-immunofluorescence protocols are excellent tools for biofilm research.
Multiple primary QD-based immunofluorescence and in vivo biofilms.
The retrievable enamel chip model permits direct observation of undisturbed human oral biofilms (29). Streptococcus spp. represent 65 to 85% of early colonizers (7, 24), and Veillonella spp. account for about 5 to 10% (7). RPS-bearing streptococci are a significant subset of the total streptococcal population in these oral biofilms and have been shown to play a role in coaggregation-mediated interactions on the enamel surface (27). They might be key players in the transformation from early pioneer communities to communities abundant with late colonizers (21).
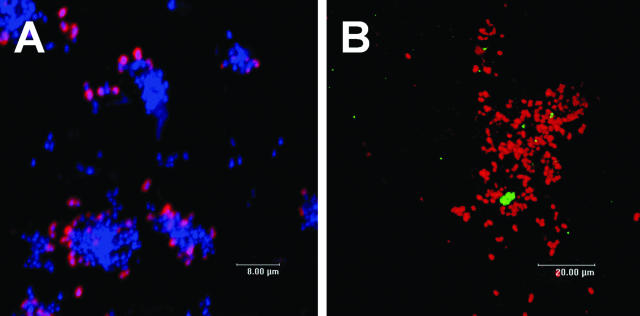
To investigate the spatial relationship of RPS-bearing streptococci to other bacteria, we conjugated antibodies against RPS of S. oralis 34 (27) to QD655. Six-hour oral biofilms were developed on retrievable enamel chips and were stained with this conjugate (Fig. 6A). Numerous clusters of RPS-bearing streptococci (red) juxtaposed with other bacteria (blue) were revealed as mixed-species communities of initial dental plaque biofilms. These results support the application of QD-based immunofluorescence as a tool for the study of in vivo biofilms, and they show multispecies communities similar to those revealed in our previous studies in which antibody-Alexa Fluor conjugates were used (26, 27).
FIG. 6.
Mixed-species communities revealed in undisturbed plaque. (A) Primary antibody anti-RPS-QD655 conjugate (red) and general nucleic acid stain 4′,6′-diamidino-2-phenylindole (blue). Spatial relationship of two species revealed in undisturbed plaque. (B) Primary antibody anti-RPS-QD655 conjugate (red) and primary antibody antiveillonella-QD605 conjugate (green). Confocal scanning laser micrographs of 6-h oral biofilms developed on an enamel surface in vivo. Maximum projection images.
Multiple QD-based immunofluorescence probes can be used simultaneously in natural oral biofilms. RPS-bearing streptococci were labeled with anti-RPS-QD655 conjugate, and veillonellae were labeled with antiveillonella-QD605 conjugate. Several antiveillonella-reactive cells were positioned within clusters of RPS-bearing streptococci (Fig. 6B). These results demonstrate that multiple QD conjugates can be excited simultaneously with a single wavelength to reveal mixed-species communities in vivo.
Bacterial single-cell resolution achievable with QD-based primary immunofluorescence is comparable to that observed with modern fluorophores, but QD conjugates offer significant advantages with standard epifluorescence microscopy. Because all QD, regardless of emission wavelength, absorb strongly in the blue/UV spectral region, mercury lamp excitation allows simultaneous excitation of several QD conjugates. In confocal microscopy, a UV laser or blue laser or multiphoton excitation is required to obtain similar flexibility. Excellent single-cell resolution occurred in both in vitro and in vivo biofilms examined in our study. One application that benefits greatly from the photostability of QD conjugates is micromanipulation of viable, spatially resolved communities from the enamel chip surface. These retrieved multispecies communities can be reconstituted and studied in an in vitro model, where the intimate mechanisms of cell-cell interspecies interactions can be discovered (19).
Acknowledgments
This research was supported by the Intramural Research Program of the NIDCR, NIH, and in part by the Colgate-Palmolive Co. CRADA.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerman, M. E., W. C. Chan, P. Laakkonen, S. N. Bhatia, and E. Ruoslahti. 2002. Nanocrystal targeting in vivo. Proc. Natl. Acad. Sci. USA 99:12617-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruchez, M., Jr., M. Moronne, P. Gin, S. Weiss, and A. P. Alivisatos. 1998. Semiconductor nanocrystals as fluorescent biological labels. Science 281:2013-2016. [DOI] [PubMed] [Google Scholar]
- 4.Chan, W. C., and S. Nie. 1998. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281:2016-2018. [DOI] [PubMed] [Google Scholar]
- 5.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 7.Diaz, P. I., N. I. Chalmers, A. H. Rickard, C. Kong, C. L. Milburn, R. J. Palmer, Jr., and P. E. Kolenbrander. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72:2837-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubertret, B., P. Skourides, D. J. Norris, V. Noireaux, A. H. Brivanlou, and A. Libchaber. 2002. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science 298:1759-1762. [DOI] [PubMed] [Google Scholar]
- 9.Edgar, R., M. McKinstry, J. Hwang, A. B. Oppenheim, R. A. Fekete, G. Giulian, C. Merril, K. Nagashima, and S. Adhya. 2006. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. USA 103:4841-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman, E. R., G. P. Anderson, P. T. Tran, H. Mattoussi, P. T. Charles, and J. M. Mauro. 2002. Conjugation of luminescent quantum dots with antibodies using an engineered adaptor protein to provide new reagents for fluoroimmunoassays. Anal. Chem. 74:841-847. [DOI] [PubMed] [Google Scholar]
- 11.Goldman, E. R., E. D. Balighian, H. Mattoussi, M. K. Kuno, J. M. Mauro, P. T. Tran, and G. P. Anderson. 2002. Avidin: a natural bridge for quantum dot-antibody conjugates. J. Am. Chem. Soc. 124:6378-6382. [DOI] [PubMed] [Google Scholar]
- 12.Goldman, E. R., A. R. Clapp, G. P. Anderson, H. T. Uyeda, J. M. Mauro, I. L. Medintz, and H. Mattoussi. 2004. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Anal. Chem. 76:684-688. [DOI] [PubMed] [Google Scholar]
- 13.Gu, F., R. Lux, M. H. Anderson, M. A. del Aguila, L. Wolinsky, W. R. Hume, and W. Shi. 2002. Analyses of Streptococcus mutans in saliva with species-specific monoclonal antibodies. Hybrid. Hybrid. 21:225-232. [DOI] [PubMed] [Google Scholar]
- 14.Gu, F., R. Lux, L. Du-Thumm, I. Stokes, J. Kreth, M. H. Anderson, D. T. Wong, L. Wolinsky, R. Sullivan, and W. Shi. 2005. In situ and non-invasive detection of specific bacterial species in oral biofilms using fluorescently labeled monoclonal antibodies. J. Microbiol. Methods 62:145-160. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, M. A., J. S. Tabb, and T. D. Krauss. 2005. Detection of single bacterial pathogens with semiconductor quantum dots. Anal. Chem. 77:4861-4869. [DOI] [PubMed] [Google Scholar]
- 16.Jaiswal, J. K., H. Mattoussi, J. M. Mauro, and S. M. Simon. 2003. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 21:47-51. [DOI] [PubMed] [Google Scholar]
- 17.Kloepfer, J. A., R. E. Mielke, and J. L. Nadeau. 2005. Uptake of CdSe and CdSe/ZnS quantum dots into bacteria via purine-dependent mechanisms. Appl. Environ. Microbiol. 71:2548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloepfer, J. A., R. E. Mielke, M. S. Wong, K. H. Nealson, G. Stucky, and J. L. Nadeau. 2003. Quantum dots as strain- and metabolism-specific microbiological labels. Appl. Environ. Microbiol. 69:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander, P. E., P. G. Egland, P. I. Diaz, and R. J. Palmer, Jr. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13:11-15. [DOI] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47-79. [DOI] [PubMed] [Google Scholar]
- 22.Lee, L. Y., S. L. Ong, J. Y. Hu, W. J. Ng, Y. Feng, X. Tan, and S. W. Wong. 2004. Use of semiconductor quantum dots for photostable immunofluorescence labeling of Cryptosporidium parvum. Appl. Environ. Microbiol. 70:5732-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansson, A., M. Sundberg, M. Balaz, R. Bunk, I. A. Nicholls, P. Omling, S. Tagerud, and L. Montelius. 2004. In vitro sliding of actin filaments labelled with single quantum dots. Biochem. Biophys. Res. Commun. 314:529-534. [DOI] [PubMed] [Google Scholar]
- 24.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 25.Otsuka, Y., K. Hanaki, J. Zhao, R. Ohtsuki, K. Toyooka, H. Yoshikura, T. Kuratsuji, K. Yamamoto, and T. Kirikae. 2004. Detection of Mycobacterium bovis bacillus Calmette-Guerin using quantum dot immuno-conjugates. Jpn. J. Infect. Dis. 57:183-184. [PubMed] [Google Scholar]
- 26.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188:4117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, R. J., Jr., R. Wu, S. Gordon, C. G. Bloomquist, W. F. Liljemark, M. Kilian, and P. E. Kolenbrander. 2001. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337:393-403. [DOI] [PubMed] [Google Scholar]
- 30.Quirynen, M., R. Vogels, M. Pauwels, A. D. Haffajee, S. S. Socransky, N. G. Uzel, and D. van Steenberghe. 2005. Initial subgingival colonization of ‘pristine’ pockets. J. Dent. Res. 84:340-344. [DOI] [PubMed] [Google Scholar]
- 31.Rosenthal, S. J., I. Tomlinson, E. M. Adkins, S. Schroeter, S. Adams, L. Swafford, J. McBride, Y. Wang, L. J. DeFelice, and R. D. Blakely. 2002. Targeting cell surface receptors with ligand-conjugated nanocrystals. J. Am. Chem. Soc. 124:4586-4594. [DOI] [PubMed] [Google Scholar]
- 32.Shi, W., A. Jewett, and W. R. Hume. 1998. Rapid and quantitative detection of Streptococcus mutans with species-specific monoclonal antibodies. Hybridoma 17:365-371. [DOI] [PubMed] [Google Scholar]
- 33.Su, X. L., and Y. Li. 2004. Quantum dot biolabeling coupled with immunomagnetic separation for detection of Escherichia coli O157:H7. Anal. Chem. 76:4806-4810. [DOI] [PubMed] [Google Scholar]
- 34.Sukhanova, A., J. Devy, L. Venteo, H. Kaplan, M. Artemyev, V. Oleinikov, D. Klinov, M. Pluot, J. H. Cohen, and I. Nabiev. 2004. Biocompatible fluorescent nanocrystals for immunolabeling of membrane proteins and cells. Anal. Biochem. 324:60-67. [DOI] [PubMed] [Google Scholar]
- 35.Thurnheer, T., R. Gmur, and B. Guggenheim. 2004. Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods 56:37-47. [DOI] [PubMed] [Google Scholar]
- 36.Thurnheer, T., R. Gmur, S. Shapiro, and B. Guggenheim. 2003. Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl. Environ. Microbiol. 69:1702-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, X., H. Liu, J. Liu, K. N. Haley, J. A. Treadway, J. P. Larson, N. Ge, F. Peale, and M. P. Bruchez. 2003. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat. Biotechnol. 21:41-46. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, Y., and P. E. Barker. 2004. Semiconductor nanocrystal probes for human metaphase chromosomes. Nucleic Acids Res. 32:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, H., M. Y. Sha, E. Y. Wong, J. Uphoff, Y. Xu, J. A. Treadway, A. Truong, E. O'Brien, S. Asquith, M. Stubbins, N. K. Spurr, E. H. Lai, and W. Mahoney. 2003. Multiplexed SNP genotyping using the Qbead system: a quantum dot-encoded microsphere-based assay. Nucleic Acids Res. 31:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, L., and Y. Li. 2005. Quantum dots as fluorescent labels for quantitative detection of Salmonella typhimurium in chicken carcass wash water. J. Food Prot. 68:1241-1245. [DOI] [PubMed] [Google Scholar]
- 41.Yang, L., and Y. Li. 2006. Simultaneous detection of Escherichia coli O157:H7 and Salmonella Typhimurium using quantum dots as fluorescence labels. Analyst 131:394-401. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, L., S. Ang, and W.-T. Liu. 2004. Quantum dots as a novel immunofluorescent detection system for Cryptosporidium parvum and Giardia lamblia. Appl. Environ. Microbiol. 70:597-598. [DOI] [PMC free article] [PubMed] [Google Scholar]