Abstract
In Vibrio cholerae, the second messenger 3′,5′-cyclic diguanylic acid (c-di-GMP) regulates several cellular processes, such as formation of corrugated colony morphology, biofilm formation, motility, and virulence factor production. Both synthesis and degradation of c-di-GMP in the cell are modulated by proteins containing GGDEF and/or EAL domains, which function as a diguanylate cyclase and a phosphodiesterase, respectively. The expression of two genes, cdgC and mbaA, which encode proteins harboring both GGDEF and EAL domains is higher in the rugose phase variant of V. cholerae than in the smooth variant. In this study, we carried out gene expression analysis to determine the genes regulated by CdgC in the rugose and smooth phase variants of V. cholerae. We determined that CdgC regulates expression of genes required for V. cholerae polysaccharide synthesis and of the transcriptional regulator genes vpsR, vpsT, and hapR. CdgC also regulates expression of genes involved in extracellular protein secretion, flagellar biosynthesis, and virulence factor production. We then compared the genes regulated by CdgC and by MbaA, during both exponential and stationary phases of growth, to elucidate processes regulated by them. Identification of the regulons of CdgC and MbaA revealed that the regulons overlap, but the timing of regulation exerted by CdgC and MbaA is different, suggesting the interplay and complexity of the c-di-GMP signal transduction pathways operating in V. cholerae.
Vibrio cholerae, the causative agent of the disease cholera, is a facultative human pathogen that is a natural inhabitant of aquatic environments. It has been speculated that to survive in different environments and adapt to fluctuating environmental parameters, V. cholerae undergoes a phase variation event that generates two morphologically different variants termed smooth and rugose. The phenotypic properties of these two phase variants, including resistance to biocides and acid, osmotic, and oxidative stresses and biofilm-forming capacity, differ greatly (13, 40, 43, 59, 63). The rugose variant has a greater capacity to produce an exopolysaccharide, termed VPS for Vibrio polysaccharide, than the smooth variant. This property is responsible for many of the phenotypic differences between the two variants (63).
Most of the vps genes are clustered on the large chromosome of V. cholerae O1 El Tor and organized into the vpsI (vpsA to -K) and vpsII regions (vpsL to -Q) (61, 63). Two positive transcriptional regulators, VpsR and VpsT, both of which exhibit homology to response regulators, are required for vps gene expression and, in turn, for VPS production and corrugated colony formation. In contrast, the HapR transcriptional regulator, which controls quorum-sensing responses, negatively regulates the expression of vps genes (20, 61, 64). Recently, it was shown that proteins responsible for the turnover of a novel nucleotide, bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), regulate the expression of the genes of two vps clusters, genes located between the vps clusters, vpsR, and vpsT (3, 4, 28, 34, 55). However, the molecular mechanisms of vps gene regulation by these proteins remain unknown.
Cyclic nucleotides are cell signaling molecules that play diverse roles in the biology of microorganisms. c-di-GMP is a ubiquitous second messenger whose production and degradation are controlled by proteins containing the GGDEF and EAL domains, respectively. The GGDEF and EAL domains, named for their common consensus sequence, were recently shown to harbor intrinsic diguanylate cyclase (DGC) and c-di-GMP phosphodiesterase (PDEA) activity, respectively (8, 41, 48, 49, 54).
c-di-GMP was initially identified in Gluconacetobacter xylinus as an allosteric activator of cellulose synthase (46). However, recent studies have recognized the involvement of c-di-GMP in regulation of metabolic processes, cell differentiation, and modulation of the cell surface properties of microorganisms. More specifically, intracellular levels of c-di-GMP have been shown to regulate the formation of rugose colony morphology, intracellular aggregation, exopolysaccharide synthesis, biofilm formation, and motility in several species, such as Pseudomonas aeruginosa, Escherichia coli, Salmonella enterica serovar Typhimurium, and V. cholerae (10, 16, 26, 44, 45). In the past several years, data have emerged on the role of c-di-GMP in regulating the pathogenic capacity of bacterial pathogens and the production of virulence factors. For example, in Bordetella pertussis, an EAL domain protein called BvgR was found to regulate the expression of virulence factors and other genes of unknown function, thereby contributing to respiratory infection in mice (37). Through an in vivo screen to identify genes contributing to the response of Salmonella enterica serovar Typhimurium to host defense mechanisms, Hisert et al. identified CdgR, an EAL domain protein (23). CdgR was shown to promote resistance to hydrogen peroxide and to affect intracellular c-di-GMP levels, thereby suppressing the killing of pathogens by macrophage (23). Recently, mutational analysis of all Pseudomonas aeruginosa genes coding for proteins with a GGDEF and/or an EAL domain revealed that intracellular c-di-GMP levels regulate virulence-related traits, such as type III secretion system-mediated cytotoxicity (32). Additionally, two studies showed that c-di-GMP levels in the cell regulate the transcription of bacterial virulence factors. In one of them, Mendez-Ortiz et al. monitored the genome-wide transcriptional profile of E. coli in response to high levels of c-di-GMP and identified a set of genes related to virulence (36). In the other, Tischler and Camilli showed that VieA, an EAL domain protein from V. cholerae, regulates the transcription of toxT, the most downstream regulator of V. cholerae virulence factors, thereby regulating the expression of the ctxA and ctxB genes, encoding cholera toxin (CT) (56). This transcriptional regulation occurred through the EAL domain's control of c-di-GMP levels in the cell (56).
There are 53 genes encoding proteins with GGDEF and/or EAL domains in the V. cholerae genome: 31 encode GGDEF proteins, 12 encode EAL proteins, and 10 encode proteins with both a GGDEF and an EAL domain on the same polypeptide (17). At present, we have a limited understanding of the functions of these proteins in V. cholerae, as only a few of them have been studied in some detail (4, 28, 30, 42, 55-57). Previously, we identified and characterized 2 of the 10 genes coding for proteins containing both GGDEF and EAL domains, cdgC and mbaA. Expression levels of both genes were higher in the rugose variant than in the smooth variant (34, 61). Phenotypic analysis classified CdgC and MbaA as negative regulators of rugose colony morphology, biofilm formation, and vpsL transcription through the known signal transduction pathway involving the response regulators VpsR and VpsT and the quorum-sensing transcriptional regulator HapR (61). In addition, epistasis analysis revealed that CdgC and MbaA regulate rugose colony morphology in a nonredundant manner in V. cholerae (34).
The goal of the current study was to determine additional biological processes controlled by these two proteins. To this end, we determined the transcriptional profile of cdgC deletion mutants generated in both the rugose (RΔcdgC) and the smooth (SΔcdgC) phase variants and of an mbaA deletion in the rugose (RΔmbaA) genetic background. We found that in addition to regulating the expression of genes that can potentially alter cell surface properties, CdgC regulates expression of genes involved in extracellular protein secretion, flagellar biosynthesis, and virulence factor production. We also observed that CdgC regulates gene expression in a phase variant-dependent manner. We carried out gene expression profile comparisons of RΔcdgC and RΔmbaA during exponential and stationary phases of growth to understand the different molecular functions of the two genes. We observed that the expression profiles of RΔcdgC and RΔmbaA overlap in the regulation of certain processes but that the timing of their regulation is unique.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. cholerae and Escherichia coli strains used in this study are listed in Table 1. E. coli DH5α and CC118λpir were used for the maintenance of plasmids. E. coli strain S17-1λpir was used to deliver plasmids to V. cholerae by conjugation. V. cholerae and E. coli strains were grown in Luria-Bertani (LB; 1% tryptone, 1% NaCl, 0.5% yeast extract, 1.5% Difco granulated agar) broth with aeration at 30°C unless otherwise noted. LB medium was supplemented with the following concentrations of antibiotics: ampicillin, 100 μg ml−1, and rifampin, 100 μg ml−1. Agar plates consisting of LB agar with 0.3% Difco agar were used to measure motility. LB broth without NaCl and with 10% sucrose was used for counterselection with sacB-containing plasmids. For virulence factor-inducing conditions, strains were cultured in AKI medium as described previously (25).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| CC118λpir | Δ(ara-leu) araD ΔlacX74galE galKphoA20 thi-1rpsE rpoBargE(Am) recA1 λpir | 22 |
| S17-1λpir | Tpr SmrrecA thiprohsdR-M+RP4: 2-Tc:Mu: Km Tn7 λpir | 11 |
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) recA1 endA1hsdR17 thi-1gyrA96 relA1 | Promega |
| V. cholerae | ||
| Rugose | El Tor; rugose variant; Rifr | 62 |
| Smooth | El Tor; smooth variant; Rifr | 62 |
| FY_Vc_1060 | Rugose ΔvpsI ΔlacZ; Rifr | 34 |
| FY_Vc_345 | Rugose ΔcdgC; Rifr | 34 |
| FY_Vc_1061 | Rugose ΔcdgC ΔvpsIΔlacZ; Rifr | 34 |
| FY_Vc_22 | Smooth ΔlacZ; Rifr | 7 |
| FY_Vc_1573 | Smooth ΔcdgC ΔlacZ; Rifr | This study |
| FY_Vc_576 | Rugose ΔtcpA; Rifr | This study |
| FY_Vc_1525 | Rugose ΔtcpP ΔvpsIΔlacZ; Rifr | This study |
| FY_Vc_1526 | Rugose ΔtcpP ΔcdgCΔvpsI ΔlacZ; Rifr | This study |
| FY_Vc_355 | Rugose ΔmbaA; Rifr | 34 |
| FY_Vc_1063 | Rugose ΔmbaA ΔvpsIΔlacZ; Rifr | 34 |
| Plasmids | ||
| pACYC177 | New England Biolabs | |
| pFY-152 | pACYC177::VCA0785 from rugose strain; Apr | 34 |
| pGP704sacB28 | pGP704 derivative; mob/oriT sacB; Apr | Gary Schoolnik |
| pCC2 | pGP704sacB28::ΔlacZ; Apr | 7 |
| pFY-222 | pGP704sacB28::ΔtcpA; Apr | This study |
| pFY-346 | pGP704sacB28::ΔtcpP; Apr | This study |
| pRS415 | lacYZA-based promoter-fusion vector; Apr | 52 |
| pCC11 | pRS415 vpsA promoter; Apr | 7 |
| pCC12 | pRS415 vpsL promoter; Apr | 7 |
| pCC10 | pRS415 vpsR promoter; Apr | 7 |
| pCC25 | pRS415 vpsT promoter; Apr | 7 |
| pFY-352 | pRS415 flaA promoter; Apr | This study |
| pFY-350 | pRS415 ctxA promoter; Apr | This study |
| pFY-351 | pRS415 tcpA promoter; Apr | This study |
| pFY-353 | pRS415 tcpP promoter; Apr | This study |
| pFY-360 | pRS415 toxT promoter; Apr | This study |
| pBAD/Myc-His B | Arabinose-inducible expression vector with C-terminal myc epitope and six-His tags | Invitrogen |
| pFY-469 | pBAD/Myc-His B::VCA0785 | This study |
| pFY-470 | pBAD/Myc-His B::VCA0785(E407A) | This study |
DNA manipulations.
All oligonucleotides used for PCR analysis and DNA sequencing were obtained from Operon Technologies (Alameda, CA) and are listed in Table S1 in the supplemental material. All PCRs were performed with the High-Fidelity (Roche) system. PCR products and plasmids were cleaned and prepared using QIAquick PCR purification and QIAprep Spin Miniprep kits from QIAGEN and the GFX PCR DNA and gel band purification kit from GE Healthcare Life Sciences. DNA sequencing was done at the UC Berkeley DNA Sequencing Facility.
In-frame deletions for all genes were carried out using the same general strategy, as previously described (14). Briefly, a ∼600-bp fragment 5′ of the gene and including several nucleotides of the gene was amplified by PCR with primers A and B (described in Table S1 in the supplemental material). A similar fragment was also amplified from the 3′ end of the gene using primers C and D (see Table S1 in the supplemental material). Purified PCR fragments from these reactions were allowed to anneal to sequences in primers B and C and amplified in a second PCR. The resulting ∼1,200-bp fragment was then amplified with primers A and D, creating the in-frame deletion construct. The purified PCR fragment was digested with two of the three restriction enzymes SacI, NcoI, and XbaI and ligated into plasmid pGP704-sacB28 digested with the same enzyme. All of the clones were sequenced to ensure that mutations were not introduced during the manipulation procedures. The in-frame deletion constructs are listed in Table 1.
Construction of overexpression plasmids.
The overexpression constructs containing cdgC and cdgC-AAL were generated in plasmid pBAD/Myc-His B. For construction of pcdgC (pFY_469), VCA0785_pBAD_A and VCA0785-pBAD-D primers (see Table S1 in the supplemental material) were used to amplify the cdgC gene. PCR products were digested with restriction enzymes XhoI and XbaI and ligated into similarly digested pBAD/Myc-His B plasmid. For construction of pcdgC-AAL (pFY_470), VCA0785_pBAD_A and VCA0785_AAL_B primers (see Table S1 in the supplemental material) were used to amplify a 1,219-bp 5′ region of the cdgC gene plus 1 bp of upstream flanking region, and VCA0785_pBAD_D and VCA0785_AAL_C primers (see Table S1 in the supplemental material) were used to amplify a 660-bp 3′ region of the cdgC gene plus 2 bp of downstream flanking region. These two fragments were joined using the splicing overlap extension technique (24, 33), and the resulting PCR product, harboring new coding sequence for the substituted amino acid residue (E407A), was digested with restriction enzymes XhoI and XbaI and ligated into similarly digested pBAD/Myc-His B plasmid. The PCR products were sequenced (UC Berkeley DNA Sequencing Facility) to ensure that no errors were introduced during PCR amplification. The overexpression constructs are listed in Table 1. Overexpression constructs were electroporated in the V. cholerae smooth wild-type strain.
Construction of lacZ transcriptional fusions.
The lacZ transcriptional fusions were generated in plasmid pRS415. Promoters of genes of interest were cloned in a nonpolar manner upstream of the promoterless lacZ gene in pRS415. Promoters were cloned using primers with EcoRI and BamHI sites that annealed about 500 base pairs upstream and about 20 base pairs downstream, respectively, of the start codon. The promoter of flaA was cloned as cited in the work of Correa and Klose and included 500 base pairs upstream and 500 base pairs downstream of the start codon (9). The primers used for cloning the promoters are listed in Table S1 in the supplemental material. All clones were sequenced to ensure that no mutations were introduced during the manipulation procedures. lacZ transcriptional fusions were electroporated in V. cholerae strains harboring a ΔlacZ deletion.
Construction of V. cholerae knockout mutants.
Strain construction with pGP704-sacB28 was performed as shown previously (14) with modifications described by Lim et al. (34).
RNA isolation.
Collection of RNA from V. cholerae in the exponential phase of growth was done as previously described (61). Briefly, cultures were grown overnight in LB medium at 30°C with shaking. Overnight-grown cultures were diluted 1:200 in fresh LB medium grown to exponential phase and then diluted again to 1:200 and grown to an optical density at 600 nm (OD600) of 0.4 to ensure homogeneity. Aliquots (2 ml) were collected when cultures reached exponential phase at an OD600 of 0.2, 0.35, and 0.6; centrifuged for 2 min at room temperature; resuspended in 1 ml of TRIzol (Invitrogen); and stored at −80°C. Total RNA from the pellets was isolated according to the manufacturer's instructions. To remove contaminating DNA, total RNA was incubated with RNase-free DNase I (Ambion), and the RNeasy Mini kit (QIAGEN) was used to clean up RNA after DNase digestion.
To obtain RNA from stationary-phase cultures, overnight-grown cultures were diluted 1:200 in fresh LB medium and incubated at 30°C with shaking (200 rpm) for 8 h to an OD600 of ∼2.0. Cell harvesting and RNA isolation procedures were performed as described above.
cDNA synthesis, microarray hybridization, and data analysis.
Whole-genome expression profiling analysis was performed using a common reference RNA that contained equal amounts of total RNA isolated from exponentially grown smooth and rugose cells. RNA from test and reference samples was used in a reverse transcription reaction. cDNA synthesis and microarray hybridization were done as described previously (3). The raw microarray data were obtained by using the software package GenePix 4.1 (Axon). Normalized signal ratios were obtained with LOWESS print-tip normalization using the Bioconductor packages (http://www.bioconductor.org) in the R environment (19). Differentially regulated genes were determined (with at least two biological and two technical replicates for each data point) with the Significance Analysis of Microarrays package (58) using 1.5-fold differences in gene expression and a 3% false-positive discovery rate as cutoff values.
Semiquantitative RT-PCR.
Semiquantitative reverse transcription-PCR (RT-PCR) was performed in one step using QIAGEN′s One Step RT-PCR kit as outlined by the manufacturer. For cDNA synthesis, 1 ng of V. cholerae RNA served as template. PCR (25-μl reaction volume) was performed as follows: 50°C for 30 min; 95°C for 15 min; and 25 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min. For visualization, 8 μl of the resulting PCR was subjected to agarose gel electrophoresis and stained with ethidium bromide. Gel band intensities were quantified using ImageQuant TL software (GE Healthcare Life Sciences).
Analysis of virulence factor production.
For toxin-coregulated pilus (TCP) detection, equivalent protein concentrations of total-cell lysates, as determined with Coomassie blue Bradford assay reagent (Pierce), were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with TcpA antibody antiserum using an ECL detection reagent from Pierce. CT was measured by a GM1-ganglioside enzyme-linked immunosorbent assay, as described previously (18).
β-Galactosidase assays.
β-Galactosidase activity was determined using a protocol similar to that described previously (38). Modifications to the procedure were carried out as described previously (34).
Motility assays.
Ampicillin-containing LB soft agar plates (0.3% agar) were used to determine the motility of bacterial strains. The diameter of the motility zone was measured after 22 h of incubation at 30°C with or without addition of arabinose.
RESULTS
CdgC acts as a phosphodiesterase.
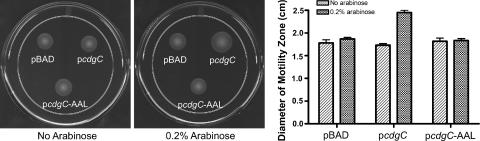
Previously, we identified CdgC as a regulator of rugose colony development, biofilm formation, and motility in both the rugose and smooth genetic backgrounds of V. cholerae (34). CdgC harbors both GGDEF and EAL domains, which were shown to have diguanylate cyclase and phosphodiesterase activities, respectively (48, 49). While CdgC has the EAL domain, the GGDEF domain of cdgC does not have conserved GG[DE]EF residues, and it is predicted that this protein mainly functions as a phosphodiesterase. In this study, we used a well-established relationship between cellular levels of c-di-GMP and flagellar motility (3, 27, 34, 51) to determine whether CdgC acts as a phosphodiesterase or a diguanylate cyclase. To this end, we overexpressed CdgC (pcdgC) and a CdgC allele harboring an E407A mutation, changing EAL to AAL (pcdgC-AAL), in the smooth wild type by using an arabinose-inducible system. We then measured the motility of these strains and the control strain harboring the vector (pBAD) under inducing and noninducing conditions. Without arabinose addition, the diameters of the motility zones for all three strains (S+pBAD, S+pcdgC, and S+pcdgC-AAL) remained similar to each other (Fig. 1). However, under inducing conditions S+pcdgC exhibited a significant increase (42%) in motility and S+pcdgC-AAL did not exhibit any significant change, indicating that the EAL domain of cdgC is in an active form and is required for increased flagellar motility (Fig. 1). It also shows that the GGDEF domain of cdgC is not active, since it does not lead to any change in flagellar motility even when the EAL domain is mutated. Previously, Tischler and Camilli also showed that the EAL domain of cdgC is active and that when it is overexpressed it leads to a decrease in vps gene expression (55). Taken together, these studies suggest that CdgC acts as a phosphodiesterase.
FIG. 1.
The EAL domain of CdgC acts as a phosphodiesterase. Shown are motility phenotypes of smooth wild-type strains harboring pBAD, pcdgC, and pcdgC-AAL. The diameters of motility zones of smooth wild-type strains harboring pBAD, pcdgC, and pcdgC-AAL were measured after 22 h of incubation at 30°C on LB soft agar plates with or without arabinose.
Transcriptome analysis of CdgC in the rugose and smooth genetic backgrounds.
To identify genes regulated by CdgC, both in general and in a phase variant-specific manner, we compared whole-genome expression profiles of the RΔcdgC and SΔcdgC mutants with respect to their wild-type parental background. Gene expression data were analyzed using the Significance Analysis of Microarrays program (58). We applied the following criteria to define significantly regulated genes: ≤3% false-positive discovery rate and ≥1.5-fold transcript abundance differences between the samples.
The transcriptomes of RΔcdgC and SΔcdgC were compared to their corresponding parental strain using total RNA isolated from cells during either the exponential (OD600, 0.3 to 0.4) or the stationary (OD600, 2.0) phase of growth in liquid LB medium. Using the selection criteria given above, a total of 490 genes were found to be differentially regulated by at least 1.5-fold in RΔcdgC compared to the wild-type rugose strain during exponential phase. Of these genes, 167 were induced and 323 repressed in RΔcdgC compared to wild type (see Tables S2 and S3 in the supplemental material). During the stationary phase, 238 genes were differentially regulated in RΔcdgC compared to wild type, with the majority being induced (224 genes) and a small number repressed (14 genes) (see Tables S2 and S3 in the supplemental material). In the SΔcdgC mutant 94 genes were differentially expressed during exponential phase, with 71 being upregulated and 23 being downregulated (see Tables S4 and S5 in the supplemental material). Four genes were differentially regulated in the SΔcdgC mutant during stationary phase.
CdgC represses expression of vpsR, vpsT, and vps region genes in both the rugose and smooth phase variants.
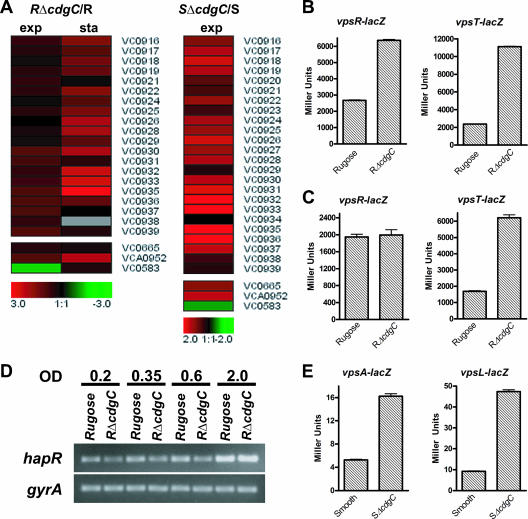
We recently showed that deletion of cdgC in the rugose background led to an increase in colony corrugation (34). Based on this result, we expected that the RΔcdgC mutant would have an induced expression of the vps region genes, VC0916, VC0917 to VC0927 (vpsI), and VC0934 to VC0939 (vpsII), compared to wild type. Expression profiling showed that mRNAs of these genes were increased by 1.5- to 7.8-fold relative to wild type (Fig. 2A) during exponential and stationary phases of growth. This finding corroborates our previous study, where we showed that RΔcdgC carrying a vpsL::lacZ fusion exhibited increased β-galactosidase activity compared to a wild-type rugose strain during both exponential and stationary phases.
FIG. 2.
Expression of vpsR, vpsT, and the vps region genes is upregulated in both the RΔcdgC and SΔcdgC mutants. (A) Expression profiles of vps region genes (vpsI, vpsII, and genes between the two clusters), vpsR (VC0665), vpsT (VCA0952), and hapR (VC0583) in the RΔcdgC mutant during both exponential (OD600, 0.3 to 0.4) and stationary (OD600, 2.0) phases and in the SΔcdgC mutant during exponential phase. The differences in the abundance of transcripts between the ΔcdgC mutants and their corresponding parental backgrounds are presented using the log2-based color scale at the bottom of the panels (red, induced; green, repressed). (B and C) The transcription of vpsR and vpsT in the rugose wild type and RΔcdgC mutant was measured by quantifying β-galactosidase activity from the lacZ fusions vpsR::lacZ and vpsT::lacZ, respectively. Strains were grown at 30°C to exponential (B) and stationary (C) phases, and transcriptional activity was measured. (D) Levels of the hapR transcript were determined using semiquantitative RT-PCR in the rugose wild type and RΔcdgC mutant during the early exponential (OD600, 0.2; lanes 1 and 2), mid-exponential (OD600, 0.35; lanes 3 and 4), late exponential (OD600, 0.6; lanes 5 and 6), and stationary (OD600, 2.0; lanes 7 and 8) phases. Bacteria were grown to identical densities. The constitutively expressed gyrA gene was used as a control. Results shown are representative of three independent experiments. (E) The transcription of vpsA and vpsL in wild-type smooth and SΔcdgC strains was measured through the β-galactosidase activity from the vpsA::lacZ and vpsL::lacZ fusion constructs. Strains were grown at 30°C to exponential phase, and transcriptional activity was measured. For all lacZ experiments, results shown are representative of three independent experiments. Error bars represent standard deviations.
Expression of the vps gene is positively regulated by VpsT and VpsR and negatively regulated by HapR (61). Analysis of expression profiling results revealed that transcription of vpsT in the RΔcdgC mutant was 1.9- and 4.5-fold higher than that in the wild type during exponential and stationary phases of growth, respectively (Fig. 2A; see also Table S3 in the supplemental material). Expression of vpsR increased slightly, by 1.4-fold, during the exponential phase and did not change significantly during the stationary phase (Fig. 2A; see also Table S3 in the supplemental material). These results were further confirmed by measuring β-galactosidase activity in RΔcdgC mutant and wild-type rugose strains carrying a vpsT::lacZ or vpsR::lacZ transcriptional fusion construct. During the exponential phase, RΔcdgC exhibited a 3.5-and 3.1-fold increase in β-galactosidase activity, compared to wild type, from the vpsT::lacZ and vpsR::lacZ fusions, respectively (Fig. 2B). During the stationary phase, RΔcdgC carrying a vpsT::lacZ fusion had 3.2-fold more β-galactosidase activity than the wild type (Fig. 2C). On the other hand, there was no significant difference in β-galactosidase activities between RΔcdgC and the wild-type rugose strain carrying a vpsR::lacZ fusion during the stationary phase.
Our transcriptome analysis also revealed a 2.7-fold decrease in the expression of the hapR gene in the RΔcdgC mutant relative to wild type during the exponential phase (see Table S3 in the supplemental material). To determine the extent to which CdgC regulates hapR expression, we performed semiquantitative RT-PCR to detect the amount of hapR transcript in both wild-type rugose and RΔcdgC strains during the early exponential (OD600, 0.2), mid-exponential (OD600, 0.35), late exponential (OD600, 0.6), and stationary (OD600, 2.0) phases of growth. Quantification of band intensities with ImageQuant software revealed that RΔcdgC exhibited decreased levels of hapR transcripts relative to the rugose variant in the early, mid-, and late exponential phases, by 1.9-, 1.3-, and 1.3-fold, respectively, but not in the stationary phase (Fig. 2D), confirming the microarray results. It should also be noted that there were no changes in the gyrA message abundance between the RΔcdgC and the rugose variant.
Expression profiling of the wild-type smooth variant and SΔcdgC mutant revealed that transcript levels for the vps genes were higher in SΔcdgC (by 1.5- to 5.0-fold; Fig. 2A; see also Table S5 in the supplemental material) during the exponential phase. Similar to the rugose genetic background, expression levels of vpsT and vpsR increased by 2.5-and 2.1-fold, respectively, in the SΔcdgC mutant relative to wild type during the exponential phase (see Table S5 in the supplemental material). Increased vps expression in the SΔcdgC mutant was confirmed by comparing the β-galactosidase activities of vpsA::lacZ and vpsL::lacZ transcriptional fusion constructs in the SΔcdgC mutant and smooth wild-type variant. The SΔcdgC mutant showed an increase in transcriptional activity for vpsA and vpsL of 3.1- and 5.3-fold, respectively (Fig. 2E).
Expression of eps, purine/pyrimidine biosynthesis, flagellar biogenesis, and chemotaxis genes is altered in the RΔcdgC mutant.
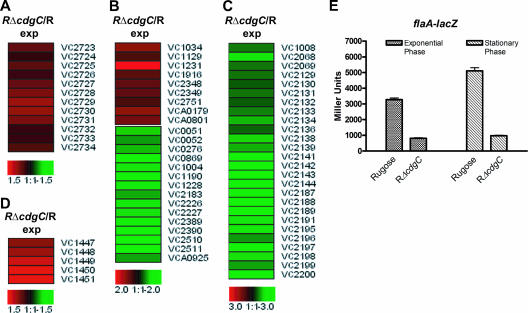
The expression of eps (extracellular protein secretion) genes, located in a 12-gene operon containing genes epsC to epsN, increased by 1.4- to 1.9-fold in the RΔcdgC mutant relative to rugose wild type during the exponential phase (Fig. 3A; see also Table S5 in the supplemental material). The products of the EPS pathway (encoded by epsD and epsE) are required for the formation of the rugose colonial morphotype (1). Thus, expression of eps genes has been shown to be consistently higher in the rugose strain than in the wild-type smooth strain (61). The EPS secretion system is predicted to be responsible for either secretion of VPS or secretion of protein(s) involved in the transport/assembly of VPS (61). Furthermore, it was recently shown that the expression of vps and eps genes is positively regulated by an increase in cellular c-di-GMP levels (3). The observed increased expression of eps genes in the RΔcdgC mutant relative to rugose wild type may be due to a minimal, but consistent, increase in c-di-GMP levels (34).
FIG. 3.
Expression of eps, purine biosynthesis, flagellar biosynthesis, and rtx genes is modulated in the RΔcdgC mutant during exponential growth phase. (A to D) Shown are expression profiles of eps (A), purine biosynthesis (B), flagellar biosynthesis (C), and rtx (D) genes in the RΔcdgC mutant compared to rugose wild type during the exponential growth phase. Differences in the abundance of transcripts between RΔcdgC and the rugose wild type are presented using the log2-based color scale at the bottom of the panels (red, induced; green, repressed). (E) The transcription of flaA (VC2188) in the rugose wild type and in RΔcdgC was measured by quantifying β-galactosidase activity from a flaA::lacZ fusion. Strains were grown at 30°C to the exponential and stationary phase, and transcriptional activity was measured. Results shown are representative of three independent experiments. Error bars represent standard deviations.
Expression of many of the genes required for de novo purine and pyrimidine biosynthesis was reduced in the RΔcdgC mutant relative to wild type during exponential phase (Fig. 3B; see also Table S1 in the supplemental material). RΔcdgC produces higher levels of cellular c-di-GMP than the wild-type rugose variant (34); these results indicate that RΔcdgC may be maximizing its guanine nucleotide pool to increase c-di-GMP production and/or may be reacting to increased cellular levels of guanine nucleotides.
Comparison of expression profiles from the RΔcdgC mutant and rugose wild type during exponential phase revealed that transcription of 24 genes required for flagellum biosynthesis was decreased in RΔcdgC (Fig. 3C; see also Table S3 in the supplemental material), suggesting that CdgC positively regulates expression of flagellar genes. Deletion of flaA, which encodes the major flagellin component of the flagellar filament in V. cholerae, leads to a nonmotile phenotype (29). To confirm the expression profile data, we engineered a flaA::lacZ transcriptional fusion construct and measured β-galactosidase activity in the RΔcdgC mutant and rugose wild type grown under the same conditions. The RΔcdgC mutant exhibited 4.4- and 5.3-fold decreases in β-galactosidase activity during exponential and stationary phases, respectively (Fig. 3E). In addition, as previously reported, there was a concomitant decrease in the motility of the RΔcdgC mutant (34).
Additionally, expression of several genes predicted to function in chemotaxis (VCA0068, VCA0663, VCA0864, VCA1031, VC1313, VC1413, VC1859, VC1898, VC2059, VC2062, VC2063, VC2064, and VC2161) was differentially regulated (see Table S3 in the supplemental material). Of these genes, nine were methyl-accepting chemotaxis proteins and four were chemotaxis proteins (Che). We have yet to determine the relative contributions of flagellar biosynthesis genes and chemotaxis genes in the RΔcdgC mutant to the observed decrease in motility.
Expression of RTX genes is increased in the RΔcdgC mutant.
The expression of genes required for producing the RTX toxin, which causes rounding of epithelial cells via its action on actin polymerization (15, 35, 50), was higher in the RΔcdgC mutant. The genes responsible for the RTX production (rtxA), transport (rtxB, rtxD, and rtxE), and activation (rtxC) are organized into two divergently transcribed operons on the large chromosome of V. cholerae O1 El Tor. Expression of the RTX genes—rtxA, rtxB, rtxC, and rtxD—was increased in the RΔcdgC mutant during the exponential phase of growth by 1.7-, 1.9-, 2.6-, and 1.8-fold, respectively, relative to wild type (Fig. 3D; see also Table S3 in the supplemental material).
CdgC regulates expression of virulence factors in V. cholerae.
The major virulence factors of V. cholerae are CT and the TCP colonization factor, encoded by ctxA/ctxB and tcpA, respectively. A complex transcriptional regulatory cascade controls CT and TCP production, whereby transcription of ctxA/ctxB and tcpA is regulated by the regulatory proteins ToxR and TcpP, respectively. Two additional proteins, ToxS and TcpH, seem to enhance transcriptional activity of ToxR and TcpP, respectively (6, 12). These two regulatory units control the expression of toxT, the most downstream regulator of virulence factors in V. cholerae (21, 39). In addition, expression of tcpP and tcpH is under the control of two other regulatory proteins, AphA and AphB. The quorum-sensing transcriptional regulator HapR acts as a negative regulator of ctxA/ctxB and tcpA expression by negatively regulating expression of aphA (31).
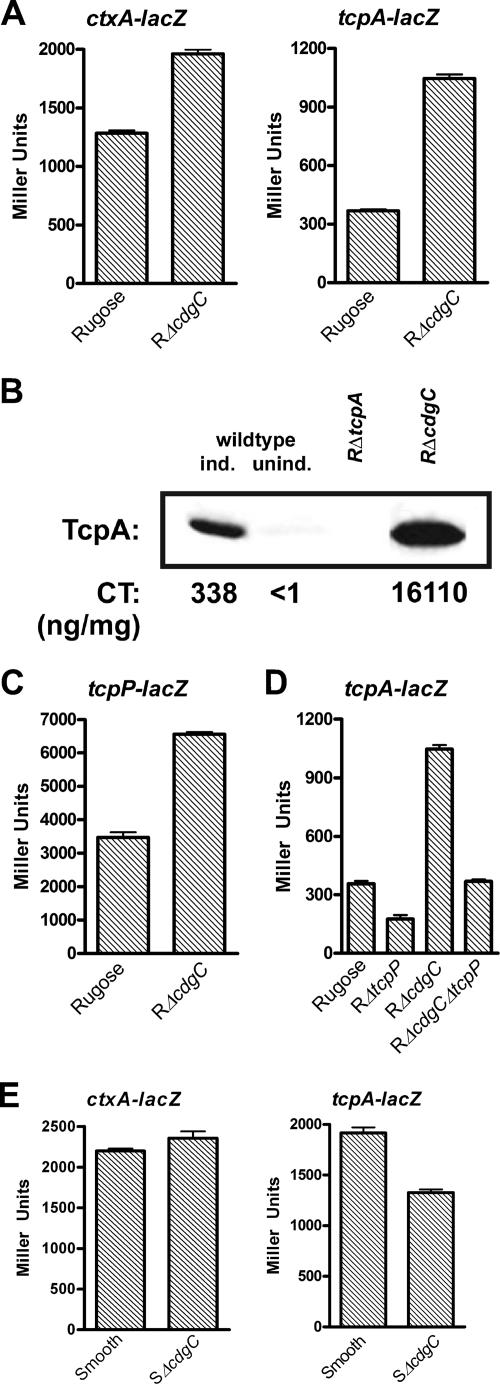
Expression analysis of the RΔcdgC mutant during stationary phase revealed a 2.0-fold increase in the transcript abundance of the tcpA gene compared to the rugose wild-type variant (see Table S3 in the supplemental material). This result was confirmed by determining the β-galactosidase activity of both rugose wild type and RΔcdgC mutant carrying transcriptional fusion constructs. During stationary phase at 30°C in LB medium, RΔcdgC carrying a ctxA::lacZ or tcpA::lacZ transcriptional fusion exhibited 1.9- and 2.8-fold increases in β-galactosidase activity, respectively, compared to the wild type (Fig. 4A). High expression levels of virulence factors among V. cholerae El Tor strains have been observed under specific in vitro growth conditions, termed AKI growth conditions. Compared to the wild-type rugose strain, TcpA production was higher in the RΔcdgC mutant grown under AKI conditions (Fig. 4B). Similarly, CT production was also greatly increased in the RΔcdgC mutant, whereby culture supernatants of RΔcdgC yielded a 48-fold-higher amount of CT than did the wild type (Fig. 4B).
FIG. 4.
CdgC negatively regulates virulence factor genes ctxAB and tcpA in the rugose variant but positively regulates tcpA in the smooth variant. (A) Expression of the ctxA::lacZ and tcpA::lacZ transcriptional fusion constructs in the rugose wild type and the RΔcdgC mutant grown at 30°C to stationary phase (OD600, 2.0). (B) In vitro production of CT and TcpA. Rugose wild type and RΔcdgC were grown under AKI inducing conditions. TcpA was detected by Western immunoblotting with anti-TcpA antiserum. CT in the supernatant was measured by GM1-ganglioside enzyme-linked immunosorbent assay. TcpA quantification and CT values are representative of three independent experiments. For the experimental controls, rugose wild-type bacteria were grown in both AKI inducing conditions and noninducing conditions (in LB medium). As an additional control, the rugose variant carrying a deletion of tcpA was also used. (C) Expression of the tcpP::lacZ fusion gene in the rugose wild type and the RΔcdgC mutant grown at 30°C to stationary phase (OD600, 2.0). (D) Expression of the tcpA::lacZ transcriptional fusion in the rugose wild type and the RΔtcpP, RΔcdgC, and RΔcdgCΔtcpP mutants grown at 30°C to stationary phase (OD600, 2.0). (E) Expression of the ctxA::lacZ and tcpA::lacZ transcriptional fusion constructs in the smooth wild-type variant and the SΔcdgC mutant grown at 30°C to exponential phase (OD600, 0.3 to 0.4). Results of all β-galactosidase assays shown are representative of at least three independent experiments. Error bars represent standard deviations.
Along with the increased transcription of tcpA in the RΔcdgC mutant, transcriptome analysis revealed a 2.6-fold increase in the transcript abundance of the aphA gene compared to the rugose wild-type variant during exponential phase (see Table S3 in the supplemental material). In addition, transcription of the tcpP regulatory gene, which is activated by aphA (53), was induced 1.7-fold during stationary phase in the RΔcdgC mutant relative to rugose wild type (see Table S3 in the supplemental material). We hypothesized that CdgC negatively regulates ctxA/ctxB and tcpA, at least in part through tcpP/tcpH transcriptional regulation. We confirmed that the transcription level of tcpP was increased in RΔcdgC relative to rugose wild type by using a tcpP::lacZ fusion construct. During stationary phase (OD600, 2.0), RΔcdgC containing the tcpP::lacZ fusion exhibited approximately 1.9-fold-more β-galactosidase activity than wild-type rugose (Fig. 4C). To determine whether CdgC regulates tcpA expression primarily by regulating tcpP expression, we compared the β-galactosidase activities of the tcpA::lacZ transcriptional fusion in rugose wild type, RΔcdgC, RΔtcpP, and the double deletion mutant RΔcdgCΔtcpP. If CdgC modulates tcpA transcription solely by regulating tcpP expression, β-galactosidase measurements in the RΔtcpP and RΔcdgCΔtcpP strains should be similar. We found that deletion of tcpP in the rugose background decreased tcpA expression by 2.0-fold (Fig. 4D). However, the double deletion mutant RΔcdgCΔtcpP exhibited β-galactosidase measurements similar to those of the rugose wild-type strain, suggesting that tcpP is not fully epistatic to cdgC. In other words, CdgC regulates tcpA expression through additional mechanisms or through a combination of the two (Fig. 4D). ToxT is believed to be the direct transcriptional activator of the ctxA and tcpA gene promoters (12). We did not, however, observe any significant differences in the transcription of toxT between mutant and wild type by either microarray analysis or the toxT::lacZ fusion (see Table S3 in the supplemental material; data not shown).
During the exponential phase of growth, we found that expression of tcpA and toxT was decreased 1.8- and 1.6-fold, respectively, in the SΔcdgC mutant relative to the smooth wild-type strain (see Table S5 in the supplemental material). To confirm these results and test whether deletion of cdgC in the smooth background selectively regulated tcpA expression, we examined the β-galactosidase activities of the ctxA::lacZ and tcpA::lacZ transcriptional fusion constructs in the smooth and SΔcdgC strains grown at 30°C to exponential phase. SΔcdgC carrying a ctxA::lacZ fusion had β-galactosidase activity similar to that of the smooth wild-type variant (Fig. 4E). However, SΔcdgC carrying a tcpA::lacZ fusion showed a 1.4-fold decrease in β-galactosidase activity compared to the smooth variant (Fig. 4E). In contrast, the RΔcdgC mutant grown at 30°C to exponential phase exhibited a 3.7-fold increase in transcriptional activity from the tcpA::lacZ fusion compared to the rugose wild type (data not shown). Altogether, these data indicate that the regulation of virulence factors by CdgC differs between the V. cholerae rugose and smooth variants.
Virulence factor expression is different between the smooth and rugose phase variants in V. cholerae El Tor A1552.
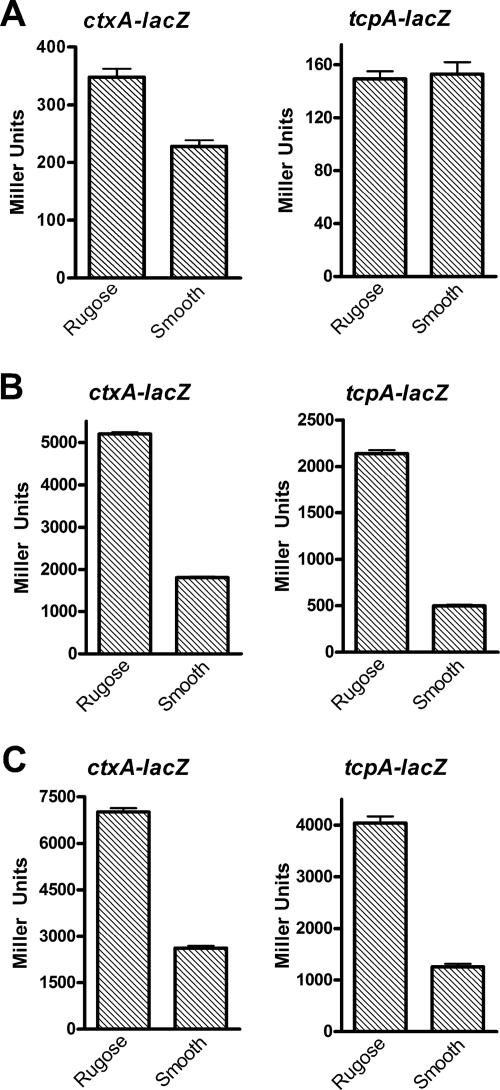
CdgC regulates the expression of virulence genes in an opposing manner in the two phase variants. To determine possible differences in the regulation of ctxA and tcpA transcription between the smooth and rugose variants, we examined the expression of ctxA and tcpA using the ctxA::lacZ or tcpA::lacZ fusion. Rugose and smooth variants carrying the fusions were grown at 30°C to exponential and stationary phases and also under AKI conditions. During the exponential phase, the rugose variant harboring the ctxA::lacZ fusion showed a minimal 1.4-fold increase in β-galactosidase activity compared to the smooth strain. No significant difference in tcpA levels between the phase variants was observed (Fig. 5A). The rugose variant carrying the ctxA::lacZ transcriptional fusion exhibited a 2.7-fold increase in β-galactosidase activity compared to the smooth variant both during the stationary phase and under AKI conditions (Fig. 5B and Fig. 5C). Similarly, the rugose variant carrying the tcpA::lacZ fusion showed a 4.3- and 3.2-fold increase in β-galactosidase activity in the stationary phase and under AKI conditions, respectively, compared to the smooth strain (Fig. 5B and Fig. 5C).
FIG. 5.
Expression of virulence factor genes ctxAB and tcpA is different between the rugose and smooth phase variants in V. cholerae El Tor A1552. Shown is the expression of the ctxA::lacZ and tcpA::lacZ fusion constructs in the wild-type rugose and smooth phase variants grown at 30°C to exponential (OD600, 0.3 to 0.4) (A) and stationary (OD600, 2.0) (B) phases in LB medium and under AKI inducing conditions (C). Results of all β-galactosidase measurements shown are representative of at least three independent experiments. Error bars represent standard deviations.
Expression analysis of MbaA in the rugose genetic background.
We previously reported that mbaA message levels are higher in the rugose variant than in the smooth one during the stationary growth phase (34, 61). Among the 10 genes predicted to encode proteins containing a GGDEF and an EAL domain in V. cholerae, the messages of only two of them, mbaA and cdgC, were increased in the rugose variant. We carried out transcriptome analysis on the RΔmbaA mutant to understand whether mbaA and cdgC regulate a similar set of genes and cellular processes. We compared gene expression patterns of the RΔmbaA mutant with those of the wild-type rugose strain during both exponential and stationary growth phases. Using the selection criteria described above, 192 genes were differentially expressed by at least 1.5-fold during exponential phase in liquid LB medium. Of these genes, 73 were induced and 119 were repressed (see Tables S6 and S7 in the supplemental material). During the stationary phase, 120 genes were differentially regulated; the majority were induced (112 genes) and a small number were repressed (eight genes) (see Tables S6 and S7 in the supplemental material).
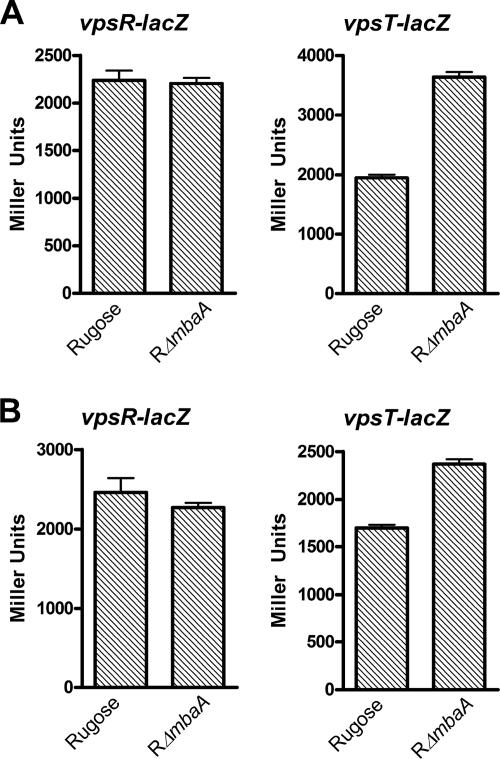
We initially identified mbaA as a negative regulator of rugose colony development. As predicted, we saw that expression of many of the genes located in the vpsI cluster (VC0917 to VC0927), in the vpsII cluster (VC0934 to VC0939), and between the vps clusters—VC0928, VC0929, VC0930, VC0932, and VC0933—was significantly induced by 1.8- to 5.1-fold in the RΔmbaA mutant compared to rugose wild type, primarily during stationary phase (see Table S7 in the supplemental material). This increase in gene expression may be responsible, at least in part, for the observed increase in the biofilm-forming capacity of the RΔmbaA mutant during later phases of growth (4, 28, 34). The induction of vps gene expression in RΔmbaA may be due to increased transcription of either vpsR or vpsT, or both. Our results showed no increase in vpsR gene expression in the RΔmbaA mutant; however, expression of vpsT was induced 2.0- and 5.1-fold relative to the wild-type rugose variant during exponential and stationary phase, respectively (see Table S7 in the supplemental material). We confirmed our array results by measuring β-galactosidase activities of vpsR::lacZ and vpsT::lacZ fusion constructs in the RΔmbaA mutant and rugose wild type. Whereas there was no significant difference in β-galactosidase activity for the vpsR::lacZ fusion, we observed a 1.5- and 1.4-fold increase in β-galactosidase activity in the RΔmbaA mutant carrying the vpsT::lacZ transcriptional fusion during exponential and stationary phases, respectively (Fig. 6A and 6B). Additionally, work from Karatan et al. identified a set of genes regulated by MbaA in the smooth variant of the O139 strain MO10 (28). This study also observed an increase in expression of vps genes and the vpsT gene in the mbaA mutant relative to wild type. Altogether, these results suggest that the increase in vps gene transcription can, in part, be due to the action of VpsT. In fact, we previously identified vpsT as being epistatic to mbaA in terms of rugose colony morphology (34). However, while vpsT expression was increased during both exponential and stationary phases in the RΔmbaA mutant, we saw a significant induction of the expression of the vps genes only during the stationary phase (see Table S7 in the supplemental material) (34). These results indicate that mbaA regulates vps gene expression temporally, through a yet-to-be-identified mechanism.
FIG. 6.
Expression of vps genes and vpsT, but not vpsR, is upregulated in the RΔmbaA mutant compared to rugose wild type. The transcription of vpsR and vpsT in the rugose wild-type variant and the RΔmbaA mutant was measured by quantifying β-galactosidase activity from the vpsR::lacZ and vpsT::lacZ fusion constructs. Strains were grown at 30°C to exponential (A) and stationary (B) phase, and transcriptional activity was measured. Results shown are representative of three independent experiments. Error bars represent standard deviations.
In addition, the RΔmbaA mutant showed a 1.6-fold repression of hapR expression during the exponential phase. This effect may be partly responsible for the induction of vpsT in the RΔmbaA mutant but cannot explain the similarity in vpsR gene expression levels between RΔmbaA and the rugose wild-type variant during exponential and stationary phases (Fig. 6A and 6B). Taken together, these results suggest that additional mechanisms are responsible for the regulation of vps transcription by mbaA and reveal the complexity of c-di-GMP signaling in V. cholerae cellular processes.
We also observed an increase (by 1.3- to 1.8-fold) in the expression of several eps genes (epsG, epsI, epsJ, and epsL) in the RΔmbaA mutant relative to wild type during exponential and stationary growth phases (see Table S7 in the supplemental material; data not shown). On the other hand, expression of several flagellar biosynthesis genes was 1.5- to 4.2-fold lower in the mutant during exponential (flaB, flaC, and flaG genes) and stationary (flaA, flaC, flaD, and flgB genes) phases. Consistent with these findings, we previously showed that the RΔmbaA mutant exhibited reduced motility compared to the wild-type rugose variant (34).
Comparison of CdgC and MbaA regulons during exponential and stationary growth phases.
Both CdgC and MbaA are negative regulators of rugose colony development (34). They contain a GGDEF and an EAL domain but differ primarily in the N-terminal domain sequence. The expression of cdgC is higher in the rugose than in the smooth variant during exponential and stationary phases, whereas expression of mbaA is higher in the rugose variant in the stationary growth phase only (61; unpublished data). These findings suggest either some redundancy in the functions of mbaA and cdgC or increased complexity in their output signal. To better understand the functions of CdgC and MbaA expression during exponential and stationary phases, we compared the transcription profiles of the RΔcdgC mutant and the rugose wild type and of RΔmbaA and the rugose wild type. Of the 165 genes found to be differentially regulated in the two mutants during the exponential phase, 15% were involved in metabolism, 8% were involved in nucleotide biosynthesis, 15% coded for transport and binding proteins, and 21% coded for hypothetical proteins. Of particular interest are the 8% of the common differentially regulated genes that have a regulatory function, including hapR and vpsT (whose expression is repressed and induced, respectively). However, during the exponential growth phase, RΔcdgC alone exhibited increased vps (vpsI and vpsII clusters) and vpsR gene transcription and a significant decrease in expression of the genes encoding flagellar and chemotaxis machinery components.
Of the 52 genes differentially regulated in both RΔcdgC and RΔmbaA mutants during the stationary phase, 31% of them consisted of vps region genes (from both clusters), genes located between the vps clusters, and vpsT. This result indicates that both CdgC and MbaA function to regulate vps transcription during stationary phase. These results all together indicate that the cdgC and mbaA regulons overlap based on several molecular phenotypes but that the timing of their transcriptional regulation differs.
DISCUSSION
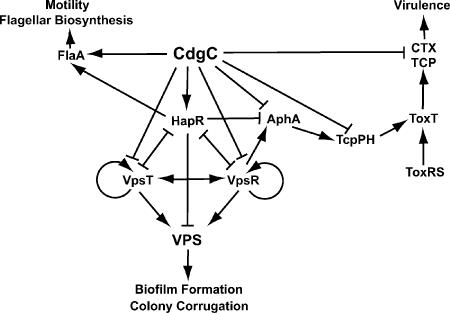
Recent studies have shown that the second messenger c-di-GMP is involved in signal transduction pathways regulating cellular processes that alter cell surface properties and, in turn, motility and biofilm formation of microorganisms (5, 10, 26, 44, 45). The abundance of c-di-GMP in cells is controlled by proteins containing GGDEF and EAL domains responsible for the generation and the degradation of c-di-GMP, respectively. Although the mechanisms by which c-di-GMP modulates cell surface properties are beginning to be understood, we have limited knowledge of the processes controlled by proteins containing GGDEF and/or EAL domains. The goal of this study was to identify genes and processes regulated by a protein, CdgC, containing both domains, previously shown to negatively regulate rugose colony development in V. cholerae. Our major findings are summarized in Fig. 7. We took a genomic approach, comparing gene expression profiles of RΔcdgC and SΔcdgC mutants to those of wild-type rugose and smooth variants, respectively, during exponential and stationary phases of growth. The analysis indicated that a large number of processes are regulated by cdgC in V. cholerae in a phase variant-specific manner. All physiological processes previously identified (34) as being modulated by cdgC—such as rugose colony development, biofilm formation and architecture, and motility behavior—could be explained by the genes being differentially regulated in the RΔcdgC and SΔcdgC mutants. Specifically, the increase in rugosity and biofilm-forming capacity of the RΔcdgC and SΔcdgC mutants correlated with the observed increase in the expression of vps and eps genes. Additionally, the decrease in motility in the RΔcdgC mutant corresponded with the repression of genes responsible for flagellar biosynthesis and chemotaxis (Fig. 7).
FIG. 7.
CdgC regulates VPS production, flagellar motility, and virulence gene expression. CdgC negatively regulates VPS biosynthesis, by inducing hapR transcription or repressing vpsR and vpsT transcription, directly or indirectly. CdgC positively regulates transcription of flaA; also, flagellar biosynthesis and motility could be mediated by HapR. Furthermore, CdgC regulates virulence gene expression by repressing transcription of aphA, tcpP, ctxA, and tcpA, in a direct or indirect manner.
Our previous (34) and current work suggests that CdgC regulates vps gene expression in V. cholerae through a regulatory network involving products of vpsT, vpsR, and hapR and other possible regulatory proteins (Fig. 7). The ΔcdgC expression analysis suggests that CdgC may negatively regulate expression of vps indirectly, by repressing vpsR and vpsT or increasing hapR expression. It was shown that VpsR and VpsT are required for the transcription of vps genes (7, 60). HapR negatively regulates vps expression either indirectly, by decreasing vpsR or vpsT expression, or directly, by physically binding upstream of the vpsI cluster (61). As HapR was shown to positively regulate expression of flagellar biosynthesis genes, the decrease in hapR transcription in the RΔcdgC mutant may be responsible for the decrease in flagellar gene expression. Consistent with this idea, in silico studies indicate the presence of a putative HapR binding site upstream of flagellar biosynthesis genes, such as VC2120, VC2129, VC2130, VC2142, VC2187, VC2195, and VC2196 (61). As several genes predicted to have regulatory functions are differentially expressed in the RΔcdgC mutant compared to wild type, we cannot rule out the possibility that CdgC may regulate vps gene expression through additional mechanisms. In addition, the regulation of a large number of differentially expressed genes in the RΔcdgC and the SΔcdgC mutants may not be directly affected by CdgC binding to their promoters but rather through the control of known regulators, either at the transcriptional or at the posttranslational level.
In this study, we observed that CdgC negatively regulates the expression of the transcriptional regulators vpsR and vpsT (Fig. 7). In contrast, CdgC positively regulates hapR expression, as the amount of hapR message was decreased in the RΔcdgC mutant compared to rugose wild type (Fig. 7). Intriguingly, in a study designed to identify the VpsR and HapR regulons, we found cdgC to be part of both regulons and its promoter region to contain potential binding sites for vpsR and hapR (61). Now, in this study, we show that regulatory proteins and GGDEF/EAL-containing proteins comprise an interrelated network that controls several cellular processes in V. cholerae. In V. cholerae, c-di-GMP and predicted DGCs and PDEAs have been implicated in the regulation of diverse cellular processes, such as vps synthesis, biofilm formation, motile behavior, virulence factor production, and phenotypic switching between the known phase variants (3, 34, 42, 55, 56). Interestingly, these physiological processes are regulated by genes that play a role in quorum sensing, which is the ability of a cell to monitor its own population as well as the abundance of other microbial populations in a bacterial community. Specifically, the quorum-sensing transcriptional regulator hapR regulates expression of several DGCs; furthermore, several DGC and PDEA genes are predicted to have hapR binding sites in their promoter regions (61, 64). As first suggested by Camilli and Bassler, extracellular quorum-sensing signaling and intracellular c-di-GMP concentrations may be linked, based on the overlap of the physiological processes that these two systems regulate (5). In this study, we reveal that DGCs and/or PDEAs can also regulate the expression of quorum-sensing regulators. Increased complexity of regulatory networks involving the c-di-GMP second messenger is likely to provide the necessary fine-tuning of cellular processes involved in environmental survival and in cellular communication.
In the current study, we observed that CdgC represses ctxA, tcpA, aphA, and tcpP expression in the rugose genetic background (Fig. 7). We previously observed that RΔcdgC exhibited a slight increase in cellular c-di-GMP levels compared to the rugose wild type (34). It is possible that such an increase could induce the expression of virulence genes. Alternatively, a decrease in the hapR transcript, as observed in the RΔcdgC mutant, could lead to an increase in virulence gene transcription (Fig. 7). We have a limited understanding of the role that c-di-GMP plays in regulating virulence factor production in V. cholerae. Previous work by Tischler and Camilli showed that increased cellular levels of c-di-GMP (due to a mutation in the response regulator VieA, an EAL domain protein) in the V. cholerae classical strain O395 led to a decrease in ctxA expression possibly through the repression of the toxT transcript (56). In contrast, the same vieA mutation in the El Tor biotype does not cause a decrease in virulence gene expression under the same growth conditions, suggesting that c-di-GMP regulates virulence gene expression in a biotype-specific manner (2, 56). Furthermore, whole-genome expression analysis of the V. cholerae classical and El Tor strains (grown at 30°C in morpholinepropanesulfonic acid minimal medium supplemented with 0.5% glycerol, 25 mM asparagine, arginine, glutamate, and serine to an OD600 of 0.3) also showed that virulence gene expression did not significantly decrease in either classical or El Tor biotypes 30 min after an increase in intracellular c-di-GMP levels (3). Taken together, these studies indicate that there are significant gaps in our knowledge of the mechanism by which c-di-GMP regulates virulence gene expression in V. cholerae.
We compared the transcriptomes of cdgC and mbaA, which were previously shown to regulate vps expression (4, 28, 34). Our transcriptome comparisons indicated that CdgC represses the expression of vps genes during both exponential and stationary phases, while MbaA represses vps gene transcription primarily in the stationary phase. Expression of mbaA is upregulated during the stationary phase in the rugose variant compared to the smooth variant, indicating that the cell temporally restricts the transcription of this GGDEF/EAL protein. Therefore, both proteins regulate vps transcription in the rugose background but in a unique temporal manner. We additionally showed that the magnitude of the increase in the expression of the vpsR and vpsT genes varies between the RΔcdgC and RΔmbaA mutants, suggesting that the mechanism by which cdgC and mbaA regulate gene expression may also be unique. This result brings into question whether the two proteins have a redundant function or an overlapping output signal activated under different cellular or environmental signals. Multiple sequence alignments of CdgC and MbaA have shown that the predicted EAL domain is enzymatically active while the GGDEF domain is predicted to be inactive, suggesting that both of the proteins function as phosphodiesterases of c-di-GMP (34). In this study, we were also able to show that the EAL domain of CdgC is active and required for a well-known c-di-GMP-associated phenotype, flagellar motility. It is also yet to be determined whether the regulatory motifs found in the N-terminal regions of CdgC and MbaA (the GAF and HAMP domains, respectively) restrict signal production to certain environmental or cellular states. It should also be noted that regulation of vps expression by MbaA could be further modulated by the presence of norspermidine in the environment (28).
Although c-di-GMP was first discovered to allosterically activate cellulose synthase in G. xylinus (46), it is becoming increasingly clear that c-di-GMP exerts its effects at levels other than the posttranslational one. Recent studies show that proteins that function as DGCs and PDEAs and proteins containing an HD-GYP domain play diverse roles in many bacterial organisms from exopolysaccharide production to the control of motility to virulence factor production and host survival (10, 26, 44, 45, 47). In order to understand the complexities of this novel signaling pathway, it will be important to further tease apart the c-di-GMP signal transduction pathways and to identify both posttranscriptional and posttranslational mechanisms by which c-di-GMP can regulate a wide range of cellular processes.
Supplementary Material
Acknowledgments
This work was supported by grants from The Ellison Medical Foundation and NIH (AI055987). F.H.Y. is a new scholar in the Ellison Medical Foundation Global Infectious Diseases Program.
We thank Lindsay Stuart Odell for creation of pFY-222 and strain FY_Vc_576 and Ron Taylor for TcpA antibody.
Footnotes
Published ahead of print on 22 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 74:3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood.1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. [DOI] [PubMed] [Google Scholar]
- 7.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829-30837. [DOI] [PubMed] [Google Scholar]
- 9.Correa, N. E., and K. E. Klose. 2005. Characterization of enhancer binding by the Vibrio cholerae flagellar regulatory protein FlrC. J. Bacteriol. 187:3158-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 11.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 12.DiRita, V. J., and J. J. Mekalanos. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29-37. [DOI] [PubMed] [Google Scholar]
- 13.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fullner, K. J., and J. J. Mekalanos. 2000. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 19:5315-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 18.Gardel, C. L., and J. J. Mekalanos. 1994. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 235:517-526. [DOI] [PubMed] [Google Scholar]
- 19.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 21.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hisert, K. B., M. Maccoss, M. U. Shiloh, K. H. Darwin, S. Singh, R. A. Jones, S. Ehrt, Z. Zhang, B. L. Gaffney, S. Gandotra, D. W. Holden, D. Murray, and C. Nathan. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234-1245. [DOI] [PubMed] [Google Scholar]
- 24.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 25.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 26.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 27.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385-407. [DOI] [PubMed] [Google Scholar]
- 28.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57:420-433. [DOI] [PubMed] [Google Scholar]
- 31.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 32.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefebvre, B., P. Formstecher, and P. Lefebvre. 1995. Improvement of the gene splicing overlap (SOE) method. BioTechniques 19:186-188. [PubMed] [Google Scholar]
- 34.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 35.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 96:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendez-Ortiz, M. M., M. Hyodo, Y. Hayakawa, and J. Membrillo-Hernandez. 2006. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J. Biol. Chem. 281:8090-8099. [DOI] [PubMed] [Google Scholar]
- 37.Merkel, T. J., S. Stibitz, J. M. Keith, M. Leef, and R. Shahin. 1998. Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis. Infect. Immun. 66:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Assay of β-galactosidase. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 40.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174:1364-1368. [DOI] [PubMed] [Google Scholar]
- 41.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227:113-119. [DOI] [PubMed] [Google Scholar]
- 43.Rice, E. W., C. J. Johnson, R. M. Clark, K. R. Fox, D. J. Reasoner, M. E. Dunnigan, P. Panigrahi, J. A. Johnson, and J. G. Morris, Jr. 1992. Chlorine and survival of “rugose” Vibrio cholerae. Lancet 340:740. [DOI] [PubMed] [Google Scholar]
- 44.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218-228. [DOI] [PubMed] [Google Scholar]
- 45.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 46.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinbergerohana, R. Mayer, S. Braun, E. Devroom, G. Vandermarel, J. Vanboom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter-xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 47.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheahan, K. L., C. L. Cordero, and K. J. Satchell. 2004. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc. Natl. Acad. Sci. USA 101:9798-9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 52.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 53.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 54.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tischler, A. D., S. H. Lee, and A. Camilli. 2002. The Vibrio cholerae vieSAB locus encodes a pathway contributing to cholera toxin production. J. Bacteriol. 184:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 62.Yildiz, F. H., and G. K. Schoolnik. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.