Abstract
Early region 1B (E1B) of adenovirus type 5 (Ad5) encodes at least five different polypeptides generated by alternative splicing of a common mRNA precursor. Two of these gene products, E1B-19K and E1B-55K, are individually capable of cooperating with the Ad5 E1A proteins to completely transform rodent cells in culture. Substantial evidence suggests that these two E1B proteins contribute to cell transformation by antagonizing growth arrest and apoptosis. Here, we performed genetic and biochemical analyses to assess the attributes of the remaining E1B proteins (E1B-156R, E1B-93R, and E1B-84R). Our results show that E1B-156R, which comprises the 79 amino-terminal and 77 carboxy-terminal amino acids of E1B-55K, also enhances focal transformation of primary rat cells in cooperation with E1A. Since E1B-156R seemed unable to relocalize p53 and inhibit its transactivating function, it must be assumed that it contributes to transformation independently of repression of p53-stimulated transcription. Furthermore, we discovered that E1B-156R contains a functional transcriptional repression domain and binds Ad5 E4orf6 and the cellular apoptosis regulator Daxx. While the ability to bind E4orf6 could indicate further biological functions of E1B-156R in viral infection, the interaction with Daxx might also be linked to its transforming potential. Taken together, these analyses introduce E1B-156R as a novel transformation-promoting E1B protein that acts without repressing p53 transactivation. Moreover, identification of the interaction partners E4orf6 and Daxx provides a first glance of E1B-156R's potential functions.
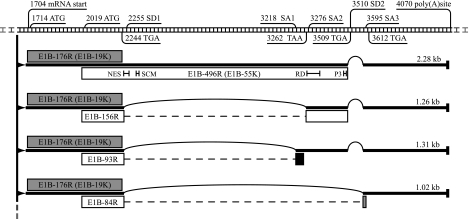
The gene products of human adenovirus early region 1B (E1B) perform functions essential for viral replication and adenovirus-mediated cell transformation. While much research has gone into revealing the functions of the E1B-19K and E1B-55K proteins, little is known about the other proteins encoded within early region 1B. The E1B gene of adenovirus type 5 (Ad5) encodes at least five gene products, utilizing a group of differentially spliced mRNAs (Fig. 1). The major 2.28-kb mRNA encodes the 176-residue E1B-19K protein (E1B-176R) in the first open reading frame and the E1B-55K protein (496 residues; E1B-496R) in the second open reading frame, overlapping with the first. The open reading frame of E1B-55K is accessed by an internal initiation site, and, since it uses an alternative reading frame, the protein is not related to E1B-19K (6, 18, 50). Apart from the major 2.28-kb mRNA, a group of shorter spliced mRNA forms are produced (1, 30, 63). While the sequence encoding E1B-19K is unaffected, the open reading frame used for E1B-55K in the 2.28-kb mRNA is altered in these transcripts. They therefore encode three additional E1B-55K-related proteins, named E1B-156R, E1B-93R, and E1B-84R after the numbers of their amino acid residues (1, 21, 30, 36, 60, 63). All contain the 79-residue amino terminus of E1B-55K, and, while E1B-93R and E1B-84R have unique carboxy termini, E1B-156R is completed by the 77 carboxy-terminal residues of E1B-55K. Another mRNA of 0.86 kb has been proposed for Ad2 (30). It utilizes a weak splice donor site and encodes proteins of 168 and 131 amino acid residues.
FIG. 1.
Predicted Ad5 E1B region and its gene products. The region and various E1B landmarks are shown at the top. Numbers refer to nucleotides in the Ad5 sequence. Four differentially spliced mRNAs are depicted below as thick lines. Their lengths are noted on the right. Thin printed arcs represent introns between splice donor (SD) and acceptor (SA) sites. The five open reading frames encoding the E1B proteins are illustrated as boxes next to their corresponding mRNA. The shading of the boxes (gray, black, and white) represents the used reading frames 1 to 3, respectively. The gene products are named with reference to their numbers of amino acid residues. E1B-176R and E1B-496R therefore represent E1B-19K and E1B-55K. The open reading frames encoding E1B-156R, E1B-93R, and E1B-84R are generated through fusion of two exons. The eliminated intron sequences are illustrated as dashed lines. Within the box representing the E1B-55K protein, some protein hallmarks are noted with black bars marking their positions within the primary sequence: the nuclear export sequence (NES) at position 83 to 93; the SUMO1 conjugation motif (SCM) at 104 to 106; the repression domain (RD) around amino acid R443; and the three phosphorylation sites at S490, S491, and T495 (P3). This figure was produced with information from reference 60. The nucleotide numbering is according to the published Ad5 sequence from GenBank accession no. AY339865.
Production of the different E1B mRNAs in infection is regulated over time. Temporal expression was originally shown by different groups for Ad2 and could also be observed with Ad5, as shown by our own unpublished work. While mainly the 2.28-kb form is produced early in infection, the proportion of the shorter spliced mRNAs increases over time and the E1B-84R-encoding transcript becomes predominant in the late phase (9, 42, 58, 63, 65). The appearance of the respective E1B proteins closely follows the transcription pattern of the mRNAs. E1B-19K and E1B-55K production commences shortly after the products of E1A and continues at high levels throughout infection. E1B-156R, E1B-93R, and E1B-84R production starts slightly later, but prior to late protein synthesis, and increases over time. While E1B-156R and E1B-84R achieve moderate-to-high expression levels, only small amounts of E1B-93R are produced in infected human KB cells (60).
An important task of E1B-19K and E1B-55K in viral infection is suppression of host cell apoptosis since premature cell death would limit viral replication (10, 64). The same functions enable both proteins to promote complete transformation of rodent cells in cooperation with E1A. E1A proteins activate cell cycle progression in nondividing cells and overrule growth inhibition through cell-to-cell contact and cyclin dependent kinase inhibitor-mediated senescence signals (reviewed in references 4 and 14). Besides this E1A function, the cellular tumor suppressor p53 is induced and metabolically stabilized. This leads to cell cycle arrest and apoptosis and therefore limits the transforming abilities of E1A (11, 20, 35, 54). The expression of E1A alone is sufficient to immortalize and partially transform primary cells, although transformation is inefficient and often incomplete (reviewed in reference 57). The E1B-19K and E1B-55K proteins can individually cooperate with Ad5 E1A to increase transformation efficiency and to convert primary rodent cells to a fully transformed tumorigenic phenotype. This is at least in part a consequence of their ability to inhibit p53-induced cell cycle arrest and apoptosis (57). E1B-19K plays an important role in the inhibition of different apoptotic pathways, since it acts as an antiapoptotic Bcl-2 family protein. E1B-19K is able to inhibit co-oligomerization of the proapoptotic Bcl-2 proteins BAK and BAX and thereby blocks the mitochondrial pathway of caspase activation (4).
The central role of E1B-55K in transformation seems to be its ability to directly counteract p53. The key function of p53 as a tumor suppressor is thought to be the transactivation of several proapoptotic and antiproliferative genes (reviewed in reference 52). E1B-55K binds the activator domain of p53 and impairs p53-mediated transactivation of genes, probably through steric blockage of coactivator interactions and through the function of its own carboxy-terminal transcription repression domain (38, 39, 61, 68, 69). It was further suggested that the ability of E1B-55K to sequester the tumor suppressor could be essential for the inhibition of p53 functions (14, 24, 71). E1B-55K relocalizes p53 into dense cytoplasmic structures, which fit the criteria for aggresomes (34).
It remains controversial which functions E1B-156R, E1B-93R, and E1B-84R might fulfill in viral infection or in transformation of rodent cells (30, 42, 60). However, the temporal regulation of their expression and the structural relation to the potent oncoprotein E1B-55K prompted us to examine these proteins more closely. E1B-156R seemed particularly promising since it contains the carboxy terminus of E1B-55K, which had been shown to harbor a transcription repression domain (Fig. 1).
MATERIALS AND METHODS
Plasmids.
pE1B-55K, which expresses Ad5 E1B-55K under the control of the human cytomegalovirus (CMV) major immediate-early promoter, was generated by inserting the nucleotide sequence 2019 to 4113 of the Ad5 genome into pcDNA3 (Invitrogen) as described previously (46). We acquired the cDNAs of E1B-156R, E1B-93R, and E1B-84R from cytoplasmic mRNA of H5dl309 (25)-infected HeLa cells (17) by reverse transcription with oligo(dT) primers. The sequences encoding E1B-156R and E1B-93R were gained by PCR from these cDNAs with the primers E1Bfwd #1003 (5′-CGGGATCCATGGAGCGAAGAAACC-3′) and E1Brev #1004 (5′-CGGAATTCCTCAATCTGTATCTTCATCG-3′), adding a 5′ BamHI site and a 3′ EcoRI site to the sequences. The PCR fragments were inserted between the BamHI and EcoRI sites of pcDNA3 (Invitrogen) to generate pE1B-156R #1326 and pE1B-93R #1327. pE1B-84R #1641 was constructed similarly, but 84Rrev #1283 (5′-CGGAATTCGAGTTGGTGCTCATGGC-3′) was used instead of E1Brev #1004.
To generate pE1B-55K ΔSD #1415, splice donor site 1 (SD1) of E1B-55K was knocked out by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit from Stratagene with the primers mSD1fwd #1142 (5′-CGG GAATGAATGTTGTACAAGTCGCAGAACTGTATCCAGAACTG-3′) and mSD1rev #1143 (5′-CAGTTCTGGATACAGTTCTGCGACTTGTACAACATTCATTCCCG-3′). pE1B-55K was used as the template. The plasmid pCMV-E1A 12S, used in transformation assays, has been described before (29).
Gal4 DNA-binding domain-E1B fusion proteins were expressed using the plasmids pGal4 E1B-55K (12) and pGal4 E1B-156R #1375. pGal4 E1B-156R #1375 was constructed by insertion of the E1B-156R BamHI/EcoRI fragment from pE1B-156R #1326 into the simian virus 40 early promoter-driven pSG424 vector (55). pC53-SN3 encodes human p53 in a pCMV/neo vector (3).
In the dual-luciferase assays, three different reporter constructs, pRE-Luc (obtained from N. Horikoshi, Washington University, St. Louis, MO), pHH0.34-TK-Luc (49), and pGalTK-Luc (43) were used to drive expression of firefly luciferase from different promoter/enhancer elements. pRL-TK (Promega) which constitutively expresses Renilla luciferase from a herpes simplex virus thymidine kinase promoter was used as an internal control.
phDaxx-HA #1607 was produced by integrating a hemagglutinin tag between the HindIII and BamHI sites of pcDNA3 using the annealed primers HAfwd #1169 (5′-AGCTTATGTACCCATACGATGTTCCAGATTACGCTG-3′) and HArev#1170 (5′-GATCCAGCGTAATCTGGAACATCG TATGGGTACATA-3′) as an insert. The cDNA of human Daxx was inserted into the vector using BamHI and SalI sites.
Cell transfections and reporter assays.
Cells were in general transfected using the calcium phosphate coprecipitation procedure of Graham and van der Eb (19) whereby salmon sperm DNA (Roche) was used as carrier DNA. The Western blot assays shown in Fig. 2, 5, and 8C were performed with cells transfected by cationic lipid-mediated transfection with Lipofectamin 2000 (Invitrogen). These transfections were carried out according to the instructions of the manufacturer.
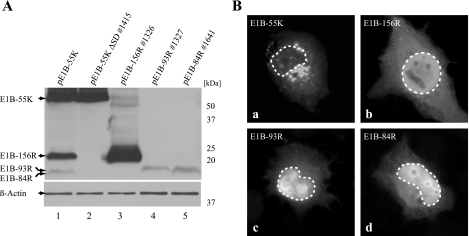
FIG. 2.
Expression and subcellular localization of E1B proteins. (A) H1299 cells were transfected with different constructs (2 μg plasmid) expressing E1B-55K, E1B-156R, E1B-93R, and E1B-84R as indicated at the top. pE1B-55K ΔSD #1415 in contrast to pE1B-55K does not express E1B-156R, E1B-93R, or E1B-84R but only E1B-55K. A Western blot assay of β-actin was included as a loading control. Molecular mass markers are indicated at the right. (B) The subcellular localization of the four E1B proteins was investigated by indirect immunofluorescence analysis of transfected H1299 cells (5 μg of pE1B-55K ΔSD #1415, pE1B-156R #1326, pE1B-93R #1327, or pE1B-84R #1641). Boundaries of nuclei are marked with dashed lines. Magnification, ×7,600.
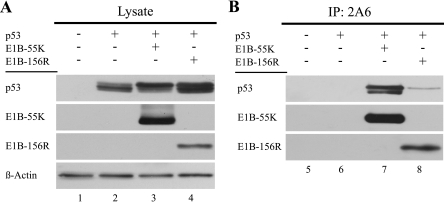
FIG. 5.
E1B-156R binds to a p53 form. (A) H1299 cells were transfected with plasmids expressing p53 (3 μg pC53-SN3), E1B-55K (7 μg pE1B-55K ΔSD #1415), and E1B-156R (7 μg pE1B-156R #1326), and total cell extracts were prepared. A plus sign indicates the use of a given plasmid in the corresponding lane. p53 and the E1B proteins were detected by immunoblotting. A Western blot assay of β-actin was included as a loading control. (B) The same lysates were used to immunoprecipitate (IP) the E1B proteins with the 2A6 antibody. The proteins could be detected by Western blot assays using 7C11. p53 was specifically coprecipitated with E1B-156R as well as E1B-55K and detected with the FL-393 antibody in Western blot assays.
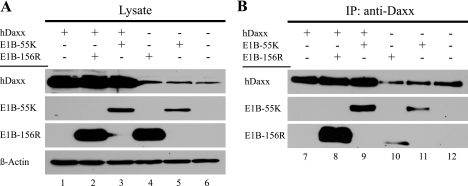
FIG. 8.
E1B-156R can bind human Daxx. (A) H1299 cells were transfected with plasmids expressing human Daxx (25 μg phDaxx-HA #1607), E1B-55K ΔSD (15 μg pE1B-55K ΔSD #1415), and E1B-156R (15 μg pE1B-156R #1326), and total cell extracts were prepared. A plus sign indicates the use of a given plasmid in the corresponding lane. Daxx and the E1B proteins were detected by immunoblotting. A Western blot assay of β-actin was included as a loading control. (B) The same lysates were used to immunoprecipitate (IP) Daxx with the anti-Daxx antibody. The protein could be detected by Western blot assays in all samples using the same antibody. E1B-156R and E1B-55K were coimmunoprecipitated with Daxx and detected by Western blot assays using the 2A6 antibody.
The effect of E1B-156R on gene transactivation was determined using the dual-luciferase reporter assay system (Promega). H1299 cells (2 × 105) were seeded into each well of a six-well plate. Cells were transfected 24 h later with the effector constructs, a reporter, and 1 μg of pRL-TK as an internal control. To avoid the effects of promoter competition, the total amount of CMV or simian virus 40 promoter was equalized by inclusion of empty pcDNA3 or pSG424 vector. Total cell extracts were prepared 48 h after transfection, and luciferase activities were assayed using an automated luminometer (Lumat LB9510; Berthold). Efficiency of transfection was calculated based on the luminescence of the Renilla luciferase control, allowing results to be standardized.
Transformation assays and cell lines.
Primary baby rat kidney (BRK) cells were obtained from kidneys of 5- to 6-day-old Sprague-Dawley rats as described previously (43, 45). They were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). For transformation assays, subconfluent cells were transfected 2 days postplating with plasmids encoding E1A and E1B proteins. To avoid promoter competition effects, the total amount of CMV promoter was equalized by addition of empty pcDNA3 vector. Five weeks after transfection cell colonies were stained with crystal violet (1% in 25% methanol) and dense foci of morphologically transformed cells were scored. Alternatively, foci were pooled or cloned and subsequently expanded into permanent cell lines.
AB605, AB606, and AB629 are neomycin (G418 from Calbiochem)-selected cell lines derived from transfections of primary BRK cells with pCMV-E1A 12S and pE1B-156R #1326, which contains the neomycin resistance cassette. While AB605 and AB629 are monoclonal, AB606 is of polyclonal origin. The BRK cell lines AB7 and AB120 have been described previously (16, 44). AB7 cells express the Ad5 E1 proteins; AB120 expresses Ad5 E1A and E1B-55K. The A4 cell line was acquired from primary BRK cells transfected with pCMV-E1A 12S by M. Nevels (Institut für Medizinische Mikrobiologie und Hygiene, Universität Regensburg, Germany). The BRK cell lines and primary BRK cells were maintained in DMEM with 10% FCS. A further 500 μg/ml of neomycin was added to the medium of all BRK cell lines except AB7. The p53-negative human H1299 cells (41) were cultivated in DMEM with 5% FCS. Growth studies were performed exactly as described previously (44).
Antibodies.
The mouse monoclonal immunoglobulin G (IgG) antibodies (MAbs) 2A6 (56) and AC-15 (Sigma) were used in this study. They recognize the amino termini of Ad5 E1B-55K and its related proteins and β-actin, respectively. The rat MAb 7C11 was used for detection of E1B-55K and E1B-156R, since it reacts with their common carboxy terminus (B. Härtl, T. Zeller, P. Blanchette, E. Kremmer, and T. Dobner, submitted for publication). In general, E1B-55K and its related proteins were detected with the antibody 2A6 unless the use of 7C11 is stated.
The polyclonal rabbit IgG antibodies anti-Daxx and anti-p53 FL-393 were obtained from Upstate and Santa Cruz, respectively. Rat-, mouse- and rabbit-specific horseradish peroxidase-coupled secondary antibodies from goat, sheep, and donkey, respectively, were acquired from Amersham. For indirect immunofluorescence mouse- and rabbit-specific fluorescein isothiocyanate (FITC)- and Texas Red-conjugated secondary donkey antibodies from Dianova were used.
Indirect immunofluorescence and phase-contrast microscopy.
For indirect immunofluorescence analysis H1299 cells were grown on coverslips to subconfluent densities in six-well dishes and transfected with plasmid DNA. Forty-eight hours posttransfection the coverslips were washed three times for 5 min in phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 4 mM Na2HPO4, 1.5 mM K2HPO4), and the cells were subsequently fixed with paraformaldehyde (2% in PBS) for 10 min at room temperature. Following a 15-min permeabilization in 0.5% Triton X-100 (Sigma) diluted in PBS, the fixation was repeated with 4% paraformaldehyde. The coverslips were washed two times for 5 min with PBS, and in a next step unspecific binding was blocked by a 1-h treatment with TBS-BG (20 mM Tris-chloride, pH 7.6, 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 5 g/liter glycine, 5 g/liter bovine serum albumin fraction V [PAA], 0.05% Tween 20 [Applichem], 0.5 g/liter sodium azide) at room temperature. After that, cells were incubated for 1 h at room temperature with hybridoma supernatant containing 2A6 antibody for detection of E1B and/or with FL-393 diluted in PBS for detection of p53. The coverslips were washed three times with PBS and incubated for another hour with 10 μg of appropriate FITC- or Texas Red-conjugated secondary antibody in PBS. Finally they were washed three times with PBS-Tween (0.1% Tween 20) and mounted in Glow medium (Energene). Digital immunofluorescence images were acquired on a DMRB fluorescence microscope (Leica) with a charge-coupled device camera (Diagnostic Instruments).
For studies on cell morphology, cells were examined by phase-contrast microscopy. Therefore subconfluent cells were washed once with PBS, treated with fresh 10% FCS-containing DMEM, and examined without fixation. All samples were analyzed and photographed with a camera-mounted Zeiss Axiovert 200 microscope. Adobe Systems Photoshop 6.0 and Illustrator CS2 were used to crop the images and design the illustrations.
Immunoprecipitation and immunoblotting.
For analysis of proteins by immunoblotting about 2 × 107 transfected H1299 cells were lysed on ice in radioimmunoprecipitation (RIPA) buffer (150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM dithiothreitol [DTT], 50 mM Tris-chloride, pH 8.0). Lysates for immunoprecipitation of Daxx were produced using NP-40 lysis buffer (50 mM Tris-chloride, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 1 mM PMSF, 2 mM DTT), while for coimmunoprecipitation of p53 RIPA light (50 mM Tris-chloride, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, 0.1% Triton X-100) was used. After normalization for protein concentration using the Bio-Rad protein assay, whole-cell extracts were used for immunoprecipitation or directly in Western blotting assays as previously described in detail (40, 45).
RESULTS
Expression and localization of E1B-55K, E1B-156R, E1B-93R, and E1B-84R in transfected H1299 cells.
For the experiments performed in this study it was necessary to generate plasmid constructs that express the different E1B proteins individually. The formerly described pE1B-55K (46) contains a 2,095-bp fragment of unspliced Ad5 sequence beginning with the ATG of the E1B-55K open reading frame under the control of a CMV promoter. This plasmid was found to express E1B-55K, but also E1B-156R, E1B-93R, and/or E1B-84R, through alternative splicing (Fig. 2A, lane 1). As it was beneficial in some experiments to express E1B-55K alone, pE1B-55K ΔSD #1415 was generated. This plasmid is identical to pE1B-55K, except that splice donor site SD1 (Fig. 1) was destroyed by site-directed mutagenesis to prevent production of differentially spliced mRNAs and the resulting E1B-55K-related proteins (Fig. 2A, lane 2). The pcDNA3 constructs pE1B-156R #1326, pE1B-93R #1327, and pE1B-84R #1641 contain the respective E1B cDNAs acquired from cytoplasmic mRNAs of Ad5-infected cells (Fig. 2A, lanes 3 to 5). In transient-transfection experiments E1B-156R, E1B-93R, and E1B-84R migrated at molecular masses of approximately 20 kDa, 17 kDa, and 16 kDa, respectively.
The subcellular localizations of E1B-55K and its related proteins in transfected H1299 cells individually expressing one of these proteins were investigated. E1B-55K showed weak, diffuse cytoplasmic and nuclear staining and brightly stained cytoplasmic structures (Fig. 2B, a), consistent with previously published reports (37, 70). E1B-156R, E1B-93R, and E1B-84R were diffusely distributed throughout the cell. The staining was most intense in the nucleus and weaker in the cytoplasm, although an accumulation in a distinct area around the nucleus could be observed (Fig. 2B, b to d).
The Ad5 E1B-156R protein cooperates with E1A to stably transform primary rat cells.
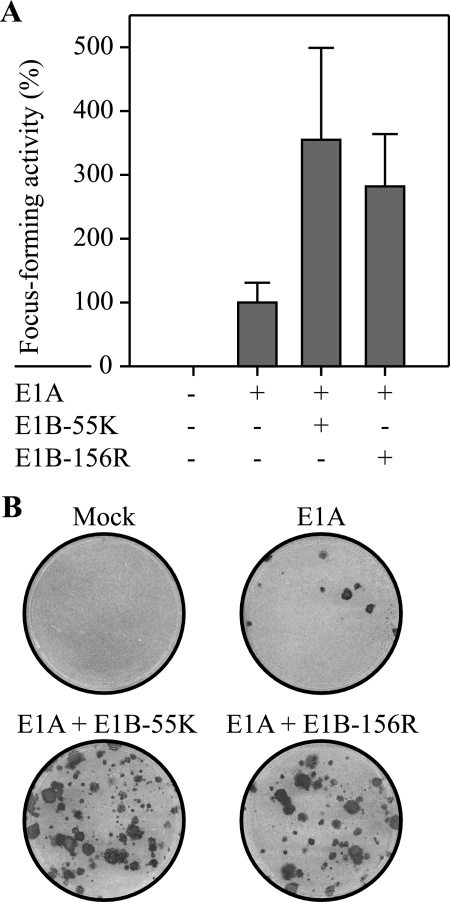
Since it was shown that the carboxy terminus of the E1B-55K oncoprotein is important for its transforming potential (61, 62), we examined if E1B-156R, which contains the same region, might also exhibit this activity. Therefore, primary BRK cells were transfected with plasmids expressing E1A 12S and E1B-55K or E1B-156R (Fig. 3A). The transfection of pCMV-E1A 12S alone yielded only a few small foci (Fig. 3B). Coexpression of E1A 12S and E1B-55K resulted in induction of dense, fast-growing foci with a three- to fourfold-higher frequency than that seen with pCMV-E1A 12S transfection alone. Similar to E1B-55K, E1B-156R in cooperation with E1A 12S was able to significantly stimulate transformation, as the rate of focus formation was almost tripled in comparison to that of E1A alone. The E1B-156R/E1A 12S-induced foci could be successfully expanded into stable cell lines. These results show that E1B-156R has transforming potential. By contrast, neither E1B-84R nor E1B-93R could significantly stimulate E1A-induced focus formation (data not shown).
FIG. 3.
E1B-156R cooperates with E1A to promote focus formation. (A) Primary BRK cells were transfected with plasmids encoding Ad5 E1A 12S (1.5 μg pCMV-E1A 12S), E1B-55K (3 μg pE1B-55K), and E1B-156R (3 μg pE1B-156R #1326). A plus sign indicates the use of a given plasmid in the corresponding column. Morphologically transformed colonies were scored 5 weeks after transfection. Focus-forming activity is represented as a percentage of pCMV-E1A 12S activity. The means and standard deviations for three independent experiments are presented. The average number of foci for pCMV-E1A 12S was 4. (B) Representative crystal violet-stained plates showing foci from transfections are included as examples.
E1A 12S/E1B-156R-transformed cell lines show alterations in morphology, and their growth rates and saturation densities are similar to those of E1A- or E1A/E1B-expressing cell lines.
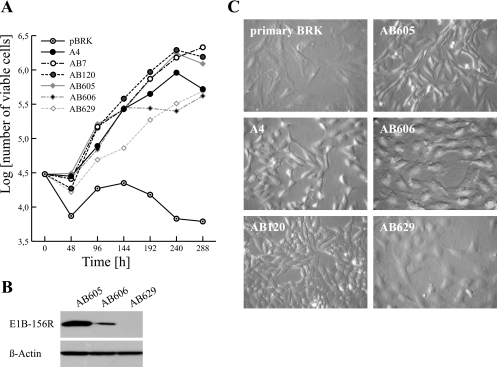
To evaluate the influence of E1B-156R on the phenotype of transformed BRK cells, we compared the growth properties and morphological appearances of three cell lines derived from E1A 12S/E1B-156R transfections with reference to primary BRK cells and cells that express only E1A or E1A plus E1B proteins. The cell lines tested in these experiments differed in their levels of E1B-156R expression. Moderate levels of E1B-156R were produced in AB605 cells, AB606 yielded only very small amounts, and in AB629 cells no E1B-156R could be detected by immunoblotting (Fig. 4B). Expression of E1B-156R correlated well with the growth rates and saturation densities, while no differences in E1A levels could be observed. In fact E1A expression levels were not detectable in standard Western blot analysis (data not shown). The moderately expressing AB605 cells were comparable to AB7 (expressing Ad5 E1A/E1B) and AB120 (expressing Ad5 E1A/E1B-55K); AB606 and AB629 grew more slowly and to lower saturation densities and were therefore similar to A4 cells, which produce only E1A (Fig. 4A). As expected all cell lines grew better than the primary BRK cells. Also the morphological appearance of the cell lines seemed to be linked to E1B-156R expression. All E1A/E1B-156R cell lines showed morphological alterations in comparison to primary BRK cells (Fig. 4C). They appeared less stretched out and smaller than their progenitors.
FIG. 4.
Growth characteristics of AB605, AB606, and AB629 cell lines. (A) The growth rates and saturation densities of E1A 12S/E1B-156R-transfected BRK cells were comparable to those of A4 (expresses E1A 12S), AB7 (expresses E1), and AB120 (expresses E1A and E1B-55K) cells and higher than that of primary BRK cells. A total of 3 × 104 cells were plated on six-well dishes in culture medium containing 5% fetal calf serum, and viable cells were counted every 48 h. The log10 means of viable cells for duplicate dishes are shown. (B) AB605, AB606, and AB629 cells show different E1B-156R steady-state levels. Whole-cell extracts of all three cell lines were prepared, and the same amounts were used in immunoblotting. A Western blot assay of β-actin was included as a loading control. (C) The morphologies of AB605, AB606, and AB629 cells are altered in comparison to primary BRK cells and similar to A4 (E1A) and AB120 (E1A and E1B-55K) cells at subconfluency. The cells were plated on coverslips and photographed. Magnification, ×1,780 (phase-contrast microscopy).
E1B-156R interacts with p53.
As the transformation-promoting activities of the Ad5 E1B-55K protein are, in general, linked to its ability to bind to p53 and to repress its functions, we tested if E1B-156R might also interact with p53 by coimmunoprecipitation assays. H1299 cells were transfected with plasmids encoding p53, E1B-55K, and E1B-156R. The steady-state levels of the exogenously expressed proteins are illustrated in Fig. 5A. p53 showed at least two closely migrating forms, likely reflecting differently modified forms of the tumor suppressor protein. Immunoprecipitation with the E1B-specific 2A6 antibody resulted in coprecipitation of p53. While E1B-55K coimmunoprecipitated two different species of p53 at comparable levels, only a weak interaction of E1B-156R with the slower-migrating form of these two could be observed. Though the possibility that E1B-156R also has some affinity for the faster-migrating p53 form cannot be completely excluded, its affinity is certainly considerably reduced in comparison to that of E1B-55K. No p53 could be precipitated in samples without E1B expression, indicating a specific interaction between p53 and both E1B proteins.
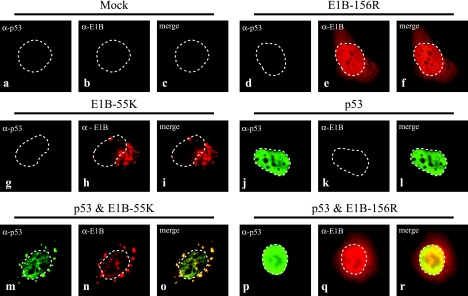
Given these findings we next tested whether E1B-156R might also have an influence on the subcellular distribution of p53. Subconfluent H1299 cells were transfected with plasmids encoding p53, E1B-55K, and E1B-156R for immunofluorescence studies. All samples were double labeled with antibodies directed against p53 and the E1B proteins. The target proteins were visualized using appropriate secondary antibodies marking p53 with FITC (green) and the E1B proteins with Texas Red (red). For analysis >100 cells were compared for each sample. In Fig. 6a to c untransfected H1299 cells were stained with antibodies directed against p53 and E1B-55K as a control for signal specificity. As illustrated (Fig. 2B, b, and 6d to f) E1B-156R is distributed diffusely in the nucleus and the cytoplasm, with an accumulation in and around the nucleus in all examined cells. E1B-55K shows a weak diffuse staining and distinct cytoplasmic bodies close to the nucleus (Fig. 6g to i) in 95% of the cells as described previously for a splicing-competent E1B-55K construct (70). The exogenously expressed p53 was in general localized diffusely in the nucleus (Fig. 6j to l); in about 13% of the cells also a weak diffuse cytoplasmic staining was observed. If p53 and E1B-55K are coexpressed, a large fraction of the tumor suppressor is always localized together with E1B-55K in the cytoplasmic aggregates (Fig. 6m to o) (70). In contrast, coexpression of p53 with E1B-156R did not alter the distribution of the two proteins (Fig. 6p to r).
FIG. 6.
E1B-156R does not relocalize p53 in transfected H1299 cells. H1299 cells were transfected with plasmids expressing p53 (1 μg pC53-SN3), E1B-55K (1 μg pE1B-55K ΔSD #1415), or E1B-156R (1 μg pE1B-156R #1326) or a combination of p53 and one of the E1B proteins. The exogenous proteins expressed in each sample are noted above the corresponding subset of triplet pictures. Images a to c were derived from untransfected (mock) cells and show the level of unspecific background staining. The cells were double labeled in situ with the anti-E1B mouse antibody 2A6 and the p53-specific antibody FL-393. These were detected with Texas Red- and FITC-conjugated secondary antibodies, respectively. Anti-p53 (green; a, d, g, j, m, and p) and anti-E1B (red; b, e, h, k, n, and q) staining patterns are visible. An overlay of both patterns (merge) is displayed in panels c, f, i, l, o, and r. Boundaries of nuclei are marked with dashed lines. Magnification was ×7,600.
E1B-156R does not repress p53-mediated transactivation.
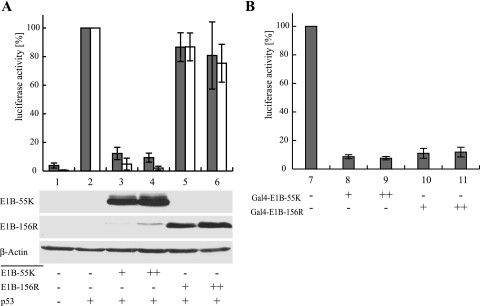
The ability of E1B-55K to repress p53-mediated transactivation has been shown to be a crucial function in adenoviral cell transformation in cooperation with E1A and to be mediated by a transcriptional repression domain located in the carboxy-terminal region of the Ad protein around amino acid residue R443 (68, 69). Since the 77 carboxy-terminal amino acids of E1B-55K, including R443, are also found in E1B-156R, this protein might possess a functional repression domain as well. Therefore, we tested the ability of E1B-156R to repress the transactivation of p53-responsive constructs expressing firefly luciferase (Fig. 7A). We used two different reporter plasmids driven by promoters containing either an artificial construct of five p53-binding sites or a sequence from the p53-responsive cyclin G promoter. While transfection of a reporter construct without p53 resulted in a low basal activity (Fig. 7A, bars 1), coexpression of the tumor suppressor led to strong induction of firefly luciferase (Fig. 7A, bars 2). Addition of increasing amounts of E1B-55K to this system caused an almost complete abolishment of p53-mediated transactivation (Fig. 7A, bars 3 and 4), while transfection of the same amounts of E1B-156R had almost no effect (Fig. 7A, bars 5 and 6). These observations were made for both the artificial and cyclin G promoters. To rule out the possibility that the lack of E1B-156R function was due to weak protein expression, Western blot assays were performed; these assays confirmed comparable steady-state levels of E1B-55K and E1B-156R (Fig. 7A, lanes 3 and 4 and 5 and 6). To preserve comparability with previously described observations made with splicing-competent E1B-55K constructs (38, 39), pE1B-55K was favored in this experimental setting instead of pE1B-55K ΔSD #1415. Therefore, E1B-55K expression was accompanied by low-level production of E1B-156R (Fig. 7A, lanes 3 and 4).
FIG. 7.
E1B-156R does not repress p53-mediated transactivation but possesses a functional repression domain. In both reporter assays “+” indicates the use of a given plasmid in the corresponding lane and “++” marks the use of the higher of two possible plasmid concentrations. The means and standard deviations for four independent experiments are presented. (A) E1B-156R does not repress p53-mediated transactivation. H1299 cells were transfected with 1 μg of a construct expressing firefly luciferase under the control of a p53-dependent promoter. Two different reporter plasmids were tested in these experiments: pHH0.34-TK Luc, containing sequences of the cellular cyclin G promoter (gray bars), and pRE-Luc, with an artificial promoter containing five copies of a p53 DNA-binding motif (white bars). Each subset of experiments further contained plasmids expressing p53 (0.025 μg pC53-SN3), E1B-55K (1 or 2 μg pE1B-55K), and E1B-156R (1 or 2 μg pE1B-156R #1326). The steady-state levels of the E1B proteins were monitored by immunoblotting of lysates used in a luciferase assay. A Western blot assay of β-actin was included as a loading control. (B) E1B-156R contains a functional repression domain. H1299 cells were transfected with 1 μg of a construct expressing firefly luciferase under the control of an artificial promoter containing five Gal4 binding sites (pGalTK-Luc). The constitutive expression of this construct was repressed by addition of the Gal4 DNA-binding domain E1B fusion proteins Gal4-E1B-55K (1 or 2 μg pGal4 E1B-55kDa) or Gal4-E1B-156R (1 or 2 μg pGal4 E1B-156R #1375).
It was shown that the E1B-55K repression domain is not restricted to repression of p53-stimulated transcription (69). Therefore it is possible that E1B-156R, though it fails to affect p53 transactivation, might still have a functional repression domain. To test this possibility, we performed another set of reporter assays using E1B-55K and E1B-156R fused to the Gal4 DNA-binding domain (Fig. 7B). The reporter construct in these experiments expressed firefly luciferase under the control of a Gal4-binding site-containing promoter (Fig. 7B, lane 7). Expression of Gal4-E1B-55K resulted in a decrease of luciferase activity to about 5% of the basal level (Fig. 7B, lanes 8 and 9). Gal4-E1B-156R also caused a dramatic decrease to about 12% luciferase activity (Fig. 7B, lanes 10 and 11), demonstrating that E1B-156R contains a functional repression domain. These findings were confirmed by data from similar experiments performed by L. Assefi (2).
E1B-156R binds to human Daxx.
As the transforming potential of E1B-156R proved to be independent of repressing p53 transactivation, we investigated other cellular factors that might mediate the transforming potential of E1B-156R. A promising candidate was the human Daxx protein, as it was shown to interact with the 58 carboxy-terminal amino acids of Ad2 E1B-55K (71). The primary sequence of this part of the Ad2 protein is identical to Ad5 E1B-55K and is also contained in Ad5 E1B-156R. Daxx was originally reported to be involved in Fas-induced apoptotic signaling. Upon binding to the death domain of an activated Fas receptor, Daxx binds and activates apoptosis signal-regulating kinase 1, which in turn activates the Jun N-terminal kinase pathway and promotes apoptosis (7, 8, 66). Furthermore, Daxx acts as a transcription regulator able to repress or activate transcription by direct interaction with different DNA-binding transcription factors, including p53 (13, 23, 27, 31, 32, 48, 71). Therefore, we tested if E1B-156R is able to interact with human Daxx (hDaxx). As illustrated in Fig. 8A, transiently transfected H1299 cells expressed human Daxx (lanes 1 to 3), E1B-55K (lanes 3 and 5), and E1B-156R (lanes 2 and 4) at appropriate levels. Immunoprecipitations of human Daxx (Fig. 8B) with anti-Daxx revealed that E1B-156R as well as E1B-55K could be coprecipitated with overexpressed (lanes 7 to 9) and also with endogenous (lanes 10 to 12) hDaxx. Therefore, it can be concluded that Ad5 E1B-156R interacts with human Daxx.
DISCUSSION
In this report we have examined the role of the 156R E1B protein in Ad5 E1A/E1B-mediated transformation of primary BRK cells. Our data show for the first time that E1B-156R promotes focal transformation of primary BRK cells in combination with Ad5 E1A (Fig. 3), demonstrating that E1B-156R possesses transforming potential. Established cell lines expressing E1B-156R exhibited similar morphological alterations, growth rates, and saturation densities compared with transformants expressing E1A plus E1B-55K (Fig. 4). Thus, E1B-156R, like E1B-55K, confers transformed in vitro properties to primary cells, typically associated with complete cell transformation.
To identify the mechanisms of transformation used by E1B-156R, we compared the protein with the highly related E1B-55K. In coimmunoprecipitation assays we found that, though E1B-156R does have some affinity for p53, the interaction is considerably altered and overall weaker than that observed for E1B-55K (Fig. 5B, lanes 7 and 8). We assume, therefore, that E1B-156R must also contain a site suitable for p53 interaction, although it is not clear if this interaction is direct or indirect. However, the site described for p53 binding by E1B-55K is located in the central part of the protein (68), which is not included in E1B-156R. The p53 binding site of E1B-156R must therefore be distinct from this central one and might also be active within the larger protein. The observation that E1B-156R predominantly binds one of the p53 forms may indicate that the affinity of the two E1B proteins for p53 is affected in different ways by posttranslational modification of the tumor suppressor. Alternatively, it is possible that the two E1B proteins actively and distinctly influence p53 modification. Indeed, it was shown that E1B-55K inhibits p53 acetylation by PCAF (33) and thus might change the proportions of differently modified p53 species. This could also explain the differences observed in the expression pattern of transfected p53 (Fig. 5A, lanes 2 to 4).
No interaction between E1B-156R and endogenous p53 could be detected in the E1A 12S/E1B-156R-transformed cells. This is probably due to the low p53 levels observed in these cells (data not shown).
The next question we addressed was if the weak and modified interaction between E1B-156R and p53 would be sufficient to inhibit the tumor suppressor's functions. It is very likely that modulation of p53 functions by E1B-156R would involve mechanisms also used by E1B-55K. It is thought that at least two mechanisms contribute to inhibition of p53 functions by E1B-55K: relocalization of the tumor suppressor into cytoplasmic bodies (15, 24, 71) and repression of p53-mediated transactivation of genes (38, 39, 61, 68, 69). It was shown that the localization of E1B-55K and also the sequestration of p53 are influenced by two structural motifs: a nuclear export signal (15, 28) and a SUMO1 conjugation motif (16). Neither motif is included in any of the E1B-55K-related proteins. This is probably also reflected in the subcellular distribution of E1B-156R and its inability to sequester p53 (Fig. 2B and 6).
Further we could show that E1B-156R does not influence the transactivating activities of p53 (Fig. 7A). Since E1B-156R apparently did not possess any of the p53-counteracting functions ascribed to E1B-55K, we conclude that the transformation-promoting function of this protein must be mediated by mechanisms that are at least independent of blocking p53-stimulated transcription. Due to the close relationship between both E1B proteins this finding might further indicate that E1B-55K also possesses these functions. However, recent data indicate that p53 can also induce apoptosis by transcriptionally independent activities. It was shown to translocate into mitochondria, directly associate with Bcl-2 proteins, and activate the intrinsic apoptotic pathway (reviewed in reference 67). Therefore, the possibility that E1B-156R might still counteract p53 by blocking this mechanism cannot be ruled out completely.
The transformation-promoting potential of E1B-156R represents the first biological activity that could be assigned to any of the E1B-55K-related proteins. Since the mechanisms underlying this function remain unclear, we further investigated the properties of E1B-156R.
The ability of E1B-55K to repress the transactivating functions of p53 depends on the activity of its carboxy-terminal repression domain, which is also contained within E1B-156R. Until now p53 is the only known target for transcriptional repression by E1B-55K (38, 39, 61, 68, 69). Although we showed that E1B-156R is unable to repress p53-mediated transactivation, one can speculate that the E1B protein might repress other yet-unidentified factors. An important prerequisite in this case would be a functional repression domain in E1B-156R. It is known that phosphorylation of E1B-55K in the proximal carboxy terminus is important for transcriptional repression by E1B-55K (61, 62). E1B-156R contains homologous phosphorylation sites, which were shown to be phosphorylated (62). While this is already a hint for functionality, we proved here that E1B-156R indeed contains a functional repression domain (Fig. 7B) and therefore might act as transcriptional repressor of factors other than p53.
While E1B-55K alone is able to only inhibit p53, the interaction with Ad5 E4orf6 enables it to efficiently promote degradation of the tumor suppressor by utilizing a Cullin5-based ubiquitin ligase complex (5, 22, 51). Testing E1B-156R for similar properties, we discovered that it also binds to E4orf6 but is unable to cooperate with it in promotion of p53 degradation (data not shown). It was recently discovered that p53 is not the only target of the E1B-55K/E4orf6 ubiquitin ligase complex since components of the cellular MRN complex, which is involved in DNA double-strand break repair, are also degraded (59). In the E1B-55K/E4orf6 complex E1B-55K acts as a substrate recognition subunit, while E4orf6 mediates the interaction with cellular factors (5). Therefore, it is tempting to speculate that E1B-156R and E4orf6 could promote degradation of proteins other than p53. The detected interaction between E4orf6 and E1B-156R was surprising, since E1B-156R does not contain the amino acid sequences of the central part of E1B-55K, which are thought to be essential for an E1B-55K/E4orf6 interaction (53). This indicates that sites distinct from those described for E1B-55K are able to mediate binding to E4orf6. The nature of this interaction is unclear and could be direct as well as indirect. It seems likely that the site within E1B-156R could also contribute to E1B-55K binding to E4orf6.
The sites in the central part of E1B-55K described to be important for interactions with p53 as well as E4orf6 were identified by using mutant E1B proteins unable to bind these interactants (26, 53). Furthermore, some of these mutations were shown to inactivate the carboxy-terminal repression domain of the protein (69). It was speculated that mutations in the central part of the protein might disrupt its tertiary structure, thereby impairing its ability to interact with other proteins and act as a transcriptional repressor (53, 69). Therefore, it is feasible that, instead of two interaction sites for p53 and two sites for E4orf6 in E1B-55K, there is only one for each and that these are located within the amino acid sequences shared with E1B-156R. This would imply that the central part of E1B-55K is of general importance for the stability and/or conformation of the protein.
Since E1B-156R shows a transforming potential that is independent of p53 transactivation repression, we tried to identify other interactants that might influence transformation. A possible candidate was the Daxx protein, an apoptotic regulator described as direct interactant of the Ad2 and Ad12 E1B-55K proteins (71). We found that this protein also interacts with Ad5 E1B-55K and E1B-156R. Although it is not certain at the moment if interaction with or modulation of Daxx contributes to the transforming potential of the E1B proteins, it was shown that modulations of Daxx affect several apoptotic pathways (reviewed in reference 47).
Apart from any individual functions E1B-156R might have, its ability to interact with Daxx and E4orf6 could also enable it to modulate E1B-55K functions requiring these proteins. Any activity of E1B-156R that depends on an interaction with these shared cellular and viral factors would lead to competition with E1B-55K. The fact that Ad2 E1B-55K, which is highly similar to the Ad5 protein, was shown to form dimers (38) opens up a further possibility: one task of E1B-156R might be the direct modification of E1B-55K functions by forming heterodimers with altered functionality. Our data and those from others show that E1B-156R and other E1B-55K-related proteins are expressed in adenovirus-infected human cells (60), E1A/E1B-transformed rat (AB120; our unpublished results) and human cells (293) (60), and H1299 cells transiently transfected with plasmids containing splicing-competent E1B-55K sequences (Fig. 2). With the latest evidence for E1B-156R functions it seems likely that E1B-55K-related proteins might contribute to the spectrum of functions accredited to E1B-55K in all these systems.
In conclusion, we have shown that E1B-156R does transform primary rat cells in cooperation with E1A 12S independently of repression of p53-mediated transactivation. This, in combination with the newly revealed interactions with E4orf6 and Daxx, indicates a functional role for E1B-156R. As the experiments of this study were performed with overexpressed E1B-156R, it will now be interesting to dissect the interactions and functions of E1B-156R in virus infections.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Anderson, C. W., R. C. Schmitt, J. E. Smart, and J. B. Lewis. 1984. Early region 1B of adenovirus 2 encodes two coterminal proteins of 495 and 155 amino acid residues. J. Virol. 50:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assefi, L. 1998. Identification of the transcriptional repression domains of the adenovirus type 5 E1B-55KDa protein. Ph.D. thesis. McGill University, Montreal, Canada.
- 3.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. [DOI] [PubMed] [Google Scholar]
- 4.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673-7685. [DOI] [PubMed] [Google Scholar]
- 5.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, J. L., L. J. Polder, R. Bernards, P. I. Schrier, P. J. van den Elsen, A. J. van der Eb, and H. van Ormondt. 1981. The 2.2 kb E1b mRNA of human Ad12 and Ad5 codes for two tumor antigens starting at different AUG triplets. Cell 27:121-131. [DOI] [PubMed] [Google Scholar]
- 7.Chang, H. Y., H. Nishitoh, X. Yang, H. Ichijo, and D. Baltimore. 1998. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281:1860-1863. [DOI] [PubMed] [Google Scholar]
- 8.Charette, S. J., H. Lambert, and J. Landry. 2001. A kinase-independent function of Ask1 in caspase-independent cell death. J. Biol. Chem. 276:36071-36074. [DOI] [PubMed] [Google Scholar]
- 9.Chow, L. T., T. R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. [DOI] [PubMed] [Google Scholar]
- 10.Cuconati, A., and E. White. 2002. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 16:2465-2478. [DOI] [PubMed] [Google Scholar]
- 11.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 12.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 13.Emelyanov, A. V., C. R. Kovac, M. A. Sepulveda, and B. K. Birshtein. 2002. The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J. Biol. Chem. 277:11156-11164. [DOI] [PubMed] [Google Scholar]
- 14.Endter, C., and T. Dobner. 2004. Cell transformation by human adenoviruses. Curr. Top. Microbiol. Immunol. 273:163-214. [DOI] [PubMed] [Google Scholar]
- 15.Endter, C., B. Hartl, T. Spruss, J. Hauber, and T. Dobner. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 24:55-64. [DOI] [PubMed] [Google Scholar]
- 16.Endter, C., J. Kzhyshkowska, R. Stauber, and T. Dobner. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. USA 98:11312-11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gey, G. O., W. D. Coffman, and M. T. Kubicek. 1952. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 12:264-265. [Google Scholar]
- 18.Gingeras, T. R., D. Sciaky, R. E. Gelinas, J. Bing-Dong, C. E. Yen, M. M. Kelly, P. A. Bullock, B. L. Parsons, K. E. O'Neill, and R. J. Roberts. 1982. Nucleotide sequences from the adenovirus-2 genome. J. Biol. Chem. 257:13475-13491. [PubMed] [Google Scholar]
- 19.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 20.Grand, R. J., M. L. Grant, and P. H. Gallimore. 1994. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology 203:229-240. [DOI] [PubMed] [Google Scholar]
- 21.Green, M., K. H. Brackmann, M. A. Cartas, and T. Matsuo. 1982. Identification and purification of a protein encoded by the human adenovirus type 2 transforming region. J. Virol. 42:30-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenbach, A. D., J. E. Sublett, C. J. McPherson, and G. Grosveld. 1999. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 18:3702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutton, F. G., A. S. Turnell, P. H. Gallimore, and R. J. Grand. 2000. Consequences of disruption of the interaction between p53 and the larger adenovirus early region 1B protein in adenovirus E1 transformed human cells. Oncogene 19:452-462. [DOI] [PubMed] [Google Scholar]
- 25.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 26.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179:806-814. [DOI] [PubMed] [Google Scholar]
- 27.Kim, E. J., J. S. Park, and S. J. Um. 2003. Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML. Nucleic Acids Res. 31:5356-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 29.Kraus, V. B., E. Moran, and J. R. Nevins. 1992. Promoter-specific trans-activation by the adenovirus E1A12S product involves separate E1A domains. Mol. Cell. Biol. 12:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, J. B., and C. W. Anderson. 1987. Identification of adenovirus type 2 early region 1B proteins that share the same amino terminus as do the 495R and 155R proteins. J. Virol. 61:3879-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, R., H. Pei, D. K. Watson, and T. S. Papas. 2000. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene 19:745-753. [DOI] [PubMed] [Google Scholar]
- 33.Liu, Y., A. L. Colosimo, X. J. Yang, and D. Liao. 2000. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol. Cell. Biol. 20:5540-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 36.Lucher, L. A., K. H. Brackmann, J. S. Symington, and M. Green. 1984. Antibody directed to a synthetic peptide encoding the NH2-terminal 16 amino acids of the adenovirus type 2 E1B-53K tumor antigen recognizes the E1B-20K tumor antigen. Virology 132:217-221. [DOI] [PubMed] [Google Scholar]
- 37.Maheswaran, S., C. Englert, S. B. Lee, R. M. Ezzel, J. Settleman, and D. A. Haber. 1998. E1B 55K sequesters WT1 along with p53 within a cytoplasmic body in adenovirus-transformed kidney cells. Oncogene 16:2041-2050. [DOI] [PubMed] [Google Scholar]
- 38.Martin, M. E., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 72:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, M. E., and A. J. Berk. 1999. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol. Cell. Biol. 19:3403-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 64:2345-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7:171-180. [PubMed] [Google Scholar]
- 42.Montell, C., E. F. Fisher, M. H. Caruthers, and A. J. Berk. 1984. Control of adenovirus E1B mRNA synthesis by a shift in the activities of RNA splice sites. Mol. Cell. Biol. 4:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 1997. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. USA 94:1206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevels, M., T. Spruss, H. Wolf, and T. Dobner. 1999. The adenovirus E4orf6 protein contributes to malignant transformation by antagonizing E1A-induced accumulation of the tumor suppressor protein p53. Oncogene 18:9-17. [DOI] [PubMed] [Google Scholar]
- 45.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nevels, M., B. Tauber, T. Spruss, H. Wolf, and T. Dobner. 2001. “Hit-and-run” transformation by adenovirus oncogenes. J. Virol. 75:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norbury, C. J., and B. Zhivotovsky. 2004. DNA damage-induced apoptosis. Oncogene 23:2797-2808. [DOI] [PubMed] [Google Scholar]
- 48.Ohiro, Y., A. Usheva, S. Kobayashi, S. L. Duffy, R. Nantz, D. Gius, and N. Horikoshi. 2003. Inhibition of stress-inducible kinase pathways by tumorigenic mutant p53. Mol. Cell. Biol. 23:322-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto, K., and D. Beach. 1994. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 13:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perricaudet, M., G. Akusjarvi, A. Virtanen, and U. Pettersson. 1979. Structure of two spliced mRNAs from the transforming region of human subgroup C adenoviruses. Nature 281:694-696. [DOI] [PubMed] [Google Scholar]
- 51.Querido, E., M. R. Morrison, H. Chu-Pham-Dang, S. W. Thirlwell, D. Boivin, and P. E. Branton. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Resnick, M. A., D. Tomso, A. Inga, D. Menendez, and D. Bell. 2005. Functional diversity in the gene network controlled by the master regulator p53 in humans. Cell Cycle 4:1026-1029. [DOI] [PubMed] [Google Scholar]
- 53.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabbatini, P., S. K. Chiou, L. Rao, and E. White. 1995. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol. Cell. Biol. 15:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadowski, I., and M. Ptashne. 1989. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 17:7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510-517. [DOI] [PubMed] [Google Scholar]
- 57.Shenk, T. 2001. Adenoviridae: the viruses and their replication, vol. 4. Raven Press, New York, N.Y.
- 58.Spector, D. J., M. McGrogan, and H. J. Raskas. 1978. Regulation of the appearance of cytoplasmic RNAs from region 1 of the adenovirus 2 genome. J. Mol. Biol. 126:395-414. [DOI] [PubMed] [Google Scholar]
- 59.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 60.Takayesu, D., J. G. Teodoro, S. G. Whalen, and P. E. Branton. 1994. Characterization of the 55K adenovirus type 5 E1B product and related proteins. J. Gen. Virol. 75:789-798. [DOI] [PubMed] [Google Scholar]
- 61.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teodoro, J. G., T. Halliday, S. G. Whalen, D. Takayesu, F. L. Graham, and P. E. Branton. 1994. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J. Virol. 68:776-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Virtanen, A., and U. Pettersson. 1985. Organization of early region 1B of human adenovirus type 2: identification of four differentially spliced mRNAs. J. Virol. 54:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, E. 2001. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene 20:7836-7846. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, M. C., and J. E. Darnell, Jr. 1981. Control of messenger RNA concentration by differential cytoplasmic half-life. Adenovirus messenger RNAs from transcription units 1A and 1B. J. Mol. Biol. 148:231-251. [DOI] [PubMed] [Google Scholar]
- 66.Yang, X., R. Khosravi-Far, H. Y. Chang, and D. Baltimore. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yee, K. S., and K. H. Vousden. 2005. Complicating the complexity of p53. Carcinogenesis 26:1317-1322. [DOI] [PubMed] [Google Scholar]
- 68.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 69.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 70.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]
- 71.Zhao, L. Y., A. L. Colosimo, Y. Liu, Y. Wan, and D. Liao. 2003. Adenovirus E1B 55-kilodalton oncoprotein binds to Daxx and eliminates enhancement of p53-dependent transcription by Daxx. J. Virol. 77:11809-11821. [DOI] [PMC free article] [PubMed] [Google Scholar]