Abstract
A set of lactobacilli were investigated by polyphasic analysis. Multilocus sequence analysis, DNA typing, microarray analysis, and in silico whole-genome alignments provided a remarkably consistent pattern of similarity within the Lactobacillus acidophilus complex. On microarray analysis, 17 and 5% of the genes from Lactobacillus johnsonii strain NCC533 represented variable and strain-specific genes, respectively, when tested against four independent isolates of L. johnsonii. When projected on the NCC533 genome map, about 10 large clusters of variable genes were identified, and they were enriched around the terminus of replication. A quarter of the variable genes and two-thirds of the strain-specific genes were associated with mobile DNA. Signatures for horizontal gene transfer and modular evolution were found in prophages and in DNA from the exopolysaccharide biosynthesis cluster. On microarray hybridizations, Lactobacillus gasseri strains showed a shift to significantly lower fluorescence intensities than the L. johnsonii test strains, and only genes encoding very conserved cellular functions from L. acidophilus hybridized to the L. johnsonii array. In-silico comparative genomics showed extensive protein sequence similarity and genome synteny of L. johnsonii with L. gasseri, L. acidophilus, and Lactobacillus delbrueckii; moderate synteny with Lactobacillus casei; and scattered X-type sharing of protein sequence identity with the other sequenced lactobacilli. The observation of a stepwise decrease in similarity between the members of the L. acidophilus group suggests a strong element of vertical evolution in a natural phylogenetic group. Modern whole-genome-based techniques are thus a useful adjunct to the clarification of taxonomical relationships in problematic bacterial groups.
Lactobacilli belong to the phylum Firmicutes (low-G+C-content gram-positive bacteria), class Bacilli, order Lactobacillales. Lactobacilli have great importance for humans as starter bacteria in food and feed fermentation (e.g., Lactobacillus delbrueckii in yogurt and Lactobacillus plantarum in sauerkraut and silage fermentation) and as commensals in the vagina (e.g., Lactobacillus jensenii) or the gut (e.g., Lactobacillus gasseri) (50). While lactobacilli play a certain role in endocarditis (45) and are occasionally found in clinical samples (6), no professional pathogens are known in the genus Lactobacillus. Consequently, lactobacilli enjoy a “generally regarded as safe” status (4). In addition, lactobacilli have attracted the attention of the medical community as vaccine carriers and as probiotic, i.e., health-promoting, bacteria (42). Immunostimulatory activity has been demonstrated in animal experiments, and antidiarrhea effects were shown in carefully conducted clinical trials (Lactobacillus rhamnosus and Lactobacillus paracasei) (46). However, the genetic basis for the probiotic properties is not well defined—for example, it is not known to what extent the different reported probiotic effects of lactobacilli are strain-, species-, or genus-specific properties (48).
Notably, the ecological and phenotypic diversity of lactobacilli is mirrored in a taxonomical diversity. Currently, 100 species are described in the genus Lactobacillus. Taxonomists realized as early as 15 years ago that the genus Lactobacillus could be subdivided into groups (for a recent review, see reference 15). Initially, three groups were proposed. Among them, the L. delbrueckii group was later renamed the Lactobacillus acidophilus group, and the Lactobacillus casei/Pediococcus group was split into further subgroups and a new genus. The overall phylogenetic structure of the rRNA tree in the genus Lactobacillus is quite complicated, leading to the suggestion of eight groups (15).
Until about 1970, new Lactobacillus isolates from mucosal surfaces were identified as L. acidophilus. Then, six homology groups were distinguished among them by DNA-DNA hybridization (23), leading to the definition of the species L. acidophilus, Lactobacillus amylovorus, Lactobacillus crispatus, Lactobacillus gallinarum, L. gasseri, and Lactobacillus johnsonii (21). Within the L. acidophilus/L. delbrueckii group, there is substantial genetic diversity, as mirrored by an overall genome G+C content ranging from 33% (L. gasseri) to 51% (L. delbrueckii). This span of G+C values is about twice as large as that normally accepted for well-defined bacterial genera (47), raising questions as to whether we are dealing with a natural group of bacteria. Taxonomists know very well that the genus Lactobacillus contains many deep branches and that its relationship to the genera Paralactobacillus and Pediococcus is not based on the phylogenetic data. However, a phylogenetic genus concept does not yet exist in bacterial taxonomy.
For some genomics researchers with an interest in bacterial phylogeny, it is a perennially vexatious question whether bacteria have species (19). Taxonomists have a more pragmatic approach to the problem and define the bacterial species as the basic unit of bacterial taxonomy and operationally as a group of strains sharing 70% or greater DNA-DNA relatedness under standardized hybridization conditions. Phenotypic and chemotaxonomic features should agree with this definition. The use of many genotypic, phenotypic, and phylogenetic data in defining bacterial species became known as polyphasic taxonomy, which has been the consensus approach to bacterial systematics for 35 years (54). Meanwhile, 16S rRNA sequencing and its cataloguing have revolutionized bacterial systematics. To this technique have been added still other DNA-based approaches, like rapid DNA typing methods, multilocus sequence analysis (MLSA) of housekeeping genes, and lately, sequence analyses of complete genomes. This technical progress led the ad hoc committee for the reevaluation of the species definition in bacteriology to the following species definition. “A species is a category that circumscribes a (preferably) genomically coherent group of individual isolates/strains sharing a high degree of similarity in (many) independent features, comparatively tested under highly standardized conditions” (49). This committee identified the sequencing of housekeeping genes, DNA profiling, and DNA arrays as methods with great promise in bacterial systematics. While the first two methods became standard tools of the bacterial taxonomist, DNA arrays are still not routinely used in bacterial taxonomy. Lactobacilli have been intensively investigated by polyphasic taxonomy (54), DNA typing (59), and rRNA analysis (9, 15) and lately also by the comparison of complete genome sequences (9, 34). Only one study with L. plantarum, using microarray technology, addressed the intraspecies diversity in lactobacilli (36). We conducted a microarray analysis in the L. acidophilus complex for intra- and interspecies diversity, which we complemented with multilocus sequence analysis, sugar fermentation analysis, DNA typing, and comparative genomics. A remarkably consistent pattern emerged from this comparative analysis, underlining the fact that we are dealing in the L. acidophilus complex with a natural group, despite the highly variable genomic G+C contents.
MATERIALS AND METHODS
Strains.
The following bacteria were used for phenotypic and molecular typing, and a subset of them was used for comparative genomic hybridization (CGH) on microarrays. Lactobacillus delbrueckii subsp. bulgaricus strain ATCC 11842, L. plantarum strain WCFS1, Lactobacillus salivarius subsp. salivarius strain UCC118, L. acidophilus strains ATCC 11975 and NCFM, L. gasseri strains ATCC 33323 (the type strain) and NCC2547, and L. johnsonii strains NCC533 (41), NCC2767, NCC2822 (both isolates from dog feces), ATCC 33200 (the type strain), and ATCC 11506. The two fecal isolates were derived from healthy dogs in France, and the species assignment was initially based on 16S rRNA data. The L. johnsonii strains were selected from a collection of 15 strains based on their extensive differences from NCC533 and each other in insertion element (IS) mapping (less than 4 insertion sites were detected out of 14 in NCC533 [unpublished data]). All lactobacilli were grown anaerobically in MRS broth (Difco Laboratories, Detroit, Mich.) at 37°C. They were characterized for their ranges of fermentable carbohydrates by using the API50-CHL kit (bioMérieux, Lyon, France).
Microarray description, labeling, and hybridization.
The NCC533 microarrays covered 96% of the open reading frames (ORFs) from the complete genome. PCR amplicons of 127 to 800 bp corresponding to 1,756 ORFs were spotted in duplicate onto slides (Eurogentec). Amplicons from the luciferase gene (pSP-luc+; Promega) were also spotted as spiking controls. Genomic DNA was prepared as previously described (16). An additional purification step on QIAGEN Genomic-tip 20/G proved to be essential to obtain the reproducibility needed for our normalization method (see below). Five nanograms of pSP-luc+ plasmid was added to 1 μg of genomic DNA and labeled with FluoroLink Cy5- or Cy3-dUTP (Amersham) using the DNA High Prime Kit (Roche Applied Science) according to the supplier's instructions. Reactions were stopped after 5 h by heating the sample to 65°C for 10 min. The combined reactions (Cy5 and Cy3) were purified with the MinElute Reaction Cleanup Kit (QIAGEN) and eluted in 10 μl 10 mM Tris-HCl, pH 8.0. The labeled DNA and 1.5 μl of 1 mg/ml MB grade fish sperm DNA in DIG Easy Hyb Buffer (Roche Applied Science) were denatured by heating them for 2 min at 95°C and were applied to the microarrays under a coverslip. Hybridization was performed overnight at 42°C. The slides were washed for 5 min at 42°C with 2× SSC (Invitrogen) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate and then at room temperature with the same solution. Additional washes for 1 min at room temperature with 0.2× SSC and then 0.1× SSC were conducted twice. The slides were immediately dried by centrifugation.
Microarray data acquisition and treatment.
Fluorescence scanning was performed on a ScanArray 4000 confocal laser scanner (Perkin-Elmer). Signal intensities for each spot were determined using Imagene software (Biodiscovery). Data analysis was performed with scripts written in Python (http://www.python.org). The local background value was subtracted from the intensity of each spot. Each spot was tagged and eliminated from the analysis if its images showed clear alteration (scratches, leaking, printing, dust, etc.) or if its signal strength was lower than twice the standard deviation of the local background when hybridized with NCC533 genomic DNA. To preserve the signal intensity differences obtained when strains from different species were hybridized, we introduced into the labeling reactions a spiking control (pSP-luc+) and used its signal ratio to correct for differences in the labeling-reaction efficiencies. The signal ratio (Cy5-labeled unknown strain DNA versus Cy3-labeled NCC533 DNA) of each spot was normalized using this spiking signal ratio as a reference (ratio = 1).
CGH was performed in triplicate, with duplicate spots on the slide. The distribution of the log2-transformed signal ratios was analyzed for each hybridization reaction separately. The mean of a normal distribution fitting the main peak was calculated. For each group of replicates, the median of these means provided the final position of the main peak for the corresponding unknown strain. The log2 signal ratio of each spot was then modified in order to shift the mean of the main peak of each hybridization reaction to this value. Finally, for each amplicon, we obtained the mean R of the ratios and the corresponding standard deviation. The distribution of these values is depicted in the inset of Fig. 2.
FIG. 2.
Genomic diversity in the Lactobacillus acidophilus group as seen from the viewpoint of L. johnsonii strain NCC533. (Left) CGH data. Each horizontal row corresponds to an amplicon on the array, and the genes are vertically ordered according to their positions on the NCC533 genome. The columns represent the analyzed strains, and the strains are identified by their code numbers. The color code corresponding to the CGH score (BlastN-like score) is given at the bottom right of the figure; the gradient goes from black to yellow to depict the presence, divergence, or absence of a gene sequence. Some relevant gene and genetic-element positions are shown on the left side along the genome. ori, origin of replication; ter, terminus of replication. (Right inset) Signal ratio distribution of the CGH data. The reference is L. johnsonii strain NCC533. Ratios are expressed in a log2 scale. See the text for details.
CGH score and gene presence determination.
For each NCC533 amplicon, the BlastN result (2) against the ATCC 33323T genome was compared to its CGH result. When the BlastN score was plotted against the CGH ratio multiplied by the amplicon length in base pairs, a good correlation (coefficient = 0.87) was observed (data not shown). This allowed the transformation of the CGH ratios into BlastN-like scores, which are more intuitive for biologists. This CGH score was calculated by the following empirically determined equation: with L being the amplicon length and R the mean CGH ratio, CGH score = (1.8327 · R · L) − [(0.0006) · (R · L)2]. For low values of R · L, a better correlation was given by the following two equations: for 187 ≥ R · L ≥ 90, CGH score = −346.13 + (5.1979 · R · L) − [(0.0087) · (R · L)2]; for R · L < 90, CGH score = 40. Similar to the bit score of a BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST), this CGH score reflects the level of conservation between the two genomes, taking into account the size of the amplicon sequence. This approach was also validated by using the sequence data from the eps-rfb clusters of three test strains (see below). The CGH scores are color coded in Fig. 2. Clustering of the CGH results was performed using the unweighted-pair group method with average linkages (UPGMA) algorithm and cosine correlation (Spotfire, Somerville, MA).
A gene corresponding to an amplicon was considered to be present in the unknown genome when the CGH score of R − standard deviation was higher than 200. Using this threshold, which is commonly used with bit scores of BLAST analysis, all the CGH results of NCC533 were qualified as present, despite a few low CGH scores.
PCR and sequencing.
The DNA sequencing of the eps regions from L. johnsonii strains ATCC 33200T, ATCC 11506, and NCC2767 was initiated from the flanking conserved regions and followed by filling the intervening gaps. This was achieved by alternating inverted PCR (35), to generate new DNA sequence data, and long-range PCR using the Expand long-template PCR system (Roche Applied Science), with primers designed from the new inverted PCR sequences, until the intervening gap was bridged. The amplicons were used as templates for DNA sequence determination from both strands. For the annotation of the eps regions, sequence similarity analyses were performed with the gapped BLAST algorithm (E value < 10−11) by using local copies of a nonredundant protein database and the Cluster of Orthologous Genes (COG) database from the National Center for Biotechnology Information. Protein two-dimensional structures were predicted with the EMBOSS software suite (43).
Multilocus sequence analysis methodology.
The different gene sequences were amplified by PCR using the primers specified in Table 1, and the PCR products were sent out for sequencing of both strands (Fasteris SA, Geneva, Switzerland). Design of the primers was performed using Primaclade (22). Alternatively, the sequences were retrieved from the published genomes (see references below). Raw sequence data were transferred into BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium), where consensus sequences were determined using two reads for each gene (one read for the 16S rRNA genes). A similarity matrix and phylogenetic trees were created based on the maximum-parsimony and neighbor-joining methods. The reliability of the groups was evaluated by bootstrap with 500 resamplings.
TABLE 1.
Primers used for PCR in DNA typing and MLSA of the investigated Lactobacillus strains
| Primer | Nucleotide sequence |
|---|---|
| ERIC-1 | ATGTAAGCTCCTGGGGATTCAC |
| ERIC-2 | AAGTAAGTGACTGGGGTGAGCG |
| REP-1 | NNNNCGNCGNCATCNGGC |
| REP-2 | NCGNCTTATCNGGCCTAC |
| rpoA-7F | GAATTTGAAAAACCAAATATTACCGT |
| rpoA-844R | CCTAAGTTACGTACRCGCATCATATC |
| pheS-456F | CCCWGCWCGTGATATGCA |
| pheS-952R | TATTTCAAAATTGCRAAACGRTC |
| tuf-181F | GATGCTGCWCCWGAAGAAAAG |
| tuf-1065R | ATAACCATTTCRGTACCTTCTGGC |
| groEL-309F | GAAGAACGTTACTGCTGGTGC |
| groEL-822R | ACCAAARCCAGGWGCCTTA |
DNA typing.
Enterobacterial repetitive intergenic consensus sequence (ERIC) and repetitive extragenic palindromic element (REP) PCRs were performed using the primers specified in Table 1. Amplification reactions were always performed in 50 μl of a solution containing 0.2 mM each of the two opposing primers, 2.5 μM of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 5 μl of 10× amplification buffer, 2.5 U of Platinium Taq polymerase (Invitrogen), and 1 μl of 10-fold-diluted bacterial DNA. All amplifications were performed in the same DNA thermocycler (GeneAmp PCR System 9700; Applied Biosystems). For REP-PCR, the PCR profile consisted of 35 cycles at 94°C for 30 s, 40°C for 30 s, and 72°C for 4 min and, finally, 1 cycle at 72°C for 6 min. For ERIC-PCR, the PCR profile consisted of 35 cycles at 94°C for 30 s, 48°C for 30 s, and 72°C for 4 min and, finally, 1 cycle at 72°C for 6 min. The PCR products were loaded on 1% agarose gels. Images of the gels were transferred into BioNumerics. Phylogenetic trees were created using UPGMA and the Dice similarity coefficient.
API 50-CHL analysis.
Data were transferred into BioNumerics. A tree was created by cluster analysis using UPGMA and a simple matching similarity coefficient.
MUMmer analysis.
Genome comparisons were performed with the MUMmer 3 package (1, 30). Only completed genome sequences were used for the alignments: L. johnsonii strain NCC533 (NC_005362), L. gasseri strain ATCC 33323 (NC_008530); L. acidophilus strain NCFM (NC_006814), L. delbrueckii subsp. bulgaricus strain ATCC 11842 (NC_008054), L. plantarum strain WCFS1 (NC_004567), Lactobacillus sakei subsp. sakei strain 23K (NC_007576), L. salivarius subsp. salivarius strain UCC118 (NC_007929), L. casei strain ATCC 334 (NC_008526), Lactobacillus brevis strain ATCC 367 (NC_008497), Pediococcus pentosaceus strain ATCC 25745 (NC_008525), Oenococcus oeni strain PSU-1 (NC_008528), and Leuconostoc mesenteroides strain ATCC 8293 (NC_008531). All of the analyses were performed with default parameters, except that the minimum length of a maximal exact match was set to 20. The results were displayed with Mummerplot script with default parameters.
Nucleotide sequence accession numbers.
The sequences of the eps regions from L. johnsonii strains ATCC 33200T, ATCC 11506, and NCC2767 were deposited in GenBank under accession numbers EF138833, EF138834, and EF138835, respectively.
RESULTS AND DISCUSSION
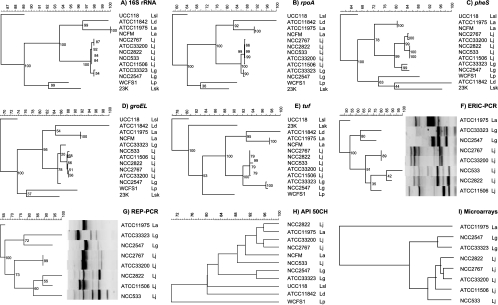
Polyphasic analysis in the L. acidophilus complex. (i) 16S rRNA sequence.
Sequencing of 16S rRNA genes from the investigated lactobacilli confirmed the clustering of the L. delbrueckii/L. acidophilus complex, which was clearly separated from L. plantarum, L. sakei, and L. salivarius (Fig. 1A). The last three species belonged to three distinct clusters in the more extensive 16S rRNA tree of Canchaya et al. (9). On our 16S rRNA tree, L. gasseri and L. johnsonii could be distinguished due to five nucleotide differences in the V1 variable region (26).
FIG. 1.
Polyphasic analysis of the indicated Lactobacillus isolates by MLSA (A to E), DNA typing (F and G), fermentation capacity (H), and clustering of the microarray analysis (I). In each tree, the strain is identified at the right end of the branch by a strain and an abridged genus/species identifier. La, L. acidophilus; Ld, L. delbrueckii subsp. bulgaricus; Lg, L. gasseri; Lj, L. johnsonii; Lp, L. plantarum; Lsk, L. sakei subsp. sakei; Lsl, L. salivarius subsp. salivarius. The numbers at the nodes give the bootstrap probabilities. The scale above the MLSA gives the percentage of base pair sequence identity.
(ii) Multilocus sequence analysis.
For lactic acid bacteria, the sequence analysis of genes encoding the RNA polymerase α subunit (rpoA) and the phenylalanyl-tRNA synthase (pheS) were useful taxonomic criteria (37, 38). Indeed on the rpoA tree (Fig. 1B), the investigated L. johnsonii strains formed a cluster distinct from L. gasseri. As in the 16S rRNA tree, their nearest neighbors were L. acidophilus, L. delbrueckii, L. plantarum/L. sakei, and then L. salivarius, in that order. On the pheS tree (Fig. 1C), L. johnsonii strains split into two subgroups, but both were clearly separated from the two L. gasseri strains. The nearest neighbor was again L. acidophilus, while L. delbrueckii took a more distant position on the pheS tree than on the rpoA trees. On the chaperonin groEL tree (Fig. 1D), the sequence from one L. gasseri strain could not be clearly separated from the L. johnsonii cluster, while for the other lactobacilli, the pattern observed for the rpoA sequences was reproduced. In contrast, the tuf gene, encoding the elongation factor Tu, again clearly separated L. johnsonii from L. gasseri in a tree analysis and defined an L. acidophilus/L. delbrueckii group (Fig. 1E) (60).
(iii) DNA typing.
Next, we applied ERIC-PCR to the strains from the L. acidophilus complex. In this analysis, the sequenced L. johnsonii strain NCC533 and the dog feces isolate NCC2822 were closely related. Two other L. johnsonii strains, namely, the type strain, NCC1680, and the other dog feces isolate, NCC2767, were nearly identical (Fig. 1F). In a tree analysis, the L. johnsonii strains could be clearly separated from L. gasseri and L. acidophilus. In REP-PCR, L. johnsonii NCC1680 and NCC2767 yielded identical patterns, while the remaining three L. johnsonii strains gave a distinct but related pattern. The two L. gasseri strains gave a distinct pattern (Fig. 1G).
(iv) Fermentation phenotype.
The sugar fermentation pattern clustered the different species of the L. acidophilus complex together and excluded L. plantarum and L. salivarius, but no species differentiation could be obtained for this metabolic phenotype in the L. acidophilus complex (Fig. 1H). This observation agrees with the analysis by taxonomists, who observed an absence of correlation between phylogenetic placement and metabolic properties in lactobacilli.
L. johnsonii intraspecies differences as revealed by microarray analysis. (i) Overall view.
Next, we explored the degree of genetic diversity within a single species of the L. acidophilus complex. For that purpose, we asked what number of ORFs from the sequenced L. johnsonii strain NCC533 failed to hybridize with the four L. johnsonii strains investigated for the multilocus sequences, DNA typing, and sugar fermentation phenotype and what their distribution was. Overall, DNA from the test L. johnsonii strains failed to efficiently hybridize with 8% to 17% of the ORFs from the reference L. johnsonii strain, NCC533. When projected on the genome map of NCC533, these CGH results showed some clustering of conserved and NCC533-specific ORFs (Fig. 2) . The region around the origin of replication represented the largest genome segment of relative gene conservation (denoted I-a and I-b in Fig. 2), followed by two shorter regions of conservation (II and III in Fig. 2). In contrast, the region around the terminus of replication was a major area of genetic diversity (IV in Fig. 2). The symmetrical orientation of this diversity region around the terminus is remarkable, since the terminus itself was asymmetrically placed with respect to the origin of replication (1, 41) (Fig. 2). Two further regions of high gene diversity were identified (V and VI in Fig. 2). In L. plantarum, the opposite was observed, as the so-called “lifestyle adaptation island,” representing the largest region of diversity, was close to the replication origin (36).
A priori, one expects two types of genomic diversity between strains belonging to the same species. One type comes from selfish mobile DNA that invaded or left the genome, which does not necessarily add to the fitness of the strain (a “mobilome”). The second type (“diversity regions”) may underlie the ecological adaptation of the investigated strains and could represent laterally acquired DNA or remnants of ancestral DNA that were not lost during genome reduction that occurred in the species. According to the gene annotation, suggestive evidence for both types of diversity was obtained (Fig. 2).
(ii) Mobilome.
Mobile DNA is suggested when a DNA segment fulfils a combination of the following criteria: association with a recombinase gene, presence of recombination sites, gene annotations compatible with known mobile DNA (phages, plasmids, transposons, IS, etc.), and restricted distribution between strains (10). Three large DNA segments, which are essentially restricted to NCC533, clearly represent mobile DNA: two prophages, Lj965 and Lj928, and a 6-kb DNA element (Fig. 2, annotated as a, b, and c), were all previously described (58, 58a). The third element (LJ01749 to LJ1755) was apparently integrated via a Campbell-like mechanism (showing an integrase and attL and attR recombination sites) but lacks further phage links. It contained a copy of the cell division gene ftsK and thus resembled three “potentially autonomous units” described in L. acidophilus (1). The attL and attR recombination sites of the two prophages exactly flanked the region of genetic diversity. In all three cases, PCRs with primers located to the left and right of the attachment sites demonstrated an unoccupied attB site in the test strains (data not shown). In some strains, stretches of hybridizing prophage genes matched individual modules of the prophages (Fig. 2, lane ATCC 33200T in Lj928), an observation which agrees with the standard model of modular phage evolution (7). This observation suggests the presence of related modules in prophages occupying distinct genomic sites in the L. johnsonii test strains. Four further integrases were observed in the NCC533 genome; only one was associated with a cluster of divergent genes. This cluster was annotated as an arsenic resistance cassette (Fig. 2, bracket d). Two fragments of an insertion element flanked a small cluster of divergent genes that lacked a bioinformatic annotation (Fig. 2, e), and several IS flanked or interrupted diversity regions, e.g., the cluster of genes involved in the synthesis of the bacteriocin lactacin F (Fig. 2, f) and the exopolysaccharide (eps) biosynthesis gene cluster (Fig. 2, g). Cumulatively, the 110 genes of the mobilome accounted for 28% (93 genes) of the 335 NCC533 variable genes and 58% (56 genes) of the 96 NCC533-specific genes.
(iii) Diversity regions.
Within the remaining variable regions of the CGH map, three categories of annotations dominated. They were (i) genes associated with bacterium-environment interaction, (ii) metabolic genes, and (iii) genes encoding unknown functions. In the first category were mucin-binding proteins, the eps cluster, a putative fimbrial-biosynthesis regulon, the lactacin F gene cluster, and other cell wall-anchored proteins, including a putative immunoglobulin A protease (Fig. 2 shows the locations and extents of the diversity regions). The eps cluster was associated with the dTDP-rhamnose biosynthesis operon (rfb) and represented the largest genome segment in this diversity group showing substantial genetic variability. Genes from this variability category were previously discussed as candidate probiotic genes (41), suggesting strain-specific rather than species-specific probiotic properties. In the second category were genes involved in peptide metabolism and acquisition of sugars from unusual polysaccharides, phosphotransferase genes, a lactose operon, a transketolase gene group, and a pentose utilization cluster.
(iv) Genetic diversity in the eps cluster.
In the first category of diversity regions, the eps cluster is of particular interest, as it is expected that non-sequence-related DNAs fulfilling comparable functions are found at the same locus in the other strains (31, 56). Therefore, we sequenced the eps-rfb regions in three L. johnsonii test strains and compared them to the eps-rfb cluster from the reference strain NCC533 and the L. gasseri type strain, ATCC 33323T (Fig. 3). Regions of sequence identity alternated with regions of sequence diversity in a more complex patchwork pattern than had previously been reported (24). Only at their 5′ ends were the variable eps core genes clearly embedded between conserved regions. Moreover, the 3′ halves of the clusters (including rfb) differed strikingly in length. Even if the eps genes did not always share DNA sequence identity, the gene order was well conserved. The glycosyltransferase genes, involved in repeating-unit assembly and therefore responsible for differences in the EPS composition at the cell surface, formed the major component of the variable eps core. This region showed a clear modular organization, as already described for other lactic acid bacteria (57). Pairs of strains shared distinct parts of the eps operon. Notably, the most similar eps operons were found in NCC533 and NCC2767, which otherwise showed the least conserved gene contents in the microarray analysis (see below). The L. johnsonii and L. gasseri comparison showed more sharing of genes than some intraspecies L. johnsonii comparisons, suggesting that this locus differentiated after speciation. The high base deviation index shown by the genes LJ1027 to LJ1047 in NCC533 (5) argues for their recent acquisition and supports this interpretation. Interestingly, the eps region is separated from the rfb region by one or several insertion sequences or their remnants, as has been frequently observed in the 3′ regions of eps operons (39, 57). However, no data clearly point to an involvement of these mobile elements in the acquisition of these diversity regions. Taken together, all these observations suggest a complicated evolutionary history for the eps cluster, which might be a hot spot of recombination that creates variability under the pressure of positive selective forces.
FIG. 3.
Alignment of the genetic maps of the exopolysaccharide and dTDP-rhamnose biosynthetic gene cluster in selected L. johnsonii and L. gasseri strains, identified by their code numbers on the left of the maps. Genes sharing high nucleotide sequence identity are linked by blue shading; the shading is striped when the identity is lower than 85%. The numbering of NCC533 genes follows that of the GenBank file. For ATCC 33323T, the genes 04 to 28 correspond to LGAS_1157 to LGAS_1133 of the GenBank file, respectively. The annotation is indicated by the color code explained below the figure. WZY proteins (polymerases) were tentatively classified on the basis of their similar hydrophobicity patterns.
Intra- versus interspecies similarities in the L. acidophilus complex by microarray analysis.
A simplified visualization of the CGH results is presented in the inset of Fig. 2. Raw CGH data come in couples of intensity values associated with each probe on the NCC533 array. These values are expressed as mean ratios of the results from the test to the reference strain, normalized so that a ratio of 1 expresses the perfect conservation of the gene. Due to technical variability, the control hybridization with the reference genome versus itself results in a normal distribution when expressed on a log2 scale. It is a sharp peak centered around zero, as depicted in Fig. 2, inset, A. When the four test L. johnsonii strains were hybridized against the reference strain NCC533, we observed a major peak at the same position, suggesting very similar DNA sequences (Fig. 2, inset, B to E). In addition, all four L. johnsonii test strains showed a reduction in peak height and a leftward spread in the distribution of the hybridization ratios, indicating decreasing DNA sequence identity between the test DNA and 13% to 19% of the ORFs from NCC533.
The two L. gasseri strains, in contrast, showed a marked leftward shift for the bulk of the DNA and substantial trailing in the display of the fluorescence ratios (Fig. 2, inset, F and G). Obviously, the majority of the reference DNA only imperfectly matched the test DNA. In addition to this shift to −1 on the log2 scale, the main peak showed a broadening corresponding to a higher variability of DNA sequence identity. When the CGH results were mapped on the NCC533 genome, we observed that the genetic diversity represented by L. johnsonii strains was a subgroup of the genetic diversity detected between L. johnsonii NCC533 and the L. gasseri strains (Fig. 2). Exceptions were several prophage Lj965 genes, part of the lactacin F operon, and rhamnose biosynthesis genes.
The L. acidophilus DNA showed a normal distribution centered around a log2 value of −3, with substantial trailing to both higher and lower values (Fig. 2, inset, H). Thus, only very few DNA sequences from L. acidophilus found a good match in the L. johnsonii reference strain. The cross-hybridizing DNA, with the exception of one cluster, was scattered over the NCC533 genome (Fig. 2). The shared genes, as expected, encoded the most conserved cellular functions and comprised genes for ribosomal proteins, aminoacyl tRNA synthetases, enzymes of the central metabolic pathway, and a few other known highly conserved genes (e.g., RNA polymerase, elongation factor, peptide release factor, DNA gyrase, ABC transporter, ATP synthase, and chaperones).
As represented in Fig. 2, the percentages of conserved NCC533 genes were 83 to 92% in the L. johnsonii strain comparisons. These percentages decreased to 67, 65, and 12% when the two L. gasseri strains and the single L. acidophilus strain, respectively, were considered. Using the previously reported COG annotation (51) of the NCC533 genome (5), we asked what functional categories significantly contributed to the L. johnsonii strain variability (see the supplemental material). As expected, the highest percentage of variable genes was detected in the mobile-DNA category (85% of the mobilome). Genes from the carbohydrate transport and metabolism category were overrepresented (P < 0.001) in the variable-gene set, as well as genes without COG attribution. Genes with unknown functions or without COG attribution differentiated the L. johnsonii and L. gasseri genomes (see the supplemental material).
In addition to the inset of Fig. 2 which globally quantifies the similarity of the test strains versus L. johnsonii NCC533, a clustering of the microarray results was performed in order to extract the qualitative information about the presence of each gene. A tree analysis of the CGH scores not only separated L. johnsonii from L. gasseri, but identified L. johnsonii strains NCC2767 and NCC1680 as nearest neighbors, which also shared nearly identical ERIC and REP patterns (Fig. 1G, H, and I).
Interspecies differences in the genus Lactobacillus as revealed by in silico genome alignments. (i) DNA level.
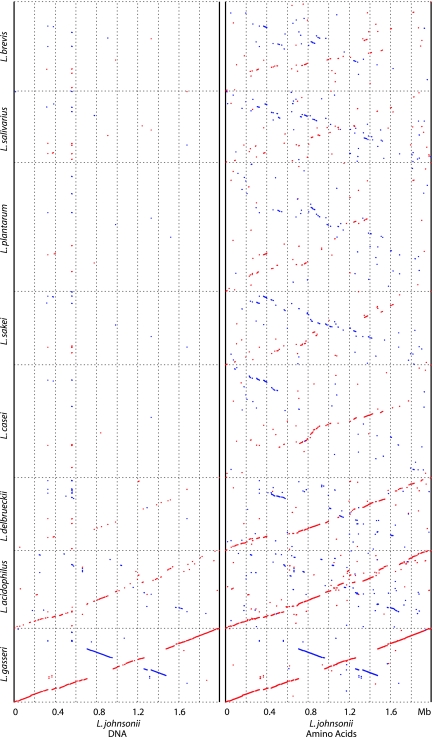
Nine species belonging to the genus Lactobacillus have been sequenced: L. plantarum, L. johnsonii, L. delbrueckii, L. acidophilus, L. sakei, L. salivarius, and, very recently, L. casei, L. gasseri, and L. brevis (1, 11, 13, 25, 41, 55). When the different genomes from the sequenced lactobacilli were plotted against the DNA sequence of L. johnsonii strain NCC533 (Fig. 4, left), L. gasseri showed the closest alignment. Only two large inversions located on either side of the terminus of replication interrupt the straight alignment of the two species genomes, demonstrating close synteny. Only a few segments of the L. gasseri genome did not find a match in the L. johnsonii sequence when the MUMmer program was applied. L. acidophilus showed the next-best alignment at the DNA sequence level. Here again, a clear diagonal line along the entire genome length was detectable. However, the extent of the alignment was substantially weaker than the L. gasseri/L. johnsonii alignment. L. delbrueckii still shared with L. johnsonii some DNA sequence identity, resulting in a faint diagonal line. L. plantarum, L. sakei, and L. salivarius shared DNA sequence identity with L. johnsonii mainly over one repetitive DNA segment (rRNA at 0.56 Mb).
FIG. 4.
DNA and protein sequence similarities between completely sequenced lactobacilli (identified on the y axis) as revealed by in silico genome alignments with L. johnsonii NCC533, which was used as the sequenced reference strain (MUMmer analysis). (Left) Alignments obtained with NUCmer script, highlighting the conserved regions at the DNA level. The dots represent the positions of conserved DNA sequences on the genomes. (Right) Alignments obtained with PROmer script, highlighting the conserved regions at the protein level. The dots represent the positions of conserved protein sequences on the genomes. Identities in direct or reverse orientation are indicated in blue and red, respectively. Note that the sequenced L. acidophilus strain does not correspond to the strain used in the CGH analysis.
(ii) Protein level.
L. gasseri and L. acidophilus share protein sequence identity with L. johnsonii along the entire genome length when analyzed by MUMmer (Fig. 4, right). The number and length of the alignment gaps were greater in the L. acidophilus than in the L. gasseri comparison. In contrast to the marked loss of sequence identity in the L. johnsonii/L. delbrueckii DNA comparison, both species still showed a straight line in the protein MUMmer display (Fig. 4, right), with the exception of one major inversion (0.4 to 0.6 Mb) and one poorly aligned segment (1.0 to 1.2 Mb). The conserved synteny of these four lactobacilli was previously observed by BlastP comparisons (24, 55). When lactobacilli outside the L. acidophilus/L. delbrueckii group were used for alignment with L. johnsonii, protein sequence identities were restricted to small genome segments, but despite all dispersion of the aligned regions, an X-shaped alignment pattern could still be clearly detected (especially pronounced with L. sakei), suggesting multiple inversions around the terminus of replication.
Evolutionary and taxonomical implications.
The sequenced L. johnsonii strain differed from the test L. johnsonii strains in up to 17% of its gene content and displayed 5% strain-specific genes. Comparable and sometimes even higher percentages of variable-gene content were reported for many sequenced bacterial strains (17, 20, 32, 33, 40, 44), including L. plantarum (36). For example, the sequencing of eight Streptococcus agalactiae strains demonstrated a core genome shared by all isolates, accounting for 80% of any single genome, and a dispensable genome part consisting of partially shared and strain-specific genes. Extrapolation of the data suggested that the S. agalactiae “pangenome” is vast and that the sum of the variable genes within the confines of a bacterial species will rapidly exceed the number of conserved genes (53). This hypothesis raises important questions with respect to the origin of these variable genes, since only a third of the variable genes and two-thirds of the strain-specific genes in L. johnsonii were identified as mobile DNA. Microbiologists are thus confronted with the problem of the roles of horizontally versus vertically inherited genes in the evolution of bacterial genomes (8). Some microbiologists have rejected the idea of a phylogenetic tree for bacteria (3, 18), while more recent large-scale genome comparisons have stressed the overwhelming dominance of vertical over horizontal gene transfers (27-29) (for a recent review, see reference 14).
What do these considerations mean for a natural system of bacteria and a taxonomy based on it? Our analyses within the acidophilus group of the genus Lactobacillus show a clear-cut concordance between the different techniques. The 16S rRNA analysis can provide an overview of the phylogenetic relationships between the investigated lactobacilli that was largely confirmed by the other techniques applied. In multilocus sequence analysis of housekeeping genes, only three of the four genes allowed a clear separation of L. johnsonii from L. gasseri (rpoA and pheS, as suggested by Naser et al. [38], and tuf, as suggested by Chavagnat et al. [12], but not groEL, contrary to the report by Teng et al. [52]). The two DNA-typing methods confirmed the close relationships within strains of the species L. johnsonii and allowed intraspecies strain differentiation, confirming earlier reports (59). As already reported (15), the metabolic phenotypes of lactobacilli have only poor taxonomical power in the genus Lactobacillus.
What is the contribution of newer whole-genome-based typing methods to the taxonomical discussion? The microarray analysis clearly classified all investigated L. johnsonii strains into a single close-knit group of genomes, which was clearly separated from L. gasseri. There was no continuous transition from L. johnsonii into L. gasseri—the investigated genomes formed two clearly separated clusters defining them as genomically coherent groups, which agrees with the current taxonomic bacterial-species definition. Although this conclusion is at the moment based on a relatively small number of investigated strains, the chosen strains likely represent a sufficient variety, as testified by their extensive IS-mapping differences (see Materials and Methods). Therefore, the gap separating L. johnsonii from L. gasseri might get smaller, but will remain, when more strains add more breadth to each species. The microarray analysis revealed, furthermore, that L. gasseri was more closely related to L. johnsonii than L. acidophilus, demonstrating a gradual loss of similarity in the group. As the microarray analysis is based on DNA-DNA hybridization, more distantly related lactobacilli could not be evaluated with this technology, since these strains lacked sufficient DNA sequence similarity with the reference strain, as confirmed by the MUMmer analysis of the whole-genome sequences. Since the whole-genome in silico analysis can be extended to the protein sequence level, one can also explore the similarity of the reference strain to even more distantly related bacteria. This analysis identified L. delbrueckii as the most closely related bacterium to the L. acidophilus complex, confirming previous taxonomical classifications (54). The most closely related lactobacilli from the perspective of L. johnsonii were L. casei and L. sakei, which agrees with a previously reported tree constructed on the basis of concatenated ribosomal protein sequences (34) and the 16S rRNA tree (9), respectively. At the next lower level of similarity was L. salivarius, and even more distantly related are L. brevis and L. plantarum. In the last case, the larger genome size of L. plantarum might dilute the similarity visually, suggesting a lower degree of similarity to L. johnsonii. This stepwise decrease in similarity in the genome alignments still approximately reflects the trends of the 16S rRNA tree.
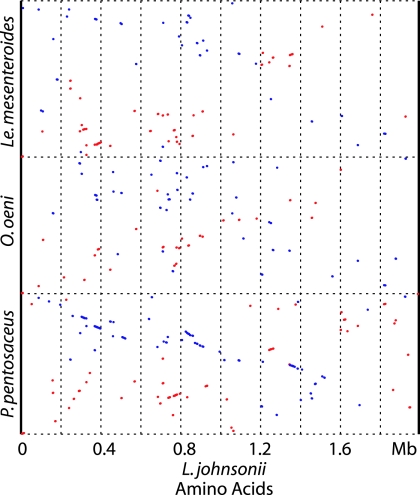
The stepwise-decreasing degrees of similarity observed in the L. delbrueckii/L. acidophilus group are a hallmark of Darwinian evolution. If strong elements of vertical evolution are observed in such a problematic taxonomical group as lactobacilli, there is hope that in many bacterial groups, whole-genome-based analyses will lead to a taxonomy that also reflects the phylogenetic relationships of the investigated bacteria, at least to a first approximation. It is currently not clear whether the observed protein sequence relationships among all the sequenced lactobacilli are a genomics argument for the coherence of the genus Lactobacillus. A critical test for a genomics-supported Lactobacillus genus concept will be the demonstration that bacteria classified outside of the genus Lactobacillus share less genomic relatedness with L. johnsonii than L. brevis, currently the most distant L. johnsonii relative among the sequenced lactobacilli. In a PROmer analysis, L. johnsonii was as closely related to L. brevis as to Pediococcus, while Oenococcus and Leuconostoc share practically no synteny with L. johnsonii (Fig. 4, right, and Fig. 5). Interestingly, trees based on ribosomal protein (34) and 16S rRNA sequences (15) place Pediococcus close to L. brevis. The congruency between the “new” whole-genome comparisons and “the old methods” used for studying intra- and interspecific variation is gratifying. With the growing bacterial-genome database, whole-genome-based comparisons will become an increasingly popular aid in settling controversial taxonomical issues at both the species and the genus levels.
FIG. 5.
Protein sequence similarities (PROmer) of Pediococcus pentosaceus, Oenococcus oeni, and Leuconostoc mesenteroides with L. johnsonii NCC533 as the sequenced reference strain.
Supplementary Material
Acknowledgments
We thank Todd Klaenhammer for providing us the unfinished L. gasseri genome sequence before its publication, Maria Karmirantzou for her work on the annotation of the L. johnsonii NCC533 genome sequence, Jean-Marc Aeschlimann for helpful discussions on statistics, Deborah Moine and Juan Olivera for help with MLSA, and Enea Rezzonico for editing the references.
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bapteste, E., Y. Boucher, J. Leigh, and W. F. Doolittle. 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12:406-411. [DOI] [PubMed] [Google Scholar]
- 4.Bernardeau, M., M. Guguen, and J. P. Vernoux. 2006. Beneficial lactobacilli in food and feed: long-term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol. Rev. 30:487-513. [DOI] [PubMed] [Google Scholar]
- 5.Boekhorst, J., R. J. Siezen, M. C. Zwahlen, D. Vilanova, R. D. Pridmore, A. Mercenier, M. Kleerebezem, W. M. de Vos, H. Brussow, and F. Desiere. 2004. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology 150:3601-3611. [DOI] [PubMed] [Google Scholar]
- 6.Brook, I., and E. H. Frazier. 1993. Significant recovery of nonsporulating anaerobic rods from clinical specimens. Clin. Infect. Dis. 16:476-480. [DOI] [PubMed] [Google Scholar]
- 7.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 8.Bushman, F. 2002. Lateral DNA transfer mechanisms and consequences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Canchaya, C., M. J. Claesson, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152:3185-3196. [DOI] [PubMed] [Google Scholar]
- 10.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaillou, S., M. C. Champomier-Verges, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongere, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 12.Chavagnat, F., M. Haueter, J. Jimeno, and M. G. Casey. 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol. Lett. 217:177-183. [DOI] [PubMed] [Google Scholar]
- 13.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dagan, T., and W. Martin. 2006. The tree of one percent. Genome Biol. 7:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dellaglio, F., and G. E. Felis. 2005. Taxonomy of lactobacillus and bifidobacteria, p. 25-49. In G. W. Tannock (ed.), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 16.Delley, M., B. Mollet, and H. Hottinger. 1990. DNA probe for Lactobacillus delbruecki. Appl. Environ. Microbiol. 56:1967-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2129. [DOI] [PubMed] [Google Scholar]
- 19.Doolittle, W. F., and R. T. Papke. 2006. Genomics and the bacterial species problem. Genome Biol. 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujisawa, T., Y. Benno, T. Yaeshima, and T. Mitsuoka. 1992. Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int. J. Syst. Bacteriol. 42:487-491. [DOI] [PubMed] [Google Scholar]
- 22.Gadberry, M. D., S. T. Malcomber, A. N. Doust, and E. A. Kellogg. 2005. Primaclade—a flexible tool to find conserved PCR primers across multiple species. Bioinformatics 21:1263-1264. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. L., C. F. Phelps, C. S. Cummins, J. London, and F. Gasser. 1980. Taxonomy of the Lactobacillus acidophilus group. Int. J. Syst. Bacteriol. 30:53-68. [Google Scholar]
- 24.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria affecting bioprocessing and health. FEMS Microbiol. Rev. 29:393-409. [DOI] [PubMed] [Google Scholar]
- 25.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kullen, M. J., R. B. Sanozky-Dawes, D. C. Crowell, and T. R. Klaenhammer. 2000. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511-516. [DOI] [PubMed] [Google Scholar]
- 27.Kunin, V., D. Ahren, L. Goldovsky, P. Janssen, and C. A. Ouzounis. 2005. Measuring genome conservation across taxa: divided strains and united kingdoms. Nucleic Acids Res. 33:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunin, V., L. Goldovsky, N. Darzentas, and C. A. Ouzounis. 2005. The net of life: reconstructing the microbial phylogenetic network. Genome Res. 15:954-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurland, C. G., B. Canback, and O. G. Berg. 2003. Horizontal gene transfer: a critical view. Proc. Natl. Acad. Sci. USA 100:9658-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamothe, G. T., L. Jolly, B. Mollet, and F. Stingele. 2002. Genetic and biochemical characterization of exopolysaccharide biosynthesis by Lactobacillus delbrueckii subsp. bulgaricus. Arch. Microbiol. 178:218-228. [DOI] [PubMed] [Google Scholar]
- 32.Le Gall, T., P. Darlu, P. Escobar-Paramo, B. Picard, and E. Denamur. 2005. Selection-driven transcriptome polymorphism in Escherichia coli/Shigella species. Genome Res. 15:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonard, E. E., L. S. Tompkins, S. Falkow, and I. Nachamkin. 2004. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barre syndrome and strains that cause enteritis by a DNA microarray. Infect. Immun. 72:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, V. J. J., and W. W. Mohn. 1999. An alternative inverse PCR (IPCR) method to amplify DNA sequences flanking Tn5 transposon insertions. J. Microbiol. Methods 35:163-166. [DOI] [PubMed] [Google Scholar]
- 36.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187:6119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naser, S. M., K. E. Hagen, M. Vancanneyt, I. Cleenwerck, J. Swings, and T. A. Tompkins. 2006. Lactobacillus suntoryeus Cachat and Priest 2005 is a later synonym of Lactobacillus helveticus (Orla-Jensen 1919) Bergey et al. 1925 (Approved Lists 1980). Int. J. Syst. Evol. Microbiol. 56:355-360. [DOI] [PubMed] [Google Scholar]
- 38.Naser, S. M., F. L. Thompson, B. Hoste, D. Gevers, P. Dawyndt, M. Vancanneyt, and J. Swings. 2005. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151:2141-2150. [DOI] [PubMed] [Google Scholar]
- 39.Pluvinet, A., F. Charron-Bourgoin, C. Morel, and B. Decaris. 2004. Polymorphism of eps loci in Streptococcus thermophilus: sequence replacement by putative horizontal transfer in S-thermophilus IP6757. Int. Dairy J. 14:627-634. [Google Scholar]
- 40.Porwollik, S., R. M. Y. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, G., J. Jass, M. T. Sebulsky, and J. K. McCormick. 2003. Potential uses of probiotics in clinical practice. Clin. Microbiol. Rev. 16:658-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 44.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salvana, E. M., and M. Frank. 2006. Lactobacillus endocarditis: case report and review of cases reported since 1992. J. Infect. 53:e5-e10. [DOI] [PubMed] [Google Scholar]
- 46.Sarker, S. A., S. Sultana, G. J. Fuchs, N. H. Alam, T. Azim, H. Brüssow, and L. Hammarstrom. 2005. Lactobacillus paracasei strain ST11 has no effect on rotavirus but ameliorates the outcome of nonrotavirus diarrhea in children from Bangladesh. Pediatrics 116:e221-e228. [DOI] [PubMed] [Google Scholar]
- 47.Schleifer, K. H., and W. Ludwig. 1995. Phylogeny of the genus Lactobacillus and related genera. Syst. Appl. Microbiol. 18:461-467. [Google Scholar]
- 48.Servin, A. L. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28:405-440. [DOI] [PubMed] [Google Scholar]
- 49.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 50.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng, L. J., P. R. Hsueh, Y. H. Wang, H. M. Lin, K. T. Luh, and S. W. Ho. 2001. Determination of Enterococcus faecalis groESL full-length sequence and application for species identification. J. Clin. Microbiol. 39:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Kranenburg, R., I. C. Boels, M. Kleerebezem, and W. M. de Vos. 1999. Genetics and engineering of microbial exopolysaccharides for food: approaches for the production of existing and novel polysaccharides. Curr. Opin. Biotechnol. 10:498-504. [DOI] [PubMed] [Google Scholar]
- 57.van Kranenburg, R., H. R. Vos, I. I. van Swam, M. Kleerebezem, and W. M. de Vos. 1999. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J. Bacteriol. 181:6347-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ventura, M., C. Canchaya, R. D. Pridmore, and H. Brüssow. 2004. The prophages of Lactobacillus johnsonii NCC 533: comparative genomics and transcription analysis. Virology 320:229-242. [DOI] [PubMed] [Google Scholar]
- 58a.Ventura, M., C. Canchaya, R. D. Pridmore, B. Berger, and H. Brüssow. 2003. Integration and distribution of Lactobacillus johnsonii prophases. J. Bacteriol. 185:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ventura, M., and R. Zink. 2002. Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 217:141-154. [DOI] [PubMed] [Google Scholar]
- 60.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.