Abstract
Beta-glucuronidase-negative, sorbitol-nonfermenting isolates of Shiga toxin-producing Escherichia coli O157 comprise part of a clone complex of related enterohemorrhagic E. coli isolates. High-resolution genotyping shows that the O157 populations have diverged into two different lineages that appear to have different ecologies. To identify genomic regions unique to the most common human-associated genotype, suppression subtractive hybridization was used to identify DNA sequences present in two clinical strains representing the human lineage I O157:H7 strains but absent from two bovine-derived lineage II strains. PCR assays were then used to test for the presence of these regions in 10 lineage I strains and 20 lineage II strains. Twelve conserved regions of genomic difference for lineage I (CRDI) were identified that were each present in at least seven of the lineage I strains but absent in most of the lineage II strains tested. The boundaries of the lineage I conserved regions were further delimited by PCR. Eleven of these CRDI were associated with E. coli Sakai S-loops 14, 16, 69, 72, 78, 82, 83, 91 to 93, 153, and 286, and the final CRDI was located on the pO157 virulence plasmid. Several potential virulence factors were identified within these regions, including a putative hemolysin-activating protein, an iron transport system, and several possible regulatory genes. Cluster analysis based on lineage I conserved regions showed that the presence/absence of these regions was congruent with the inferred phylogeny of the strains.
Enterohemorrhagic Escherichia coli (EHEC) serotype O157:H7 is a major cause of both large-scale epidemics and sporadic cases of diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome in many countries around the world (13, 34, 36, 54, 56). The annual incidence of reported E. coli O157:H7 infections in Canada and the United States ranges from 1.7 to 5.3 per 100,000 persons and may be much higher in certain regions (14, 56). Within the United States alone, it has been estimated that approximately 73,000 cases of E. coli O157:H7 infection occur annually (33). The most common E. coli O157:H7 isolates are motile, non-sorbitol fermenting (SOR−), and β-glucuronidase negative (GUD−), while a nonmotile SOR+ GUD+ O157:H− clone has also been isolated in Germany (20) and a nonmotile SOR− GUD− O157:H− clone is commonly isolated in Australia (46).
Population genetic analysis has shown that E. coli O157:H7 and O157:H− isolates belong to a geographically disseminated clone complex that acquired virulence genes independently from other EHEC isolates (31, 45, 61, 62). Despite the clonal nature of E. coli O157:H7 and O157:H− isolates, significant variability was observed when they were tested by high-resolution genomic typing methods, such as pulsed field gel electrophoresis and octamer-based genome scanning (OBGS) (12, 22, 23, 53), implying that subpopulations are diverging quite rapidly.
OBGS is a large-scale genome comparison method based on pattern analysis of PCR amplification products generated using overrepresented octamers as primers. Recent studies using OBGS suggest that extant populations of O157:H7 isolates have diverged through two primary lineages, lineage I and lineage II, and that these lineages can be detected in geographically unlinked regions, such as the United States and Australia (22, 23). The origin of these two lineages, therefore, appears to predate the geographical spread of E. coli O157:H7 and the regional evolution of the SOR− GUD− O157:H− clone commonly isolated in Australia (23). More recently, the lineage-specific polymorphism assay (LSPA-6) was developed, based on six loci that show bias in their allelic distribution between the two lineages. The LSPA-6 is therefore a more efficient alternative for inferring lineage assignments compared to laborious OBGS typing (63). The two methods were demonstrated to generate highly concordant data (63). All lineage I isolates were LSPA-6 genotype 111111 (lineage I allele at each locus), while the majority of lineage II isolates were LSPA-6 genotypes 222222, 211111, and 212111.
In the initial OBGS studies and in the LSPA-6 study, a low proportion of human strains were observed in OBGS lineage II and LSPA-6 genotype 222222, respectively (22, 63). The paucity of OBGS lineage II and LSPA-6 genotype 222222 human isolates led workers to postulate that these E. coli O157:H7 isolates may be deficient in their abilities either to be transmitted to humans or to cause clinically significant human infections (22, 63). Several other studies also suggest that there are clear differences in the expression of virulence attributes, such as Shiga toxin and the locus for enterocyte effacement (LEE) proteins, by E. coli O157:H7 isolates from humans and from cattle (27, 32, 47, 48). These latter studies, however, did not consider the population structure of E. coli O157:H7 (e.g., lineage of descent) as a variable.
In this study, suppression subtractive hybridization (SSH) was used to identify genomic regions present in E. coli O157:H7 lineage I (LSPA-6 111111) strains but absent from lineage II (LSPA-6 222222) strains. We show that lineage I strains do indeed share a set of unique genes that are largely absent in lineage II strains. Several of these genes encode proteins that could contribute to virulence characteristics or which are known to regulate expression of virulence genes.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains included in this study are listed in Table 1. OBGS type strains 93-001, FDA 516-520, and FRIK 523-2001 have previously been described by Kim et al. (22). ZAP strains were obtained from David Gally at the University of Edinburgh (32). Escherichia coli O157:H7 strains EDL933 (ATCC 700927) and Sakai (ATCC BAA-460) were obtained from American Type Culture Collection (Manassas, VA). The remaining strains were from our culture collection and were isolated from humans or cattle in Canada. LSPA-6 genotyping of these strains was performed as described previously (63).
TABLE 1.
Escherichia coli O157:H7 strains included in the study
| Strain | LSPA-6 genotype | Source | Country of origin | Phage type | Reference(s) |
|---|---|---|---|---|---|
| 93-001 | 111111 | Human | United States | 14 | 22, 23 |
| EC19950361 | 111111 | Bovine | Canada | 87 | |
| EC20000122 | 111111 | Bovine | Canada | 31 | |
| ECI-577 | 111111 | Bovine | Canada | 4 | |
| EDL933 | 111111 | Human | United States | 21 | 43 |
| FDA 516 | 111111 | Human | United States | 21 | 22, 23 |
| FDA 518 | 111111 | Human | United States | 21 | 22, 23 |
| FDA 520 | 111111 | Human | United States | 1 | 22, 23 |
| FRIK 523 | 111111 | Human | United States | 34 | 22, 23 |
| Sakai | 111111 | Human | Japan | 32 | 16 |
| EC19920027 | 222222 | Bovine | Canada | 34 | |
| EC19920283 | 222222 | Human | Canada | 23 | |
| EC19970520 | 222222 | Bovine | Canada | 67 | |
| ECI-1433 | 222222 | Bovine | Canada | 23 | |
| ECI-633 | 222222 | Bovine | Canada | 23 | |
| ER6554 | 222222 | Human | Canada | 23 | |
| FRIK 1990 | 222222 | Bovine | United States | 54 | 22, 23 |
| FRIK 1999 | 222222 | Bovine | United States | 23 | 22, 23 |
| FRIK 920 | 222222 | Bovine | United States | 23 | 22, 23 |
| EC20011139 | 222222 | Bovine | Canada | 82 | |
| ECI-882 | 211111 | Human | Canada | 1 | |
| 278F1 | 211111 | Human | Canada | 2 | 21 |
| Zap0032 | 211111 | Bovine | Scotland | 8 | 32, 47, 48 |
| Zap0054 | 211111 | Bovine | Scotland | 32 | 32, 47, 48 |
| Zap0058 | 211111 | Human | Scotland | 87 | 32, 47, 48 |
| ECI-241 | 222212 | Human | Canada | 74 | |
| ER4511 | 222212 | Human | Canada | 74 | |
| ECI-240 | 222213 | Human | Canada | 54 | |
| FRIK 2001 | 222213 | Bovine | United States | 54 | 22, 23 |
| FRIK 1985 | 222223 | Bovine | United States | 45 | 22, 23 |
Preparation of suppression subtractive hybridization DNA libraries.
Bacteria were grown overnight in brain heart infusion broth (Difco, Becton Dickinson Microbiology Systems, Sparks, MD) in a 37°C shaker-incubator (200 rpm), and genomic DNA was extracted from harvested cells using the DNeasy tissue kit (QIAGEN, Valencia, CA).
SSH was performed using the Clontech PCR-Select bacterial genome subtraction kit (BD Biosciences, Palo Alto, CA). Advantage polymerase mix (BD Biosciences) was used during amplification steps. Two SSH experiments were performed, the lineage I strain (LSPA-6 genotype 111111) E. coli O157:H7 Sakai was subtracted with the bovine-derived lineage II strain FRIK920 (LSPA-6 genotype 222222), and a second human-derived lineage I strain 93-001 (LSPA-6 genotype 111111) was subtracted with bovine-derived lineage II strain EC19970520 (LSPA-6 genotype 222222). In addition to the RsaI-digested DNA recommended in the Clontech kit, each SSH experiment was also performed on AluI- and HaeIII-digested DNA to increase the diversity of DNA fragments obtained. The SSH DNA fragments isolated in these experiments were cloned into the pCR2.1-TOPO plasmid vector, using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and plated on LB agar (Difco) containing 50 μg/ml of ampicillin or kanamycin (Sigma-Aldrich Canada, Oakville, ON, Canada).
Sequence analysis of suppression subtractive hybridization DNA libraries.
Cloned DNA inserts were amplified by PCR using the primers M13 Forward, 5′-GTAAAACGACGGCCAG-3′, and M13 Reverse, 5′-CAGGAAACAGCTATGAC-3′ (Invitrogen). The amplified DNA was purified by passage through superfine Sephadex G-50 (Sigma) packed into a multiscreen 96-well filtration plate (Millipore, Billerica, MA). Sequencing was performed using M13 Forward and M13 Reverse primers with the DYEnamic ET terminator cycle sequencing kit (GE Healthcare, Piscataway, NJ), and sequencing products were purified by passage through superfine Sephadex G-50 plates as before. Sequence analysis was performed on a MegaBACE 500 capillary sequencer (GE Healthcare).
Base calling of sequence trace data was performed using the PHRED algorithm as implemented in the Interphace program (CodonCode Corporation, Dedham, MA). Base-called sequences from both SSH DNA libraries were pooled and analyzed with the SeqMan 5.05 sequence analysis program (DNAStar Inc., Madison, WI). This program removed vector and adaptor sequences and grouped sequences displaying at least 90% sequence homology into “contigs.” BlastN (1) searches were then performed on contig consensus sequences using the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and The Institute for Genomic Research (http://www.tigr.org/) BLASTN servers. Sequences that displayed at least 90% sequence identity to published chromosomal and plasmid DNA sequences of the lineage I E. coli O157:H7 Sakai strain (16, 29) were included in the final analysis. Genomic regions within the E. coli Sakai chromosome and plasmids that contained localized clusters of ORFs represented in the SSH libraries at high frequency were identified. The representation of these regions was significantly higher than the background frequency across the chromosomal and plasmid sequences, although the frequency at which individual ORFs within these regions were identified varied from 1 to 24 times.
PCR screening.
PCR assays were used to determine the distribution of these highly represented genome regions within E. coli O157:H7 strains of different LSPA-6 genotypes. Primers for these assays were designed to detect unique DNA sequences from ORFs within each region (see Table S1 in the supplemental material). All primer sequences were queried by BLASTN to ensure that they would not amplify multiple sequences within the E. coli Sakai genome. PCR screening reactions were performed against genomic DNA isolated from 10 LSPA-6 genotype 111111 E. coli O157:H7 strains, 10 LSPA-6 genotype 222222 strains, and 10 strains of other LSPA-6 genotypes. This strain collection is representative of the diversity within E. coli O157:H7 populations but is not representative of the proportions of LSPA-6 genotypes that naturally occur in humans and cattle. Larger numbers of LSPA-6 111111 and 222222 strains were included, as these genotypes were previously reported to display a biased distribution between human and bovine strains (63), and the proportions of human and bovine strains within each major genotype are different than previously described (63). Genome regions that were observed in these initial PCR screening reactions to be conserved in most or all LSPA-6 genotype 111111 strains but rare or absent in LSPA-6 genotype 222222 strains were tested by additional PCR assays against this same strain collection to identify the boundaries of the lineage I-specific regions.
All PCR assays were performed in duplicate in a 20-μl reaction volume containing 1× buffer II (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphates, 1 U AmpliTaq DNA polymerase (Applied Biosystems), 0.2 μM primers, and 0.5 ng genomic DNA template. All PCRs included an initial 2-min denaturation step at 94°C, followed by 30 cycles of 1-min denaturation at 94°C, 1-min annealing, and 1-min extension at 72°C, and a final 10-min extension at 72°C. Each PCR was tested against genomic DNA of E. coli O157:H7 Sakai strain as a positive control, E. coli K12 MC1061 as a negative control, and a no-template blank.
Cluster analysis of the distribution of lineage I-specific regions in E. coli O157:H7 strains by UPGMA.
Results of the preliminary PCR screening assays were converted into binary data to indicate the presence or absence of a single PCR marker for each of the different lineage I conserved regions within the strains tested. A distance matrix of the binary data was created in PAUP version 4.0b10, and the distance matrix was imported into MEGA 3.1 (26) to generate an unweighted-pair group method using average linkages (UPGMA) dendrogram.
RESULTS AND DISCUSSION
Analysis of SSH-derived sequences.
Our experimental approach was to first identify candidate genomic regions that are present in lineage I strains but absent in lineage II strains. Candidate regions were identified using SSH from representative strains, and the candidate regions were then distinguished as strain specific versus lineage specific based on their conservation across a larger, representative strain set. To this end, two SSH experiments were conducted. In one library, the lineage I strain (LSPA-6 genotype 111111) E. coli O157:H7 Sakai was subtracted with the bovine-derived lineage II strain FRIK920 (LSPA-6 genotype 222222), and the second library used a human-derived lineage I strain 93-001 (LSPA-6 genotype 111111) subtracted with bovine-derived lineage II strain EC19970520 (LSPA-6 genotype 222222). A total of 1,155 clones prepared from SSH DNA libraries of RsaI-, AluI-, and HaeIII-digested DNA were sequenced. Sequences were assembled into 754 contigs, with each sequence possessing 90% or greater sequence identity to other contributing sequences in the same contig. BlastN search results of the contig consensus sequences revealed that 85.1% of the contigs possessed sequences with greater than 90% sequence identity to one or more of the ORFs present in the lineage I representative strain E. coli O157:H7 Sakai chromosome or pO157 and pOSAK1 plasmid sequences (16, 29). This was expected, since E. coli Sakai was used as the tester for preparing one of the SSH DNA libraries. The frequency at which sequences possessing 90% or greater sequence identity to each E. coli Sakai chromosomal ORF was identified, either partly or wholly, within the SSH libraries was enumerated, and 24 putative regions of genomic difference for lineage I (RDI) identified on the E. coli Sakai chromosome and pO157 and pOSAK1 plasmids were selected for further testing (Fig. 1). These regions consisted of clusters of 1 to 47 ORFs that were each identified in the SSH libraries from 1 to 24 times. The locations of SSH sequences within pO157 and pOSAK1 plasmids are not shown, but plasmid ORFs with 90% or greater sequence identity to SSH library sequences were evenly distributed across pOSAK1 (GenBank accession number NC_002127) and concentrated in the region of a putative reverse transcriptase within pO157 (GenBank accession number AB011549).
FIG. 1.
Frequency of detection of sequences homologous to Escherichia coli Sakai (GenBank accession no. BA000007) ORFs within SSH libraries.
Identification of lineage I conserved regions.
Having identified candidate regions of the genome that are unique to lineage I strains, the next step was to identify which of the candidate RDI were conserved across multiple lineage I strains and likewise absent in multiple lineage II strains. While SSH is a versatile tool for identification of sequence differences between bacterial strains, the time required to obtain and analyze the subtracted sequences is substantial, thus limiting the number of strains that can be analyzed. To confirm the lineage-specific distribution of the RDI, 30 different E. coli O157:H7 strains were subsequently tested for the presence of the RDI by PCR. This confirmatory strain set comprised 10 lineage I strains of LSPA-6 genotype 111111, 10 lineage II strains of LSPA-6 genotype 222222 strains, and 10 lineage II strains having other LSPA-6 genotypes. Each PCR-based RDI detection assay was performed using a single PCR assay designed to detect DNA sequences within the 24 RDI. Twelve of the 24 RDI were conserved in at least seven of the LSPA-6 genotype 111111 strains but were found in two or fewer of the genotype 222222 strains (Table 2). Amplicons for each PCR assay corresponded in size to that predicted for E. coli Sakai. These conserved RDI (CRDI) were also absent from other LSPA-6 genotypes, except for genotype 211111 strains. Their presence within LSPA-6 genotype 211111 strains was variable.
TABLE 2.
Results of preliminary screening of ORF clusters identified in SSH libraries
| RDI | ORF(s) represented in SSH libraries | ORF(s) targeted by primers | No. positive for LSPA-6 genotype
|
Genotype 111111 conserved? | |||
|---|---|---|---|---|---|---|---|
| 111111 (n = 10) | 222222 (n = 10) | 211111 (n = 5) | Other (n = 5) | ||||
| 14 | ECs0237-ECs0241 | ECs0238 and -9 | 10 | 0 | 5 | 0 | Yes |
| 16 | ECs0275-ECs0284 | ECS0281 | 10 | 0 | 5 | 0 | Yes |
| 27 | ECs0377-ECs0379 | ECs0377 | 10 | 10 | 5 | 5 | No |
| 52 | ECs0759-ECs0760 | ECs0760 | 10 | 9 | 5 | 5 | No |
| 67 | ECs1114-ECs1121 | ECs1120 | 10 | 10 | 5 | 5 | No |
| 69a | ECs1158-ECs1173 | ECs1164 | 8 | 0 | 3 | 0 | Yes |
| 69b | ECs1186-ECs1195 | ECs1191 | 7 | 0 | 0 | 0 | Yes |
| 69c | ECs1205-ECs1251 | ECs1223 | 10 | 2 | 3 | 0 | Yes |
| 72 | ECs1377-ECs1393 | ECS1382 | 10 | 0 | 4 | 0 | Yes |
| B1 | ECs1425 | ECs1425 | 10 | 10 | 5 | 5 | No |
| 78 | ECs1576-ECs1600 | ECs1588 and -9 | 8 | 0 | 0 | 0 | Yes |
| 82/83 | ECs1687-ECs1705 | ECs1698 | 10 | 0 | 3 | 0 | Yes |
| 91/92/93 | ECs1929-ECs1958 | ECs1954 | 10 | 0 | 0 | 0 | Yes |
| B2 | ECs1997-ECs1998 | ECs1998 | 10 | 10 | 5 | 5 | No |
| 126/127 | ECs2775-ECs2776 | ECs2775 | 10 | 10 | 5 | 5 | No |
| 153 | ECs2976-ECs2981 | ECs2979 | 10 | 0 | 2 | 0 | Yes |
| 274a | ECs4942-ECs4946 | ECs4943 | 3 | 5 | 0 | 3 | No |
| 274b | ECs4957-ECs4960 | ECs4960 | 1 | 4 | 0 | 2 | No |
| 274c | ECs4998-ECs5005 | ECs5000 | 10 | 9 | 5 | 5 | No |
| 286 | ECs5242-ECs5252 | ECs5250 | 9 | 0 | 4 | 0 | Yes |
| pO157-RTa | pO157, sopB-RT (p36) | Upstream of RT | 9 | 0 | 0 | 0 | Yes |
| pO157-katP | pO157, katP | katP | 9 | 10 | 5 | 5 | No |
| pO157-etpF | pO157, etpF | etpF | 10 | 10 | 5 | 5 | No |
| pOSAK1 | pOSAK1 | Unknown | 5 | 0 | 1 | 0 | No |
RT, putative reverse transcriptase.
To determine if the lineage-conserved distribution of these regions was maintained when tested over a larger population of E. coli O157:H7 strains, the representative PCR assay from each CRDI (except CRDI 78) was tested against a larger set of 119 E. coli O157:H7 isolates of different LSPA-6 genotypes. Sequences recognized by these assays were present in 81 to 100% of LSPA-6 genotype 111111 strains but were found in ≤7% of LSPA-6 genotype 222222 strains, confirming the earlier screening results for these regions. Complete PCR assay results for the preliminary screening of 30 strains for the presence of the 24 RDI regions and the subsequent testing of 119 strains for the presence of the 12 CRDI regions are shown in Tables S2 and S3, respectively, of the supplemental material.
Localization of genomic boundaries of CRDI.
While the SSH and PCR screening results suggest 12 CRDI exist across the E. coli Sakai chromosome and pO157 plasmid, it was not known if the E. coli Sakai ORFs identified by SSH accurately reflected the boundaries of these CRDI as they exist in lineage I strains. Subsequent PCR screening assays were performed to confirm the boundaries of the CRDI by testing for the presence of E. coli Sakai DNA segments flanking the CRDI among lineage II strains (Table 3) (16, 29). All but one of the estimated full-length CRDI were localized within E. coli Sakai S-loops and the corresponding E. coli EDL933 O-islands. These E. coli O157:H7 chromosomal regions are absent from E. coli K-12 and are thought to have arisen by horizontal gene transfer (16). The remaining CRDI was located on the pO157 virulence plasmid of E. coli O157:H7 (7, 29). Complete PCR results for these assays are shown in Table S2 of the supplemental material. Amplicons from each of these additional PCR assays corresponded in size to that predicted for E. coli Sakai.
TABLE 3.
Results of PCR screening to determine boundaries of CRDI in E. coli O157:H7 strains
| CRDI | ORF(s) targeted by primers | No. positive for LSPA-6 genotype
|
LSPA-6 genotype 111111 conserved? | |||
|---|---|---|---|---|---|---|
| 111111 (n = 10) | 222222 (n = 10) | 211111 (n = 5) | Other (n = 5) | |||
| 14 | ECs0234 and -5 | 10 | 10 | 5 | 5 | No |
| ECs0238 and -9 | 10 | 0 | 5 | 0 | Yes | |
| ECs0240 | 10 | 0 | 5 | 0 | Yes | |
| ECs0243 and -4 | 10 | 10 | 5 | 5 | No | |
| 16 | ECs0270 | 10 | 10 | 5 | 5 | No |
| ECs0271 and -2 | 10 | 0 | 5 | 0 | Yes | |
| ECs0276 | 10 | 0 | 3 | 0 | Yes | |
| ECS0281 | 10 | 0 | 5 | 0 | Yes | |
| ECs0282 and -3 | 10 | 10 | 5 | 5 | No | |
| ECs0284 | 10 | 10 | 5 | 5 | No | |
| 69a | ECs1158 | 10 | 10 | 5 | 5 | No |
| ECs1162 and -3 | 10 | 0 | 0 | 0 | Yes | |
| ECs1164 | 8 | 0 | 3 | 0 | Yes | |
| ECs1165 | 9 | 0 | 3 | 0 | Yes | |
| ECs1168 | 10 | 1 | 1 | 0 | Yes | |
| 69b | ECs1180 and -2 | 3 | 10 | 3 | 4 | No |
| ECs1186 and -8 | 3 | 0 | 0 | 0 | No | |
| ECs1191 | 7 | 0 | 0 | 0 | Yes | |
| ECs1192 and -4 | 10 | 9 | 3 | 3 | No | |
| 69c | ECs1217 and -8 | 10 | 10 | 5 | 5 | No |
| ECs1219 | 10 | 2 | 3 | 0 | Yes | |
| ECs1223 | 10 | 2 | 3 | 0 | Yes | |
| ECs1251 | 10 | 0 | 0 | 0 | Yes | |
| ECs1252 | 10 | 10 | 5 | 5 | No | |
| 72 | ECs1374 and -6 | 10 | 5 | 0 | 5 | No |
| ECs1377 | 10 | 0 | 3 | 0 | Yes | |
| ECS1382 | 10 | 0 | 4 | 0 | Yes | |
| ECs1389 and -90 | 10 | 0 | 5 | 0 | Yes | |
| ECs1394 and -5 | 10 | 10 | 5 | 5 | No | |
| 78 | ECs1573 and -4 | 8 | 10 | 5 | 5 | No |
| ECs1575 and -6 | 8 | 10 | 5 | 5 | No | |
| ECs1582 and -3 | 8 | 3 | 1 | 0 | Yes | |
| ECs1584 | 8 | 0 | 1 | 1 | Yes | |
| ECs1588 and -9 | 8 | 0 | 0 | 0 | Yes | |
| ECs1590 | 8 | 0 | 1 | 1 | Yes | |
| ECs1595 | 8 | 0 | 1 | 1 | Yes | |
| ECs1598 | 8 | 0 | 1 | 1 | Yes | |
| ECs1599 and -1600 | 8 | 0 | 0 | 0 | Yes | |
| 82/83 | ECs1687 | 10 | 10 | 5 | 5 | No |
| ECs1691 | 10 | 0 | 4 | 0 | Yes | |
| ECs1698 | 10 | 0 | 3 | 0 | Yes | |
| ECs1705 | 10 | 0 | 4 | 0 | Yes | |
| ECs1706 | 10 | 10 | 4 | 5 | No | |
| ECs1707 | 9 | 9 | 4 | 5 | No | |
| 91/92/93 | ECs1927 | 10 | 10 | 5 | 5 | No |
| ECs1931 and -2 | 10 | 0 | 0 | 0 | Yes | |
| ECs1937 to -40 | 10 | 0 | 0 | 0 | Yes | |
| ECs1945 | 10 | 2 | 0 | 1 | No | |
| ECs1953 | 10 | 0 | 0 | 0 | Yes | |
| ECs1954 | 10 | 0 | 0 | 0 | Yes | |
| ECs1957 | 10 | 0 | 0 | 0 | Yes | |
| 153 | ECs2979 | 10 | 0 | 2 | 0 | Yes |
| 286 | ECs5240 and -1 | 7 | 10 | 5 | 5 | No |
| ECs5242 | 9 | 0 | 5 | 0 | Yes | |
| ECs5250 | 9 | 0 | 4 | 0 | Yes | |
| ECs5252 | 9 | 0 | 5 | 0 | Yes | |
| ECs5253 | 10 | 10 | 5 | 5 | No | |
| ECs5254 to -6 | 10 | 10 | 5 | 5 | No | |
| PO157-RT | pO157, sopB | 9 | 10 | 5 | 5 | No |
| pO157, upstream of reverse transcriptase | 9 | 0 | 0 | 0 | Yes | |
| pO157, reverse transcriptase | 9 | 0 | 0 | 0 | Yes | |
| pO157, p36 | 9 | 10 | 5 | 5 | No | |
Relationships between CRDI and inferred phylogeny of different Escherichia coli O157:H7 LSPA-6 genotypes.
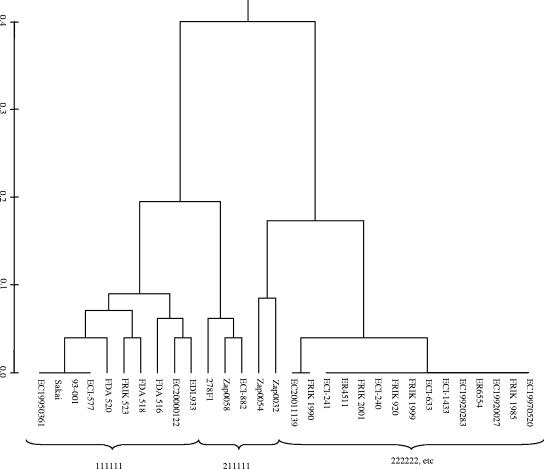
To test for congruence between presence/absence of CRDI and phylogeny of the strains, cluster analysis was performed on the different strains using a distance matrix developed from comparison of the presence/absence of the CRDI. The LSPA-6 genotype was then superimposed onto the corresponding neighbor-joining dendrogram (Fig. 2). Strains of LSPA-6 genotypes 111111 and 222222 clustered separately on the basis of the CRDI identified in this study. This would be expected based on the definition of the CRDI, which were selected on the basis of their presence in LSPA-6 111111 strains but not in LSPA-6 222222 strains. LSPA-6 genotype 211111 strains, however, clustered with both of the major groups, a result that is consistent with their variable CRDI content. This variability suggests that the folD2 allele (the only lineage II allele in the 211111 genotype) may have arisen independently in different genomic backgrounds or that the CRDI identified in this study emerged differentially in descendants of the original LSPA-6 211111 strains. Given that the folD2 allele is thought to be the result of a tandem duplication event (23), it seems more plausible that that it is a polyphyletic allele that arose from a simple mutation or lateral transfer event as opposed to the hypothesis that multiple events resulted in differential CRDI distribution within isolates of this genotype. Strains from the remainder of the lineage II LSPA-6 genotypes had profiles that were very similar to genotype 222222 strains.
FIG. 2.
UPGMA dendrogram displaying relationship between LSPA-6 genotype and the presence of CRDI in Escherichia coli O157:H7 strains.
Functional categorization of genes within CRDI.
The results of the PCR assays indicated that the CRDI were strongly associated with LSPA-6 111111 strains and rare or absent in LSPA-6 222222 strains. Because the LSPA-6 genotypes 111111 and 222222 show statistically significant biases in their distribution among strains isolated from human clinical samples and bovine fecal samples (63), it is possible that genes within the CRDI could contribute to the apparently different ecological distributions of the LSPA-6 genotypes that have been observed. To identify genes within CRDI that could contribute to host specificity, we conducted functional classification of the ORFs present within the CRDI, based on annotation entries, identification of homologous DNA sequences by BLASTN searches, and Pfam identification (4) of common protein domains and families using hidden Markov models (http://www.sanger.ac.uk/Software/Pfam/). The results from this set of experiments are summarized in Table 4.
TABLE 4.
Summary of potential virulence factors contained within CRDI in E. coli O157:H7 strains
| CRDI | Genetic environment | Potential virulence factor(s) identified within LSPA-6 genotype 111111 CR |
|---|---|---|
| 14 | Rhs elements | None identified |
| 16 | Sp1 prophage | None identified |
| 69a | Sp5 prophage | ECs1170, gene for putative protein with DksA/TraR C4-type zinc finger domain (42) |
| 69b | Sp5 prophage | None identified |
| 69c | Sp5 prophage, transposon | ECs1236, gene for putative outer membrane precursor protein with Ail/Lom protein domain (3, 35); ECs1250, gene for putative protein with DksA/TraR C4 type zinc finger domain (42) |
| 72 | SpLE1 prophage-like element, transposon | ECs1382, similar to hemolysin-activating protein hecB gene of Neisseria meningitidis (57); ECs1386, similar to immunoglobulin-binding regulator ibrA gene of E. coli ECOR-9 (50); ECs1387, similar to immunoglobulin-binding regulator ibrB gene of E. coli ECOR-9 (50); ECs1388, similar to plasmid-encoded regulator perC gene of EPEC (11); ECs1391, similar to bundle-forming pilus bfpM gene of EPEC EAF plasmid (55) |
| 78 | Sp7 prophage | ECs1588, similar to plasmid-encoded regulator perC gene of EPEC (11) |
| 82/83 | Transposon | ECs1693, homologue of prrA of a proposed iron transport system in E. coli CFT073 (15); ECs1694, homologue of the modD of a proposed iron transport system in E. coli CFT073 (15); ECs1695, homologue of yc73 of proposed iron transport system in E. coli CFT073 (15); ECs1696, homologue of yc73 of proposed iron transport system in E. coli CFT073 (15); ECs1697, homologue of fepC of proposed iron transport system in E. coli CFT073 (15); ECs1698, homologue of iron transport permease FecD of E. coli CFT073 (60); ECs1699, homologue of putative ATP-binding protein of ABC transporter in Shigella flexneri 2a strain 301 (18) |
| 91/92/93 | Sp10 prophage, transposon | None identified |
| 153 | Sp15 prophage | None identified |
| 286 | SpLE5 prophage-like element, transposon | ECs5250, required for colonization of calves (9); ECs5252, similar to BamHI control element in Bacillus amyloliquefaciens (6) |
| pO157-RTa | pO157 plasmid | None identified |
RT, putative reverse transcriptase.
The most striking characteristic of the ORFs contained within the CRDI is their location with respect to mobile genetic elements in E. coli Sakai (16). Nine of the 12 CRDI were within Sp1, Sp5 (stx2), Sp7, Sp10, and Sp15 prophage and SpLE1 and SpLE5 prophage-like elements (Table 4). One of the CRDI was surrounded by Rhs element genes, mobile genetic elements that were originally identified based on their association with recA-dependent intrachromosomal recombination (2, 10, 28). Five of the CRDI contained or were located in close proximity to transposase genes. One CRDI was found on the pO157 virulence plasmid (7, 29). Not surprisingly, a large proportion of the ORFs identified within the CRDI were bacteriophage structural and regulatory genes, Rhs genes, and transposases. These elements would be expected to promote genomic recombination and rearrangement and may contribute to bacterial virulence through associated structural and regulatory functions (58, 59).
Lineage I-specific CRDI 69 and 153 are located within E. coli Sakai Sp5 and Sp15 prophages, respectively (Table 4). These two prophages contain the structural genes for Stx2 and Stx1, respectively. The Sp5 prophage has been shown to be highly variable in both its genomic structure and integration sites (37, 38, 51). The presence of the stx2 gene is strongly associated with virulence in E. coli O157:H7 (5, 16, 24, 41). In a recent study, it was reported that the Q antiterminator gene, which regulates toxin gene expression in stx2 prophage, differs between E. coli O157:H7 strains of different OBGS lineages and that these differences are related to toxin production (27). However, the Q antiterminator gene was not among the ORFs identified within the SSH libraries in this study. Only a small region located upstream of the stx1 and Q antiterminator genes in the Stx1-converting Sp15 prophage was identified in the SSH libraries. PCR assays confirmed that at least one of these ORFs, ECs2979, was conserved in lineage I strains and absent in lineage II strains.
A cluster of lineage I-specific ORFs within CRDI 82/83 may represent an iron uptake system. Virulence genes, such as E. coli O157:H7 stx1 genes (8), are often iron regulated. The ORFs ECs1693 to ECs1697 are highly homologous to the prrA-modD-yc73-fepC gene cluster located on the pyelonephritis and cystitis pathogenicity island of uropathogenic E. coli CFT073 (15). This gene cluster was proposed to be involved in iron uptake in E. coli CFT073 (64). The fepC gene was reported to be present in O157 EHEC isolates and absent from non-O157 isolates, although the OBGS lineage of the O157 EHEC strains tested in this study was not examined (40). This gene is also found in EAEC2 and DAEC2 phylogenetic groups of enteroaggregative E. coli (EAEC) and diffusely adherent E. coli (DAEC), respectively, and it has been proposed that these groups might represent hypervirulent isolates of EAEC and DAEC (40). Immediately following the prrA-modD-yc73-fepC gene cluster are ECs1698, which is homologous to the Fe(III) dicitrate transport system permease FecD protein of E. coli CFT073 (60), and ECs1699, which is homologous to a putative ATP-binding protein of the ABC transporter in Shigella flexneri 2a strain 301(18).
The ORF ECs1382 within CRDI 72 has 34% amino acid similarity at its amino terminus to the gene for the hemolysin activation protein HecB of Neisseria meningitidis (57), which directs the translocation of hemolysin A (HlyA) across cytoplasmic and cell membranes (25), and it possesses two copies of a 20-residue repeat found in the Bordetella pertussis filamentous hemagglutinin family of adhesins (49). These homologies suggest that ECs1382 might contribute to virulence. In a recent investigation of the distribution of Z1640 (the E. coli EDL933 homologue of ECs1382) within different E. coli serotypes, intact ECs1382/Z1640 was associated with serotypes that cause hemolytic uremic syndrome and outbreaks of human illness, while fragmented ECs1382/Z1640 was found in nonepidemic human disease-associated strains of serotypes O91:H21 and O113:H21 and animal-associated Shiga toxin-producing E. coli serotypes not associated with human disease (52).
A number of ORFs were identified within the SSH libraries that displayed homology to regulatory genes. These genes could affect virulence of E. coli O157:H7 by regulating expression of effector genes directly involved in pathogenesis. The genome sequence of E. coli O157:H7 Sakai contains five genes homologous to the plasmid-encoded LEE regulatory protein PerC, which is produced by certain enteropathogenic E. coli (EPEC) strains (16, 17, 39, 44). Two of these perC homologue (pch) genes, ECs1388 of CRDI 72 and ECs1588 of CRDI 78, encode putative proteins with 25% and 39% sequence identity, respectively, to EPEC PerC. ECs1388 and ECs1588 were identified within the SSH libraries and confirmed by PCR to be present in lineage I strains and absent from lineage II strains. While other pch genes have been demonstrated to modulate expression of LEE transcription units in E. coli O157:H7 (17), deletion or overexpression of ECs1388 or ECs1588 has not, and their function, if any, in gene regulation remains to be determined (44).
Other potential regulatory genes identified within the SSH libraries include ECs1386 and ECs1387 of CRDI 72, which display 80 and 85% identity to immunoglobulin-binding regulator genes ibrA and ibrB of E. coli ECOR-9. These genes regulate production of Escherichia coli immunoglobulin-binding (eib) genes in this strain (50). The ORF ECs5252 of CRDI 286 encodes a putative transcriptional regulator with homology to the BamHI control element of Bacillus amyloliquefaciens (6). Finally, the ORFs ECs1170 of CRDI 69a and ECs1250 of CRDI 69c encode putative proteins with a pfam01258:Zn-dskA_traR domain (30). The dskA gene of E. coli regulates rRNA transcription (19, 42).
Several additional ORFs were identified within the CRDI with the potential to act as virulence factors. The ORF ECs1236 of CRDI 69c encodes a putative outer membrane precursor protein with a Pfam06316:Ail_Lom domain (30). Proteins with this domain include the Ail protein of Yersinia enterocolitica, which contributes to invasion of cultured cell lines (35), and the Lom bacteriophage protein, which has been shown to confer serum resistance (3). E. coli O157:H7, however, is not invasive, and so the role of this invasin-like protein is unclear. The amino terminus of ORF ECs1391 of CRDI 72 displayed high sequence homology to the bundle-forming pilus (BfpM) protein of EPEC (55). This gene is not required for formation of bundle-forming pili in EPEC and no other bfp homologues exist in the E. coli O157:H7 genome, and so it is difficult to postulate what its role might be (55). The ECs5250 of CRDI 286 was shown to be required for colonization of calves (9), but its function is unknown. The CRDI pO157-RT includes a putative reverse transcriptase on the pO157 virulence plasmid of E. coli O157:H7 (7, 29), but the function of the putative reverse transcriptase in E. coli O157:H7 is also unknown. Lastly, several hypothetical protein genes were also identified on prophage or prophage-like element-associated CRDI. Again, the effects of these genes on bacterial virulence, if any, are unknown.
The paucity of human clinical isolates within E. coli O157:H7 LSPA-6 genotype 222222 and some subsets of OBGS lineage II suggests but does not prove that these strains lack virulence factors present in other E. coli O157:H7 lineages (22, 63). The identification of potential virulence factors in LSPA-6 111111 strains that are lacking in LSPA-6 222222 strains supports this hypothesis, but there is no direct evidence that these regions are involved in pathogenicity. Further functional and epidemiological analyses of genes within the CRDI are essential to elucidate the impact that genome evolution has had on the virulence characteristics of populations emerging within the O157:H7 clonal complex.
CRDI support the hypothesis that lineage I may be the ancestral state of contemporary O157:H7.
Based on previous studies, it has been proposed that lineage I isolates represent the ancestral state of O157:H7, whereas lineage II isolates are derived (22, 63). This conclusion was based largely upon findings that several lineage I-specific genome segments or alleles are shared with K-12 and other E. coli strains (63). Our findings with CRDI are consistent with this hypothesis. As discussed above, many of the CRDI can be found in other E. coli strains, including distantly related K-12 and UPEC strains. The simplest explanation for their absence in lineage II strains is that they were lost during the divergence of lineage II populations.
Many of the ORFs associated with the CRDI regions were found in close proximity to bacteriophage, transposon, and Rhs element genes. Given the presence of numerous copies of these elements in E. coli O157:H7 genomes (16, 43), it seems highly likely that genome evolution occurred rapidly by movement of these elements or by recombination events occurring within or near these elements. Genomic diversity in human strains of E. coli Sakai was previously shown by comparative genomic hybridization and whole-genome PCR scanning to be strongly associated with the presence of bacteriophage (37, 38).
In a recent publication by Wick et al. (62), a microarray was used to study the conservation of E. coli O157:H7 genes in strains representing intermediates in the proposed evolution of E. coli O157:H7 and E. coli O55:H7. The microarray results indicated that the majority of the ORFs identified in this study within CRDI 16, 69a, 69b, 69c, 72, 78, 91/92/93, 153, and 286 were acquired recently by E. coli O157:H7 SOR− GUD− strains. All of these CRDI were located on bacteriophage, suggesting that these differences were a result of bacteriophage excision or recombination.
In conclusion, several CRDI that are conserved in LSPA-6 genotype 111111 strains but are not found in genotype 222222 strains were identified, supporting the existence of two genomic lineages of E. coli O157:H7 strains. These CRDI contain a number of potential virulence factors, including a putative hemolysin activation protein, a possible iron transport system, and several regulatory genes. These potential virulence factors warrant further study to determine their contribution to the pathogenicity of E. coli O157:H7 strains.
Supplementary Material
Acknowledgments
We thank Mohamed Karmali of the Public Health Agency of Canada in Guelph, Ontario, and Bruce Ciebin of the Ministry of Health in Toronto, Ontario, for their contributions of human E. coli O157:H7 strains.
This research was supported by Health Canada's Genomics Initiative and Office of Biotechnology and Science and by the Public Health Agency of Canada.
Footnotes
Published ahead of print on 20 October 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacheller, S., E. Gilson, M. Hofnung, and C. W. Hill. 1996. Repeated sequences, p. 2012-2040. In C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 3.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871-874. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, J. E., P. D. Nathan, D. Landry, L. A. Sznyter, P. Waite-Rees, C. L. Ives, L. S. Moran, B. E. Slatko, and J. S. Benner. 1991. Characterization of the cloned BamHI restriction modification system: its nucleotide sequence, properties of the methylase, and expression in heterologous hosts. Nucleic Acids Res. 19:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziva, F., P. M. van Diemen, M. P. Stevens, A. J. Smith, and T. S. Wallis. 2004. Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology 150:3631-3645. [DOI] [PubMed] [Google Scholar]
- 10.Feulner, G., J. A. Gray, J. A. Kirschman, A. F. Lehner, A. B. Sadosky, D. A. Vlazny, J. Zhang, S. Zhao, and C. W. Hill. 1990. Structure of the rhsA locus from Escherichia coli K-12 and comparison of rhsA with other members of the rhs multigene family. J. Bacteriol. 172:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouveia, S., M. E. Proctor, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1998. Genomic comparisons and Shiga toxin production among Escherichia coli O157:H7 isolates from a day care center outbreak and sporadic cases in southeastern Wisconsin. J. Clin. Microbiol. 36:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin, P. M. 1998. Epidemiology of Shiga toxin-producing Escherichia coli infections in humans in the United States, p. 15-22. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 14.Gross, E., and S. Slauson. 2004. Update on emerging infections: news from the Centers for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food-selected sites, United States, 2003. Ann. Emerg. Med. 44:532-536. [DOI] [PubMed] [Google Scholar]
- 15.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 17.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357-2571. [DOI] [PubMed] [Google Scholar]
- 18.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 185:3558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karch, H., and M. Bielaszewska. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 39:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA. 96:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J., J. Nietfeldt, J. Ju, J. Wise, N. Fegan, P. Desmarchelier, and A. K. Benson. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, beta-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183:6885-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleanthous, H., H. R. Smith, S. M. Scotland, R. J. Gross, B. Rowe, C. M. Taylor, and D. V. Milford. 1990. Haemolytic uraemic syndromes in the British Isles, 1985-8: association with verocytotoxin producing Escherichia coli. Part 2: microbiological aspects. Arch. Dis. Child. 65:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koronakis, V., P. Stanley, E. Koronakis, and C. Hughes. 1992. The HlyB/HlyD-dependent secretion of toxins by gram-negative bacteria. FEMS Microbiol. Immunol. 5:45-53. [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 27.Lejeune, J. T., S. T. Abedon, K. Takemura, N. P. Christie, and S. Sreevatsan. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 10:1482-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, R. J., M. Capage, and C. W. Hill. 1984. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J. Mol. Biol. 177:1-18. [DOI] [PubMed] [Google Scholar]
- 29.Makino, K., K. Ishii, T. Yasunaga, M. Hattori, K. Yokoyama, C. H. Yutsudo, Y. Kubota, Y. Yamaichi, T. Iida, K. Yamamoto, T. Honda, C. G. Han, E. Ohtsubo, M. Kasamatsu, T. Hayashi, S. Kuhara, and H. Shinagawa. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 30.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 32.McNally, A., A. J. Roe, S. Simpson, F. M. Thomson-Carter, D. E. Hoey, C. Currie, T. Chakraborty, D. G. Smith, and D. L. Gally. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect. Immun. 69:5107-5114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michino, H., K. Araki, S. Minami, T. Nakayami, Y. Ejima, K. Hiroe, H. Tanaka, N. Fujita, S. Usami, M. Yonekawa, S. Sadamoto, and N. Sakai. 1998. Recent outbreaks of infections caused by Escherichia coli O157:H7 in Japan, p. 73-81. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 35.Miller, V. L., K. B. Beer, G. Heusipp, B. M. Young, and M. R. Wachtel. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 41:1053-1062. [DOI] [PubMed] [Google Scholar]
- 36.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogura, Y., K. Kurokawa, T. Ooka, K. Tashiro, T. Tobe, M. Ohnishi, K. Nakayama, T. Morimoto, J. Terajima, H. Watanabe, S. Kuhara, and T. Hayashi. 2006. Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai oligo DNA microarray and the whole genome PCR scanning. DNA Res. 13:3-14. [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA. 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okeke, I. N., J. A. Borneman, S. Shin, J. L. Mellies, L. E. Quinn, and J. B. Kaper. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okeke, I. N., I. C. Scaletsky, E. H. Soars, L. R. Macfarlane, and A. G. Torres. 2004. Molecular epidemiology of the iron utilization genes of enteroaggregative Escherichia coli. J. Clin. Microbiol. 42:36-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostroff, S. M., P. I. Tarr, M. A. Neill, J. H. Lewis, N. Hargrett-Bean, and J. M. Kobayashi. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J. Infect. Dis. 160:994-998. [DOI] [PubMed] [Google Scholar]
- 42.Paul, B. J., M. M. Barker, W. Ross, D. A. Schneider, C. Webb, J. W. Foster, and R. L. Gourse. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311-322. [DOI] [PubMed] [Google Scholar]
- 43.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 44.Porter, M. E., P. Mitchell, A. Free, D. G. Smith, and D. L. Gally. 2005. The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J. Bacteriol. 187:458-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 46.Robins-Browne, R., E. Elliott, and P. Desmarchelier. 1998. Shiga toxin-producing Escherichia coli in Australia, p. 66-72. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 47.Roe, A. J., S. W. Naylor, K. J. Spears, H. M. Yull, T. A. Dransfield, M. Oxford, I. J. McKendrick, M. Porter, M. J. Woodward, D. G. Smith, and D. L. Gally. 2004. Co-ordinate single-cell expression of LEE4- and LEE5-encoded proteins of Escherichia coli O157:H7. Mol. Microbiol. 54:337-352. [DOI] [PubMed] [Google Scholar]
- 48.Roe, A. J., H. Yull, S. W. Naylor, M. J. Woodward, D. G. Smith, and D. L. Gally. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71:5900-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA. 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandt, C. H., J. E. Hopper, and C. W. Hill. 2002. Activation of prophage eib genes for immunoglobulin-binding proteins by genes from the IbrAB genetic island of Escherichia coli ECOR-9. J. Bacteriol. 184:3640-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen, S., M. Mascarenhas, K. Rahn, J. B. Kaper, and M. A. Karmali. 2004. Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect. Immun. 72:1496-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, H. R., B. Rowe, G. K. Adak, and W. J. Reilly. 1998. Shiga toxin (verocytotoxin)-producing Escherichia coli in the United Kingdom, p. 49-58. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 55.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C. Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spika, J. S., R. Khakhria, P. Michel, D. Milley, J. Wilson, and J. Waters. 1998. Shiga toxin-producing Escherichia coli infections in Canada, p. 23-29. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 57.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 59.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA. 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, Z., J. Kovar, J. Kim, J. Nietfeldt, D. R. Smith, R. A. Moxley, M. E. Olson, P. D. Fey, and A. K. Benson. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye, C., and J. Xu. 2001. Prevalence of iron transport gene on pathogenicity-associated island of uropathogenic Escherichia coli in E. coli O157:H7 containing Shiga toxin gene. J. Clin. Microbiol. 39:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.