Abstract
Paramyxoviruses are a leading cause of childhood illness worldwide. A recently discovered paramyxovirus, human metapneumovirus (hMPV), has been studied by our group in order to determine the structural relevance of its fusion (F) protein to other well-characterized viruses utilizing type I integral membrane proteins as fusion aids. Sequence analysis and homology models suggested the presence of requisite heptad repeat (HR) regions. Synthetic peptides from HR regions 1 and 2 (HR-1 and -2, respectively) were induced to form a thermostable (melting temperature, ∼90°C) helical structure consistent in mass with a hexameric coiled coil. Inhibitory studies of hMPV HR-1 and -2 indicated that the synthetic HR-1 peptide was a significant fusion inhibitor with a 50% inhibitory concentration and a 50% effective concentration of ∼50 nM. Many viral fusion proteins are type I integral membrane proteins utilizing the formation of a hexameric coiled coil of HR peptides as a major driving force for fusion. Our studies provide evidence that hMPV also uses a coiled-coil structure as a major player in the fusion process. Additionally, viral HR-1 peptide sequences may need further investigation as potent fusion inhibitors.
Viruses are a leading cause of lower respiratory tract infection in children worldwide, with significant associated morbidity and mortality. Previously identified major pathogens include respiratory syncytial virus (RSV), parainfluenza viruses (PIVs), and influenza and measles viruses, all of which are associated with clinical syndromes of severe lower respiratory tract disease, such as bronchiolitis, pneumonia, and laryngotracheobronchitis. In 2001 human metapneumovirus (hMPV) was discovered by Dutch scientists (35). Samples collected longitudinally from 1976 to 2001 at the Vanderbilt Vaccine Clinic showed that 12% of lower respiratory tract diseases were attributable to hMPV (42). Additional studies have further established the importance of hMPV as a respiratory pathogen (3, 11). Subsequent genetic analysis classified hMPV as a member of the Pneumovirus subgroup within the Paramyxoviridae family (34).
The Paramyxoviridae family includes two subfamilies: the paramyxoviruses and the pneumoviruses. Paramyxoviruses include all four parainfluenza virus types, Sendai virus, mumps virus, Hendra virus, Newcastle disease virus, simian virus 5 (SV5), the morbilliviruses measles virus and canine distemper virus, and others. The Pneumovirus subfamily consists of the pneumoviruses RSV and pneumonia virus of mouse and the metapneumoviruses avian metapneumovirus and hMPV (16). Paramyxoviruses contain two major surface glycoproteins critical for viral replication and survival. The attachment protein (G, HN, or H) locates and binds to the cellular target. Presumably following target cell binding, multiple homotrimers of the fusion (F) protein take part in anchoring the viral membrane to the host cell membrane, allowing viral entry to the cell.
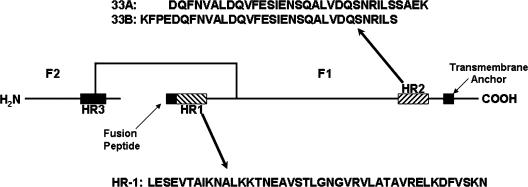
Paramyxovirus F proteins are type I integral membrane viral fusion proteins that are synthesized as inactive precursors (F0). Type I glycoproteins are found in a variety of other viruses including: influenza virus (5), simian immunodeficiency virus (22), human immunodeficiency virus (HIV) (21), and Ebola virus (23). They present on the viral membrane surface as enzymatically cleaved homotrimers (29). Cleavage is performed by host cell proteases into the fusion-active F1 and F2 domains (Fig. 1). F2 is extracellular and disulfide linked to F1.
FIG. 1.
The F1 and F2 subunits of the ectodomain are linked by a disulfide bond. The relative positions of the heptad repeat regions, fusion peptide, and transmembrane anchor are illustrated. The synthetic peptides used in this study are from the HR-1 and HR-2 regions, and their sequences are listed. Peptides do not have modified N or C termini.
A C-terminal hydrophobic transmembrane (TM) domain anchors the F protein in the plasma membrane, leaving a short cytoplasmic tail. There are two 4-3 heptad repeat (HR) domains at the N- and C-terminal regions of the protein (designated HR-1 and HR-2), which form coiled-coil alpha-helices following target cell binding (Fig. 1). These coiled coils become apposed in an antiparallel fashion when the protein undergoes a conformational change into the fusogenic state (45). A hydrophobic fusion peptide N proximal to the N-terminal heptad repeat is thought to insert into the target cell membrane, while the association of the heptad repeats brings the TM domain into high proximity, inducing membrane fusion (for a review, see reference 13). This process has been postulated to provide ample energy to drive fusion in HIV infection (24).
Given the putative role of the F protein in type I viral membrane fusion, an attractive strategy to probe the function of the coiled-coil motif and design new antiviral inhibitors is to create antagonists that bind one of the heptad repeat regions and trap the structure prior to bundle formation. Synthetic peptides corresponding to both HR-1 and -2 of the HIV gp41 protein have been shown to inhibit viral fusion (31, 40, 41). Due to the relatively high potency of HR-2 peptides, this approach has been extended to peptides derived from the fusion proteins of several viruses including RSV (17), hPIV3 (17, 43), avian pneumovirus (37), Newcastle disease virus (46), measles virus (17), and SV5 (25). It also has been noted that HR peptides are virus specific; no peptide has been shown to inhibit multiple viruses.
To further study the molecular concatenation of the hexameric fusion core, researchers examined the crystal structures presumed to be fusion cores (1, 22, 23, 44, 47). These structures revealed two important structural features: (i) hydrophobic pockets and grooves on the surface of the inner trimer and (ii) key contact points lining the pockets. These two features likely are responsible for the extraordinary binding strength of HR peptides and their specificity. Here we report the first structural studies of the hMPV F protein hexameric core and the in vitro inhibition of hMPV using synthetic peptides. Moreover, we found that for hMPV, HR-1 peptides exhibited greater inhibitory potency than did HR-2 peptides.
MATERIALS AND METHODS
Coiled-coil model.
The F protein sequences of 12 paramyxoviruses were compared using a global PAM250 scoring matrix with a gap start of 10 (MOE 2005.06). HR regions of viruses with available coiled-coil crystal structures were aligned further with hMPV using a Blosum80 matrix for the HR regions. RSV (Protein Data Bank [PDB] identification 1G2C) was found to be the most homologous sequence, with 50% identity in HR-1 and 35% in HR-2, and was chosen as a template. Fifty models were generated and evaluated based upon probability density function and overall energy using the modeler package within InsightII (Accelrys). The best model was refined further in AMBER 7 in a generalized Born solvent model minimization. Minimization involved 2,500 cycles with protein backbone atoms fixed, allowing side chains to relax, followed by 2,500 cycles allowing all atoms to relax and produce the final structure. CARNAL was used to convert the AMBER structure to PDB.
Peptide synthesis.
Peptides were synthesized on an Apex 396 synthesizer (Advanced Chemtech) equipped with a 96-well reaction block capable of vortex mixing. Customized Tentagel resin was swollen in dichloromethane (Fisher) prior to synthesis. 9-Fluorenylmethoxycarbonyl amino acids (Synpep) were coupled using O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluor-phosphate (HBTU; 5 eq with respect to resin; Synpep), 1-hydroxybenzatriazole (5 eq; Synpep), diisopropylethylamine (10 eq; Advanced Chemtech) in N,N-dimethylformamide (Fisher). Peptides were cleaved with 95% (vol/vol) trifluoroacetic acid and 5% triisopropylsilane and desalted on a G-25 Sephadex column before final purification on a C4 semipreparation reverse-phase high-pressure liquid chromatography column using water and acetonitrile gradients.
CD.
Fusion core complexes were made at 12.5 μM total peptide in 0.15 M phosphate-buffered saline (PBS) at pH 7.4 (Invitrogen) by mixing equimolar amounts of each HR peptide followed by addition of trifluoroethanol (TFE) or a freeze-thaw cycle. Spectra were collected on an Aviv 215 circular dichroism (CD) spectrophotometer equipped with variable temperature control over the wavelength range of 190 nm to 260 nm with a resolution of 0.5 nm and a bandwidth of 1 nm. The spectrometer is equipped with variable temperature control, an automatic titration system, and a pH probe. Samples were analyzed in a 300-μl strain-free quartz cell with a 1.5-s averaging time. The coiled-coil complex was examined for thermal stability by CD every 5°C from 25°C to 100°C.
SEC.
The molecular weight of the coiled-coil complex was examined by size-exclusion chromatography (SEC). Peptides from both HR regions were mixed in equimolar amounts, and the degree of coiled-coil formation was determined by CD spectroscopy. Coiled-coil complex and sodium polystyrene sulfonate weight standards were run in triplicate isocratically in 0.15 M PBS at pH 7.4 on a Waters Ultrahydrogel high-pressure liquid chromatography size-exclusion column with a flow rate of 0.3 ml/min. Standards had average masses (g/mol) of 4,950, 7,950, 16,600, 34,700, and 57,500. Eluants were monitored at wavelengths of 210, 220, and 257 nm. Coiled-coil complex elution peak masses were determined from linear calibration of the logarithmic weight values.
Plaque inhibition assay.
Serial dilutions of the 1:1 peptide solutions in OPTI-MEM medium (Invitrogen) were added to plates containing 50 to 60 PFU of hMPV per well. Concentrations tested were 25, 12.5, 6.25, 3.12, 1.56, 0.781, 0.390, 0.195, 0.0977, 0.0488, 0.0244, and 0.0122 μM. Additional HR-1 peptide testing was performed at 6, 3, 1.5, 0.76, 0.38, 0.19, 0.0095, and 0.0048 nM. Peptide and virus mixtures were incubated for 1 h at 37°C. Subconfluent cell monolayers of LLC-MK2 cells were washed twice with PBS, and 100 μl of the peptide-virus mixtures was added to the cells. Cells were incubated with the peptide-virus mixture for 1 h at 37°C with gentle rocking every 15 min and then overlaid with methylcellulose, maintaining the peptide concentration in the overlay. Monolayers were incubated for 4 days at 37°C and fixed with formalin at room temperature for 1 h. Plates were rinsed with running water followed by incubation for 30 min at 37°C with blocking buffer (PBS-0.1% Tween-5% nonfat dried milk). Blocking buffer was removed, and the plates were incubated with a 1:1,000 dilution of guinea pig anti-hMPV serum at 37°C for 2 h. The plates were washed and incubated with goat anti-guinea pig immunoglobulin-horseradish peroxidase conjugate (Southern Biotechnology) at 1:1,000 at 37°C for 2 h. Secondary antibody was removed by washing, followed by the addition of TrueBlue peroxidase substrate (Kierkegaard and Perry Laboratories) and incubation for 10 min. Plates were rinsed with water and dried overnight. Plaques were counted using a dissecting microscope, and the average between triplicate wells was calculated.
RESULTS
Analysis of the hMPV F amino acid sequence.
To compare the hMPV F protein to the F proteins of related paramyxoviruses, the sequences of 11 F proteins were aligned with that of hMPV using a PAM250 scoring matrix (7) in order to compare the hMPV F protein to the F proteins of related paramyxoviruses. The results indicated that hMPV had several key local alignments, including predicted alpha-helical structure in the suspected HR regions (unpublished data). The HR regions then were aligned locally using a Blosum80 matrix (12), revealing a 3-4-4-4-3 stutter pattern characteristic of paramyxoviruses (unpublished data) (1, 4). The cysteine alignment patterns for the 12 sequences revealed eight conserved cysteines that were previously shown to be involved in disulfide bonds within the PIV3 fusion protein (44). Within the pneumoviruses, four of the sequences (hMPV, RSV, pneumonia virus of mouse, and avian pneumovirus) displayed six additional conserved cysteine residues. These alignments suggested that the hMPV F protein may have a structure and function similar to those of previously studied paramyxovirus F proteins.
Homology model of hMPV F.
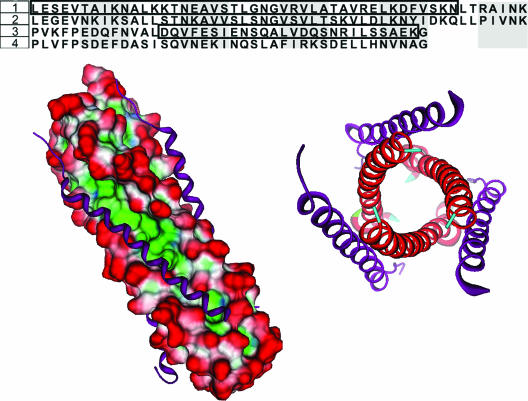
In order to generate a computational model of the hypothesized hMPV fusion core, we first identified a suitable template by comparing amino acid sequences between paramyxovirus F proteins. The available hexameric crystal structures of paramyxovirus F proteins were examined for sequence similiarity with hMPV's heptad repeat regions (1, 6, 22, 23, 33, 44, 47). The hMPV F protein HR regions showed the greatest sequence identity to RSV F protein HR regions (50% in HR-1 and 35% in HR-2, Fig. 2). These values exceed the reasonable threshold of 25% sequence identity commonly employed to obtain reasonable homology models (27).
FIG. 2.
(Top) Sequence alignment of the HR regions of RSV and hMPV. RSV sequences were obtained from the crystal structure (PDB identification 1G2C). The HR-1 region shows an identity of 50% between the two viruses while the HR-2 region has 35% identity. 1, hMPV HR-1 (L130 to K179); 2, RSV HR-1; 3, hMPV HR-2 (P448 to G487); 4, RSV HR-2. LearnCoil-VMF-generated sequences for hMPV are boxed. (Bottom) The sequence of the RSV fusion core crystal was subjected to a strong local alignment with the hMPV sequence to determine appropriate homology sequences for the hMPV fusion core model. The alignment produced two hMPV peptides with 50% (HR-1) and 35% (HR-2) identity with the RSV peptides. Using the RSV structure as a template, the hMPV peptides were grafted onto the RSV Cα backbone and allowed to minimize. (Left) Shown is the surface of the HR-1 trimeric stalk (red, solvent exposed; blue, hydrophilic; green, hydrophobic) with HR-2 (purple ribbons) filling the hydrophobic grooves. (Right) Shown is an axial view of the hexameric core. The hexameric coiled-coil formation is the major structural feature in the postfusion conformation of the F protein.
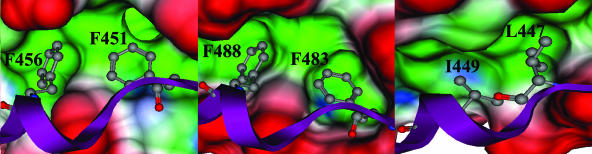
Several models were produced using the modeler package of InsightII. Subsequently, these models were scrutinized for overall energy and probability density function values to isolate the most stable model intermediate. We further subjected this intermediate to rigorous Born solvent model minimization for a final model of the hMPV hexameric F protein core (Fig. 2). The model displays several features similar to those of other hexameric fusion cores. Three HR-1 peptides form an inner helical bundle that presents three long hydrophobic grooves along the ∼72-Å stalk. Corresponding HR-2 peptides assume a helical structure, offering a hydrophobic face that packs in an antiparallel fashion into the HR-1 hydrophobic grooves. This configuration would allow for the fusion peptide, found near the N terminus of the HR-1 stalk, to be in high proximity with the transmembrane anchor located at the C terminus of the HR-2 peptide. The presence of a hydrophobic pocket and key salt bridges also was identified. The hydrophobic pocket of the hMPV model is comprised of residues V162, L165, and V169 from one HR-1 peptide and A161, L165, and F168 of a second HR-1 fragment (Fig. 3). Several polar residues contributed from HR-1 line the immediate exterior of the pocket: R163, K166, D167, S170, and K171 with E164 from a second HR-1 fragment. The polar residues likely contribute specificity among HR sequences (17, 47), as seen by the potential formation of a strong ionic network between K166 and K171 from HR-1 and E453 of HR-2.
FIG. 3.
(Left) Surface image of the hMPV hydrophobic pocket filled by phenylalanines 451 and 456 of the HR-2 peptide (red, solvent exposed; green, hydrophobic). (Center) RSV phenylalanines 483 and 488 complementing the hydrophobic cavity. (Right) Congruent leucine 447 and isoleucine 449 of SV5 packing a hydrophobic pocket. Images were produced using MOE 2005.06 (Chemical Computing Group).
The final refined hMPV model was superimposed upon the RSV fusion core crystal structure, which has been shown to be similar to that of other paramyxovirus cores (47). Comparison of the overlaid Cα and backbone nitrogen of the RSV and hMPV HR-1 peptide strands suggests a root mean square deviation (RMSD) of 0.53 Å for HR-1 and an average of 0.45 Å for HR-2. Using the same superimposition technique, RSV and SV5 HR-1 peptides have an RMSD of 1.5 Å, and the HR-2 peptides result in a 2.78-Å RMSD. Similar comparison of hMPV and SV5 peptides shows a 1.48-Å RMSD for HR-1 and a 3.55-Å RMSD for HR-2. The combined molecular surface of the hMPV model and RSV crystal structure maintained the hydrophobic cavity filled by the phenylalanine residues of the RSV and hMPV HR-2 peptides (RSV, F483 and F488; hMPV, F451 and F456). Overall, the hMPV fusion core model is consistent with known type I fusion protein core structures.
Characterization of synthetic hMPV F HR peptides.
In the absence of hMPV F protein crystallographic data, peptide sequences for synthesis and study were determined by computational methods. The homology model of the hMPV HR regions that we constructed suggested peptide lengths of 50 and 40 amino acids for HR-1 and HR-2, respectively. We analyzed the hMPV F protein sequence using LearnCoil-VMF (2, 32) to further define the critical regions of the HR peptides (Fig. 4). LearnCoil-VMF has previously been used successfully to predict HR peptides of other type I viral fusion proteins, including HIV gp120 and RSV F (43). The target peptides were synthesized, and their solution-phase secondary structure was examined using circular dichroism spectroscopy in 0.15 M PBS (pH 7.4) at 25°C. Unexpectedly, the HR-1 peptide showed a lack of helical structure (<25%). Previous HR-1 peptides from other viral fusion proteins have been shown to contain a significant alpha-helical content (1, 8, 17, 18, 26, 37, 43). The spectra of 33A and 33B also displayed little secondary structure content, consistent with previously reported HR-2 peptides. When the HR-1 and HR-2 peptides were combined at a 1:1 molar ratio, an additive spectrum was obtained indicative of no higher-order complex formation (Fig. 5).
FIG. 4.
Comparison of the LearnCoil-VMF heptad repeat peptides (green) with the heptad repeat regions predicted from the computational model (HR-1, red; HR-2, purple).
FIG. 5.
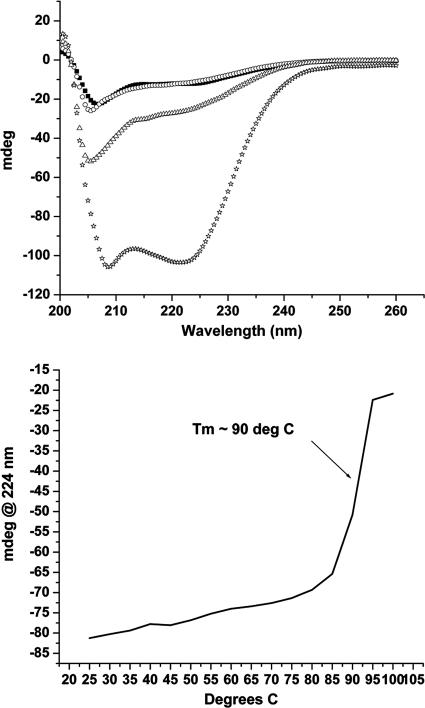
(Top) Both the HR-1 and HR-2 (33A) peptides show little secondary structure at 25°C (HR-1 43-mer, ○; HR-2 33A, ▪). Mixing of the two HR peptides produced only an additive spectrum indicative of no interaction (equimolar mixture at 25°C ▵). The same equimolar mixture at 25°C following a freeze-thaw cycle adopted the putative coiled coil (⋆). (Bottom) Melting point curve for the HR-1 and 33A peptide mixture following a freeze-thaw cycle.
These results were unexpected, considering that the fusion model and sequence analysis suggest that the HR-1 peptide should possess a propensity to adopt an alpha-helical structure. We next tested the ability of the synthetic HR peptides to form alpha-helices under different experimental conditions. TFE and thermal modulation (19) provide reaction conditions that have been shown to favor alpha-helix confirmation. The HR-1 peptide showed a strong helical spectrum with as little as 2.5% (vol/vol) TFE that reached a plateau at 5% TFE (data not shown). In contrast, HR-1 showed no increase in observed intensity at 224 nm following a freeze-thaw cycle. The HR-2 peptides were unaffected by the addition of small volumes of TFE (up to 5% [vol/vol]) or thermal cycling.
Peptides from both HR-1 and -2 regions were mixed in the presence of 5% TFE, and the intensity at 224 nm was greater than the additive spectra of the individual peptides in TFE (data not shown). The enhancement of helicity in the presence of both HR-1 and HR-2 peptides indicated the formation of the predicted coiled-coil assembly. Higher concentrations of TFE did not cause an additional increase in the 224-nm signal, suggesting that the maximal amount of peptide had transitioned to the putative hexameric core. In contrast to the effect of thermal cycling on HR-1 peptide alone, thermal cycling of an equimolar mixture of HR-1 and 33A yielded a CD spectrum that exhibited the predicted enhanced coil-coiled tracing similar to that of the mixture in the presence of TFE (Fig. 5). Thermal cycling of a mixture of HR-1 peptide and 33B yielded a CD spectrum that demonstrated similar coiled-coil formation.
The known viral F protein fusion cores have been shown to have unusually high melting temperatures (Tms) compared to similar-length helical bundles (15, 18, 22, 23, 37, 47). The melting curve of the hexameric core of hMPV formed by HR-1 and 33A, obtained from a freeze-thaw cycle, gave a Tm of ∼90°C (Fig. 5). This is consistent with the previously reported Tm of 88°C for the RSV F protein hexameric core (47). The melting temperature obtained from the 33B and HR-1 complex was found to be ∼85°C.
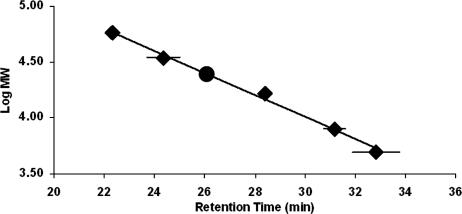
We used size-exclusion chromatography to investigate the aggregation state of the putative hexameric core. The chromatograms of the peptide solutions yielded two pronounced and well-resolved peaks. The major peak corresponded to a mass of 24,802 ± 135 g/mol (retention time, 26.1 ± 0.02 min; Fig. 6), almost identical to the theoretical mass of the hexameric core formed from the LearnCoil-VMF HR-1 and HR-2 sequences in a 1:1 ratio (24,933 g/mol). Due to the mass difference between the HR-1 and HR-2 peptides, nonhexameric structures and structures with a ratio other than 1:1 for HR-1 to HR-2 would fall outside of the standard deviation of this experiment. The second peak at 23.4 min (±0.03) corresponded to a mass consistent with 12 peptides in a 1:1 ratio of the HR-1 and -2 peptides. The fractions containing the hexameric and dodecameric peptide assemblies were isolated, and both were found to contain helical content consistent with the coiled-coil core. Time course experiments showed that the relative composition of the sample shifted towards the dodecamer over a 24-h period, suggesting slow dimerization of the hexameric core (data not shown).
FIG. 6.
Size-exclusion chromatography calibration curve used to obtain the mass of the equimolar mixture of the HR-1 and HR-2 peptides. ⧫, synthetic calibrants; •, HR-1 and 33A mixture following a freeze-thaw.
In vitro inhibition of hMPV by synthetic HR peptides.
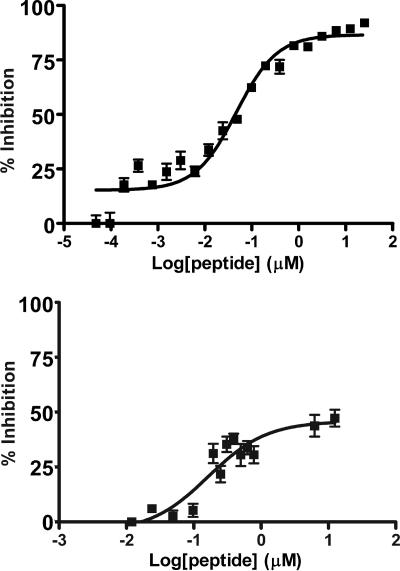
Peptides corresponding to the HR sequences of other hexameric F protein cores have been shown to be potent inhibitors of viral propagation (1, 8, 9, 17, 18, 22, 30, 31, 37, 40, 41, 43, 46). The hMPV F protein HR sequences predicted by LearnCoil-VMF and the 33B peptide were synthesized, purified, and tested in vitro for their ability to inhibit hMPV infectivity. We tested peptides from 25 μM to 12 nM in initial experiments and found peptide 33A to inhibit hMPV entry up to ≤50%. Peptide 33B mediated very little inhibition at the maximum test concentration of 25 μM. However, HR-1 inhibited infectivity by ≥95% at 25 μM and still displayed substantial activity at 12 nM. Therefore, we conducted further experiments to determine the dose-response curve of the peptides and the MIC of each peptide down to 4.8 pM. Peptide 33A caused substantial viral inhibition with an EC50 of ∼165 nm, where EC50 is defined as the concentration of peptide required to elicit 50% of maximal inhibition. Surprisingly, HR-1 resulted in an EC50 of ∼46 nM (Fig. 7), making it at least 1,000 times more potent than previously reported HR-1 peptides (21, 28, 36).
FIG. 7.
Data were obtained by adding virus and peptide to confluent cells for 1 h. A semisolid overlay of methylcellulose was added that maintained the peptide concentration. Anti-hMPV guinea pig primary antibodies then were utilized followed by an anti-guinea pig secondary antibody linked to horseradish peroxidase in order to detect foci. Data were fitted using the Hill-Slope model. The HR-1 peptide (top) was found to be a very potent inhibitor (EC50, ∼46 nM) while the 33A peptide failed to achieve greater than 50% inhibition (bottom).
To obtain insight into the inhibitory action of the HR-1 peptide, plaque assays were performed as described above save the addition of HR-1 peptide to the cell medium or overlay. The resulting conditions have peptide concentration significantly reduced during infection and are more comparable to neutralization assays testing the HR-1 peptide's ability to inhibit viral activity prior to cell exposure. The results were nearly identical to those of the assays maintaining peptide concentration throughout (EC50, ∼13 nM), thus indicating that the HR-1 peptide is displaying most of its activity prior to infection. The prophylactic nature of the hMPV HR-1 peptide is the first report of a potential peptide-based antiviral therapeutic.
DISCUSSION
The recently discovered paramyxovirus hMPV is one of the most common causes of serious lower respiratory tract illness in infants and children (35, 42). Our studies show that the fusion protein of hMPV possesses a high level of similarity at the amino acid sequence level and at the level of predicted protein structure to other type I integral membrane viral fusion proteins. Peptides from the HR regions of the hMPV F protein inhibited virus-cell fusion, supporting the hypothesis that the hMPV F protein functions through an HR peptide-sensitive membrane fusion mechanism similar to that of SV5 (1), RSV (18), and HIV-1 (41).
Homology modeling suggested the presence of key electrostatic contacts and a hydrophobic pocket similar to that seen in the SV5, RSV, and hPIV3 F proteins that have been characterized structurally (1, 44, 47). Several polar groups line the hydrophobic cavity in the hMPV fusion core model. Analogous residues in related paramyxoviruses appear to convey specificity among virus-derived HR-2 peptides (17, 47). The hydrophobic pocket predicted by the hMPV F protein model is complemented by two phenylalanines (F451 and F456) of an HR-2 strand and exhibits homology with the hydrophobic F483 and F488 residues found in RSV F protein and the L447 and I449 residues of SV5 F protein. Docking of small-molecule antagonists within the hydrophobic cavity or interference with critical ion pairs has been shown previously to inhibit fusion of HIV (10).
Based on the ability of LearnCoil-VMF to predict sequences that encompass well-studied fusogenic core peptides, the HR-1 43-mer and 33A peptides were selected for synthesis. Additionally, the homology fusion core model also suggested study of peptide 33B to examine any potential advantages of targeting the hydrophobic cavity. The characterization of synthetic HR-1 and HR-2 peptide interaction was consistent with a hexameric coiled-coil fusion core for hMPV. Following helical induction by either TFE or thermal modulation, assembly of the hexameric coiled coil resulted in CD spectra containing alpha-helical content substantially greater than the additive spectra of the individual peptides. This alpha-helical species also was determined to have a melting temperature (Tm ∼ 90°C) consistent with that of other previously described viral fusogenic cores (1, 18, 22, 23, 47). SEC determined the mass of the induced samples to be consistent with the expected hexameric moiety in a 1:1 HR-1-to-HR-2 ratio. This ratio is consistent with analytical sedimentation analysis of RSV HR peptides described by Lawless-Delmedico et al. (18). The sedimentation analysis of the RSV peptides found evidence of monomers, HR-1 trimers, and a hexameric structure with a 1:1 ratio of RSV's HR-1 and HR-2 peptides (18). In contrast, the peptides of hMPV did not exhibit self-trimer aggregation by SEC analysis, consistent with the absence of alpha-helical content observed for HR-1.
The experimental induction of the hMPV HR-1 peptide from a predominantly random conformation to an alpha-helix illustrates subtle differences between experimentally determined HR sequences and those identified by LearnCoil-VMF. One such difference between previously reported HR-1 peptides determined from enzymatic footprinting experiments and the hMPV LearnCoil-VMF HR-1 peptide is the lack of helical structure or aggregation of the hMPV HR-1 peptide (1, 17, 18, 47). A small volume of TFE produced a strong alpha-helix signal from the HR-1 solution, thus demonstrating the propensity for aggregating into the three-helix bundle comprising the inner stalk of the fusion core. The subsequent addition of an HR-2 peptide caused the hexameric assembly to form. These data suggest that the HR-1 helical transition prior to exposure to HR-2 peptides contributes significantly to the mechanism of the fusion process.
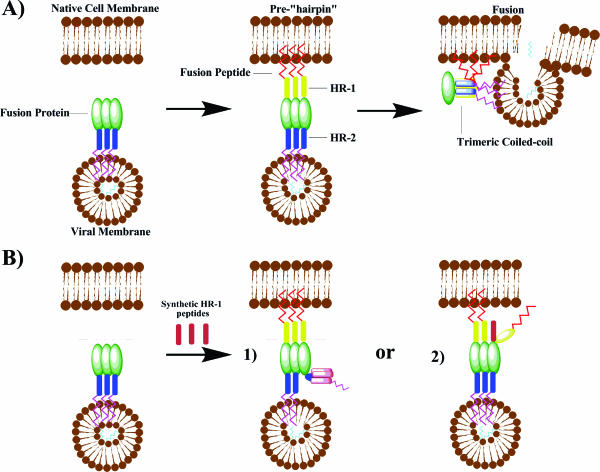
Fusion of paramyxoviruses with their target cellular membrane requires protein-driven cellular docking, activation of the prefusion F protein, and a conformational change of the F protein into the postfusion state containing a hexameric coiled coil (9, 13, 14). Following activation, the HR-1 peptides of the trimeric F protein aggregate into the central helical bundle of the hexameric core. The membrane targeting fusion peptide presents on the tip of the trimeric HR-1 peptide bundle. The fusion peptide inserts into the target cellular membrane, resulting in a pre-“hairpin” intermediate (Fig. 8). This intermediate presents the hydrophobic grooves of the HR-1 bundle to the HR-2 peptides for binding in an antiparallel fashion, resulting in the formation of a thermostable hexameric coiled-coil core. This conformational juxtaposition of the HR regions results in high proximity of the viral membrane to the target host cell, which facilitates subsequent fusion of the virus and cell membranes. The conversion from the prefusion to postfusion structures has become a popular target of peptide antifusion antagonists.
FIG. 8.
(A) Proposed mechanism of fusion between type I integral membrane viral fusion proteins and cellular membranes. Following cellular docking, a transient prehairpin complex arises as a result of a conformational change in the F protein. This results in the insertion of the fusion peptide into the cellular membrane and exposure of the HR-1 trimer. The previously constrained and sequestered HR-2 peptides migrate to pack hydrophobic channels of the HR-1 trimer, resulting in formation of the coiled-coil complex and membrane fusion. (B) Possible mechanisms of inhibition by synthetic HR-1 peptides: monomers aggregate into a synthetic trimer sequestering a native HR-2 peptide (1) or monomeric synthetic units of HR-1 intercalate into a native trimer stalk (2).
Recent work involving type I integral membrane fusion protein inhibition has focused on peptides derived from HR-2 regions due to their remarkable potency against viral fusion (9). The postulated mechanism of inhibition for the HR-2 peptides involves binding of the helical monomers to the hydrophobic groove presented along the surface of the HR-1 bundle. Binding of the synthetic HR-2 peptides prevents native HR-2 peptides from reaching their target. The synthetic peptides thus are able to arrest the fusion protein intermediate. The hMPV HR-2 peptides, 33A and 33B, failed to achieve antifusion activity comparable to that of optimal HR-2 peptides previously described (17, 37, 40, 41, 43). Peptide 33B inhibited fusion minimally even at 25 μM. The LearnCoil-VMF-predicted peptide 33A exhibited a titratable response but failed to reach >50% inhibition up to 25 μM. Previous work has shown significant variability in antifusion activity between peptides offset by as little as a single residue in the HR sequence. In studying the HR-2 region of the RSV F protein, Lambert et al. examined a single offset scanning library of potential inhibitors and found that EC50 values differed more than fivefold between the consecutive peptides T-106 and T-107 (17). Ongoing work towards development of an optimal peptide inhibitor for hMPV involves investigation of a more exhaustive set of peptides from the HR-1 and HR-2 regions.
Strikingly, we found that the HR-1 peptide exhibited much more potent in vitro inhibition of hMPV, with an EC50 and 50% inhibitory concentration (IC50) of ∼46 nM. This compares to a 90% inhibitory concentration (IC90) of the HIV HR-1 peptide of 1.3 μM and IC50 of 16 μM for SV5's HR-1 (Table 1). The current proposed model of HR-1 peptide inhibition suggests that the synthetic HR-1 peptides present as a helical trimer and then ensnare one or more of the native HR-2 peptides (Fig. 8) (8, 28, 38, 40). All previous studies of fusion protein inhibition for other viruses have reported that HR-2 peptides exhibit fusion-inhibiting activity that is orders of magnitude more potent than that of HR-1 peptides. Thus, our results suggest that either the stoichiometry or kinetics of the HR-1 inhibitor may differ from previous heptad repeat-based inhibitors of type I viral fusion. It has been hypothesized that strong HR-1 aggregation is partly responsible for the weak inhibitory properties of HR-1-based peptides. Inhibitory interactions between native and synthetic HR-1 peptides would require at least three times the concentration of HR-1 peptides relative to HR-2 peptides, which are thought to inhibit in a monomeric state (21). For this to explain our findings, a required helix-inducing molecule would need to be present on the cellular or viral surface. While our study does not exclude the presence of such an inducer, the phenomenon would likely be highly concentration dependent. This postulated mechanism is not supported by the strong fit of the dose-response model (Hill-Slope fit) to our data (39).
TABLE 1.
Comparison of inhibitory HR peptides
An alternate mechanism of inhibition for HR-1 peptides postulates that monomeric HR-1 peptide could insert and substitute for native HR-1 peptide in the trimeric bundle. Our results suggest a possible intercalation of the HR-1 peptide into the fusion protein prior to or during fusion (Fig. 8). The absence of interaction among the HR-1 and HR-2 peptides in the CD analysis suggests that the HR-1 peptide may not inhibit as a dimeric or trimeric unit by ensnaring the HR-2 peptide. Furthermore, the SEC data do not indicate the presence of an HR-1 helical bundle, unlike the trimeric HR-1 bundle seen in RSV sedimentation data (18).
Yin et al. recently solved the prefusion structure of the parainfluenza virus 5 F protein (45). The crystal displays the HR-2 peptides in a helical bundle associating the transmembrane domain with the viral membrane, while the HR-1 and fusion peptide sequences are sequestered within the globular head. This structure suggests a possible intermediate target for an intercalating HR-1 peptide. In order to expose the fusion peptide for insertion in the target membrane, the HR-1 peptides must extend into a helical bundle outward from the viral membrane. During stalk extension, incorporation of a synthetic HR-1 peptide could occur. This would result in failure of the fusion peptide to reach its final position within the target membrane and would likely arrest subsequent folding and fusion.
The data presented in this study provide evidence for the structural and functional relationship of the hMPV F protein to that of other paramyxoviruses. The study identifies the first HR-1-derived peptide with potent fusion inhibition activity. While previous research concluded that HR-1 peptides could be effective inhibitors of type I integral membrane fusion proteins when constrained and presented as trimeric helical bundles (8, 28), our data suggest that individual HR-1 peptides can lead to effective viral inhibition. Furthermore, recent studies of HR-2 peptides suggest the potential for multiple viral targets for inhibitory peptides (20). The prophylactic nature of the HR-1 peptide could be explained by interaction with additional targets or strong binding to the metastable prefusion state of the F protein, possibly preventing stalk extension. Thus, future dye transfer experiments, intermediate capture experiments, and peptide labeling will help to elucidate which critical intermediates and functions are being arrested by the hMPV HR-1 peptide inhibitor and will shed new light on the mechanism of viral fusion.
Acknowledgments
This work was supported in part by NIH grant AI-56170 to J.V.W. James E. Crowe, Jr., holds a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund. This work was also supported by NIAID, NIHR01 AI 59597.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Berger, B., and M. Singh. 1997. An iterative method for improved protein structural motif recognition. J. Comput. Biol. 4:261-273. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. H., C. Cohen, and D. A. Parry. 1996. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins 26:134-145. [DOI] [PubMed] [Google Scholar]
- 5.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 7.Dayhoff, M., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins, vol. 5, suppl. 3. National Biomedical Research Foundation, Silver Spring, MD.
- 8.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, J. T., O. Kutzki, A. K. Debnath, S. Jiang, H. Lu, and A. D. Hamilton. 2002. Design of a protein surface antagonist based on α-helix mimicry: inhibition of gp41 assembly and viral fusion. Angew. Chem. Int. Ed. 41:278-281. [DOI] [PubMed] [Google Scholar]
- 11.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 14.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, R. A., S. B. Joshi, and R. E. Dutch. 1999. The paramyxovirus fusion protein forms an extremely stable core trimer: structural parallels to influenza virus hemagglutinin and HIV-1 gp41. Mol. Membr. Biol. 16:11-19. [DOI] [PubMed] [Google Scholar]
- 16.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, 3rd ed. Raven Press, New York, NY.
- 17.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erkckson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawless-Delmedico, M. K., P. Sista, R. Sen, N. C. Moore, J. B. Antczak, J. M. White, R. J. Greene, K. C. Leanza, T. J. Matthews, and D. M. Lambert. 2000. Heptad repeat regions of respiratory syncytial virus F1 protein form a six-membered coiled-coil complex. Biochemistry 39:11684-11695. [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. S., G. G. Wood, and D. J. Jacobs. 2004. Investigations on the alpha-helix to coil transition in HP heterogeneous polypeptides using network rigidity. J. Phys. Condens. Matter 16:S5035-S5046. [Google Scholar]
- 20.Liu, S., H. Lu, J. Niu, Y. Xu, S. Wu, and S. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259-11273. [DOI] [PubMed] [Google Scholar]
- 21.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 22.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malashkevich, V. N., B. J. Schneider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9- Å resolution. Proc. Natl. Acad. Sci. USA 96:2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 26.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 27.Rost, B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12:85-94. [DOI] [PubMed] [Google Scholar]
- 28.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyoxviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell, R., R. G. Paterson, and R. A. Lamb. 1994. Studies with crosslinking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199:160-168. [DOI] [PubMed] [Google Scholar]
- 30.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75:7934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sia, S. K., P. A. Carr, A. G. Cochran, V. N. Malashkevich, and P. S. Kim. 2002. Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. USA 99:14664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, M., B. Berger, and P. S. Kim. 1999. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 290:1031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, B. J., M. C. Lawrence, and P. M. Colman. 2002. Modelling the structure of the fusion protein from human respiratory syncytial virus. Protein Eng. 15:365-371. [DOI] [PubMed] [Google Scholar]
- 34.van den Hoogen, B. G., T. M. Bestebroer, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 35.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. M. Fouchier, and A. D. M. E. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, E., X. O. Sun, Y. Qian, L. Zhao, P. Tien, and G. F. Gao. 2003. Both heptad repeats of human respiratory syncytial virus fusion protein are potent inhibitors of viral fusion. Biochem. Biophys. Res. Commun. 302:469-475. [DOI] [PubMed] [Google Scholar]
- 37.Wang, X.-J., Y.-D. Bai, G.-Z. Zhang, J.-X. Zhao, M. Wang, and G. F. Gao. 2005. Structure and function study of paramyxovirus fusion protein heptad repeat peptides. Arch. Biochem. Biophys. 436:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whiting, B., and A. W. Kelman. 1980. The modelling of drug response. Clin. Sci. 59:311-315. [DOI] [PubMed] [Google Scholar]
- 40.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. M. Pingsterhaus, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 44.Yin, H.-S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin, H.-S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 238:291-304. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion core protein. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]