Abstract
Recent murine studies have demonstrated that the role of response regulator 09 (RR09) of Streptococcus pneumoniae in virulence is different in different strains. In the present study, we used a murine pneumonia model of infection to assess the virulence of a TIGR4 rr09 mutant, and we found that TIGR4Δrr09 was attenuated after intranasal infection. Furthermore, we investigated the in vitro transcriptional changes in pneumococcal rr09 mutants of two strains, D39 and TIGR4, by microarray analysis. The transcriptional profiles of the rr09 mutants of both strains had clear differences compared to the profiles of the parental wild-type strains. In D39Δrr09, but not in TIGR4Δrr09, genes involved in competence (e.g., comAB) were upregulated. In TIGR4, genes located on the rlrA pathogenicity islet, which are not present in the D39 genome, appeared to be regulated by RR09. Furthermore, several phosphotransferase systems (PTSs) believed to be involved in sugar uptake (e.g., the PTS encoded by sp0060 to sp0066) were strongly downregulated in D39Δrr09, while they were not regulated by RR09 in TIGR4. To examine the role of one of these PTSs in virulence, D39Δsp0063 was constructed and tested in a murine infection model. No difference between the virulence of this strain and the virulence of the wild type was found, indicating that downregulation of the sp0063 gene alone is not the cause of the avirulent phenotype of D39Δrr09. Finally, expression of rr09 and expression of three of our identified RR09 targets during infection in mice were assessed. This in vivo experiment confirmed that there were differences between expression in wild-type strain TIGR4 and expression in the rr09 mutant, as well as differences between expression in wild-type strain D39 and expression in wild-type strain TIGR4. In conclusion, our results indicate that there is strain-specific regulation of pneumococcal gene expression by RR09.
Streptococcus pneumoniae (pneumococcus) is a common asymptomatic commensal of the nasopharynx in healthy individuals. However, in young children, in the elderly, and in immunocompromised people, this gram-positive bacterium is a major cause of otitis media, pneumonia, and septicemia. To persist at these various sites, the pneumococcus needs to adapt and orchestrate its gene expression. Two-component signal transduction systems (TCSs) play a central role in bacterial survival by regulating various cellular processes, such as osmoregulation, sporulation, genetic competence, and chemotaxis, in response to environmental changes (1, 27). TCSs typically consist of a membrane-associated sensory protein called a histidine kinase (HK) and a cognate cytosolic DNA-binding response regulator (RR), which acts as a transcriptional regulator. When an external signal is sensed, a histidine residue of the histidine kinase autophosphorylates, after which this phosphogroup is transferred to the response regulator. This results in a conformational change in the regulatory protein, which can then perform its regulatory function.
The pneumococcal genome contains 13 putative complete TCSs plus one orphan response regulator (15, 28). Ten of the pneumococcal TCSs have been shown to be important for virulence (for a review, see reference 22). For instance, CiaR/CiaH has been demonstrated to contribute to virulence (25, 29), probably in part through control of expression of htrA, which encodes a serine protease that is a major virulence factor (11, 19). Recently, TCS06 has been found to regulate the expression of cbpA, which codes for a major adhesin that is also a protective antigen (26).
Relatively little is known about TCS09, which consists of an RR encoded by sp0661 (spr0578 in the R6 genome) and an HK encoded by sp0662 (spr0579). Amino acid sequence homology data suggest that the sensory domain of the HK is related to the extracellular part of McpA and McpB of Bacillus subtilis (15). These proteins are believed to be involved in the control of chemotaxis through sensing of environmental nutrient concentrations. The extracellular stimulus of TCS09, however, is not known.
Several in vivo studies have demonstrated that RR09 has a role in virulence. Interestingly, the contribution of RR09 to virulence varies with the bacterial strain and the site of infection (3, 6, 15, 29). It has been suggested that in strain 0100993 (serotype 3) RR09 is involved in the dissemination from the lung to the systemic circulation, since attenuation of 0100993Δrr09 was observed in a murine pneumonia model but not in a sepsis model of infection (3). In D39 (serotype 2) the lack of RR09 led to an avirulent phenotype upon intranasal, intravenous, or intraperitoneal infection (3). The genetic differences between strains are likely to have a significant impact on the repertoire of genes regulated by RR09. In line with this, TCS04 mutants were recently found to confer strain-dependent phenotypes in a murine infection model, possibly caused by differential regulation of pneumococcal surface antigen A (20).
So far, no gene targets of RR09 that could account for the observed in vivo phenotypes of rr09 mutants have been identified, although sequence homology data have suggested that TCS09 is involved in nutrient perception (3, 15). To investigate strain differences in the role of TCS09 in pneumococcal virulence further, we examined the phenotype of a TIGR4 rr09 mutant using a murine pneumonia model. In addition, we used DNA microarray technology to examine how transcriptional patterns are affected by a lack of rr09. To this end, global expression profiles of wild-type and rr09 mutants were analyzed during different stages of in vitro growth. We assessed strain-specific regulation by RR09 by comparing transcriptional profiles of mutants with two different genetic backgrounds, namely, strains D39 and TIGR4. In vitro expression experiments and data analysis were performed independently in two laboratories (Rotterdam and Glasgow) using slightly different methods, after which the data were validated and combined. Finally, we assessed expression of rr09 and three of its identified in vitro targets during experimental virulence in mice.
MATERIALS AND METHODS
Pneumococcal strains.
Wild-type S. pneumoniae strains D39 (= NCTC 7466; serotype 2) and TIGR4 (= ATCC BAA-334; serotype 4) were used in this study. The Δrr09 derivative of TIGR4 was constructed by insertional inactivation using an erythromycin resistance cassette, as described previously for D39Δrr09 (3). Pneumococcal strains were grown on Columbia blood base agar supplemented with 5% (vol/vol) defibrinated sheep blood (and supplemented with 1 μg/ml erythromycin for the rr09 mutant strains). For RNA isolation analyses, cultures were grown in Todd-Hewitt broth supplemented with 5 g/liter yeast extract (THY broth) or in brain heart infusion broth (BHI broth) without erythromycin until they reached the desired turbidity. For construction of the sp0063 mutant (annotated spr0062 in R6), approximately 100-bp portions of up- and downstream regions of the targeted sequence were amplified by PCR using primers 50L (TCTATGATTGGTATTTCTATCGTAGG) and 50M (GGCGCGCCTGAGGTAAGATCATGTAAAGGTAACC) and primers 50N (TCTTACCTCAGGCGCGCCACTGCCTTTATCTTCTGGTTGCTTGG) and 50O (CAAATTTAGCAGTAAATTCTTCTGGG), respectively. These fragments were joined by overlap extension PCR due to overlap in the 50 M and 50N primers. This procedure also introduced an AscI site between the upstream and downstream sequences via the primers. The fusion PCR product was cloned into TOPO-pCR4 (Invitrogen) and confirmed by sequencing. The spectinomycin resistance cassette from pDL278 was cloned into the AscI site, and the resulting construct was used for transformation of D39. Spectinomycin-resistant colonies were verified by PCR. This mutation resulted in replacement of nucleotides 176 to 676 of sp0063 with the spectinomycin resistance cassette.
Mice and infections.
Female outbred MF1 mice (body weight, 25 to 30 g) were purchased from Harlan Olac, Bicester, United Kingdom. For pneumonia infection, 9-week-old mice were lightly anesthetized with 2.5% (vol/vol) halothane, after which an infection dose consisting of 1.0 × 106 CFU of wild-type strain TIGR4 and the Δrr09 mutant resuspended in 50 μl of sterile phosphate-buffered saline (PBS) was administered in the nostrils of mice held vertically (12). At predetermined times after infection, groups of mice were sacrificed by cervical dislocation, and blood samples were removed by cardiac puncture using a 1-ml syringe. Bronchoalveolar lung lavage and sampling of the lungs were performed as described previously (13). The viable bacteria in lung and blood samples were counted by plating serial 10-fold dilutions on blood agar base no. 2 (Oxoid, United Kingdom) supplemented with 5% (vol/vol) defibrinated horse blood (13). Mice infected with wild-type strain D39 and the Δsp0063 mutant were inoculated intraperitoneally with 100 μl PBS containing 1 × 106 CFU.
Survival times were analyzed using the Mann-Whitney U test. The bacteriology results are expressed below as geometric means ± standard errors of the means. Bacterial loads obtained in the time experiment were compared using Student's t test. In all analyses, a P value of <0.01 was considered statistically significant.
RNA extraction.
Five hundred milliliters of THY broth was inoculated with 10 to 20 colonies from agar plates, and the cultures were statically grown at 37°C. Samples for RNA isolation were removed when the cultures reached an optical density at 600 nm (OD600) of either 0.1 (early log phase) or 0.2 (mid-log phase), and the pneumococcal cells were harvested by centrifugation for 10 min at 3,300 × g and 4°C. Subsequently, the cells were resuspended in 400 μl of THY broth supplemented with 1.5 g of glass beads (diameter, 0.1 mm; Sigma), 500 μl of phenol, 50 μl of 10% (wt/vol) sodium dodecyl sulfate (SDS), and 50 μl of 3 M sodium acetate (pH 5.2). Each mixture was then snap-frozen in liquid nitrogen and stored at −80°C until it was used. The cells were lysed by vigorous shaking for 8 min at 4°C and subsequently centrifuged for 10 min at 9,300 × g and 4°C. The upper phase was mixed with an equal volume of chloroform and centrifuged for 2 min at 16,000 × g and 4°C. After this, RNA was purified using a High Pure RNA isolation kit (Roche Diagnostics). Contaminating genomic DNA was removed by treatment with RNase-free DNase I (Roche Diagnostics). RNA was isolated from three replicate cultures of D39 and TIGR4.
Bacterial RNA isolated at an OD600 of 0.6 (mid-log phase) was obtained from two independent cultures grown in BHI broth at 37°C. Bacteria were lysed three times for 20 s in the presence of 200 μl of lysozyme (15 mg/ml) and 50 mg of glass beads (100 μm; Sigma) using a Hybaid Ribolyser (Hybaid) set at speed 4. Subsequently, RNA was isolated using RNeasy Midi columns (QIAGEN) with an on-column DNase digestion step.
cDNA labeling.
Synthesis and subsequent labeling of cDNA for microarray hybridization were performed essentially as described previously (31). Briefly, 20 μg of total RNA was incubated for 16 h at 42°C in the presence of 400 U Superscript III RNase H− reverse transcriptase (Invitrogen), 0.2 mM aminoallyl dUTP (Amersham), 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.3 mM dTTP, and 3.2 μg random nonamers. The synthesized cDNA was then labeled by coupling either Cy3 or Cy5 to the dUTP aminoallyl reactive group (CyScribe postlabeling kit; Amersham Biosciences) and was purified using a GFX PCR, DNA, and gel band purification kit (Amersham Biosciences).
Microarray construction.
Two different DNA microarray platforms were used for analysis of samples isolated at low and high OD600; these platforms are referred to below as array 1 for an OD600 of 0.1 or 0.2 and array 2 for an OD600 of 0.6. Array 1 was constructed as described previously (14, 31) and contained amplicons representing 2,087 open reading frames (ORFs) of S. pneumoniae TIGR4 and 184 unique S. pneumoniae R6 ORFs, all spotted in duplicate.
Array 2 was designed at the Pathogen Functional Genomics Resource Centre at TIGR (http://www.pfgrc.tigr.org/). The complete genome array consisted of amplicons representing segments of 2,131 open reading frames of S. pneumoniae reference strain TIGR4 spotted in quadruplicate on glass slides. In addition, the array contained 563 open reading frames amplified from strains R6 (164 ORFs) and G54 (399 ORFs).
Microarray hybridization.
For array 1, labeled wild-type and Δrr09 cDNA were combined and dried using a SpeedVac. The cDNA was dissolved in 10 μl of Slidehyb #1 hybridization buffer (Ambion Europe Ltd.), boiled for 3 min, and kept on ice until hybridization. Prewarmed (68°C) Slidehyb #1 hybridization buffer was added to obtain a final hybridization volume of 60 μl, after which the sample was applied to a prewarmed array and incubated in a hybridization incubator (ISO20; Grant) overnight at 42°C. Slides were removed from the incubator and washed with 2× sodium chloride-sodium citrate buffer (SSC) containing 0.5% (wt/vol) SDS for 5 min, followed by two washes in 1× SSC containing 0.25% SDS and in 1× SSC containing 0.1% SDS for 5 min each. Subsequently, the slides were dried by centrifugation. In all cases, dye swapping was performed with one of the three biological replicates.
For array 2, denatured labeled wild-type and Δrr09 probes were mixed and dried using a SpeedVac (Savant DNA 110 SpeedVac; Global Medical Instruments). The labeled probes were dissolved in 30 μl of filtered hybridization buffer (50% formamide, 5× SSC, 0.1% SDS, 300 μg salmon sperm DNA) and heated to 95°C for 5 min. The samples were applied to glass slides and incubated for 18 h at 42°C in a GeneChip 640 hybridization oven (Affymetrix). After hybridization, the slides were washed in 2× SSC-0.1% SDS for 5 min at 55°C, and this was followed by two washes in 0.1× SSC-0.1% SDS for 5 min and in 0.1× SSC for 5 min. For each independent RNA sample one dye swap was performed.
Microarray data analysis.
For array 1, dual-channel array images were acquired with a GeneTac LS IV confocal laser scanner (Genomics Solutions) and were analyzed with the ArrayPro 4.5 software (Media Cybernetics Inc.). Spots were screened visually to identify the low-quality spots. Slide data were processed using MicroPreP as described previously (4, 31, 32). Prior to analysis, automatically and manually flagged spots and spots with very low background-subtracted signal intensities (5% of the weakest spots [sum of Cy3 and Cy5 net signals]) were filtered out of all data sets. Net signal intensities were calculated using grid-based background subtraction. In Prep, a grid-based Lowess transformation was performed for slide normalization, with an f value (percentage of the spots used for curve fitting) of 0.5 and an nSteps value (number of iterations) of 5. In Postprep, negative and empty values were removed, and outliers were removed by the deviation test. Further analysis was performed using a Cyber-T Student's t test for paired data (18). This web-based program lists the ratios of all intrareplicates (duplicate spots) and interreplicates (different slides), the mean ratios per gene, and standard deviations and P values assigned to these mean ratios. For identification of differentially expressed genes, only genes with a minimum of three reliable measurements (i.e., data from at least two different slides) and a P value of <0.05 were included. Since this P value is purely a statistical measure of differential gene expression and reproducibility across replicates, an additional change cutoff of twofold was applied.
Hybridized array 2 images were acquired with a Perkin-Elmer Scan Array Express. The spot intensities were defined and quantified using BlueFuse for Microarray 3.1 (BlueGnome Ltd.). The data were analyzed further with GeneSpring 7.0 (Silicon Genetics). Lowess intensity-dependent normalization was used to perform per-spot and per-array normalization, and the cross-gene error model was based on the replicate measurements. Statistically significant differences were defined as differences with a Student's t test P value of <0.05 and a ratio change threshold of at least 2 standard deviations over the median ratio for each strain.
Isolation of pneumococcal RNA during experimental infection.
Female outbred CD-1 mice (body weight, 20 to 30 g) were purchased from Harlan Olac, Bicester, United Kingdom. Nine-week-old mice were lightly anesthetized with 2.5% (vol/vol) halothane, after which 1.0 × 107 CFU wild-type D39, wild-type TIGR4, or TIGR4Δrr09 resuspended in 50 μl of sterile PBS was administered in the nostrils of mice held vertically. Control mice were inoculated with sterile PBS alone. Twenty-four hours postinfection, mice were sacrificed by cervical dislocation, and nasopharyngeal lavage fluid, bronchoalveolar lavage fluid (BALF), lungs, and blood were collected. After collection of 2 ml nasopharyngeal lavage fluid or BALF, 20 μl was used for determination of the bacterial load, and the remaining fluid was mixed with 4 ml RNAprotect (QIAGEN) and incubated for 5 min at room temperature. Bacteria were collected by centrifugation (16,000 × g, 5 min, 4°C), and the pellets were snap-frozen in liquid nitrogen. After collection of blood, 20 μl was used for determination of the bacterial load, and the remaining blood was added to 5 ml RNAprotect (QIAGEN). To separate pneumococci from host mouse cells, the mixtures were centrifuged for 10 min at 825 × g and 4°C. Each supernatant was transferred to a new tube and centrifuged for 5 min at 16,000 × g and 4°C. The pelleted bacteria were snap-frozen in liquid nitrogen. The lungs that were collected were homogenized in 2 ml RNAprotect. The lung samples were handled as described above for the blood samples. RNA of all samples was isolated using an RNeasy kit (QIAGEN) with on-column DNase treatment (QIAGEN). Subsequently, RNA isolated from homogenized lungs and blood were enriched for bacterial RNA using a MicrobEnrich kit (Ambion). Finally, all RNA samples were amplified using a SenseAmp kit (Genisphere). Pneumococcal gene expression was measured in samples obtained from three individual mice.
Real-time PCR.
Real-time PCR was used to validate the microarray data (in vitro) and to investigate expression of several genes during experimental virulence in mice (in vivo). DNA-free total RNA (2.5 μg) was reverse transcribed using 1 μg (for the in vitro experiments with array 1) and 0.5 μg (for the in vivo experiments) of random hexamers and Superscript III reverse transcriptase (Invitrogen). To confirm the absence of genomic DNA, reactions without reverse transcriptase were performed. Subsequently, 1 μl of cDNA diluted 1:10 (for in vitro experiments) or 1:4 (for in vivo experiments) was used as the template in real-time PCR. Duplicate quantitative PCR assays were performed using a DyNAmo HS SYBR green quantitative PCR kit (Bioke) with an ABI Prism 7700 according to the manufacturer's instructions. Primers (sequences are available on request) were designed using the Oligo 6.22 software (Molecular Biology Insights) and were used at a concentration of 300 nM for 40 cycles of amplification (15 s at 95°C and 1 min at 60°C), which was followed by a melting curve analysis to verify the product homogeneity.
Validation of selected targets from array 2 was performed in essentially the same manner, with a few minor differences. Two micrograms of RNA was reverse transcribed using 6 μg of random hexamers and Superscript III reverse transcriptase (Invitrogen). Serial dilutions of cDNA were used as templates in real-time PCR assays with a DNA Engine Opticon 2 (MJ Research), using the following reaction conditions: 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. The relative quantitation method (17) was used to evaluate the quantitative variation between wild-type and Δrr09 strains for each gene examined. The gyrA (sp1219) amplicon was used as an internal control for normalization of data.
Cyber-T website.
The Cyber-T website is http://visitor.ics.uci.edu/genex/cybert/index.shtml.
Nucleotide sequence accession numbers.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under GEO Series accession numbers GSE6137 (array 1) and GSE6139 (array 2).
RESULTS AND DISCUSSION
In vivo characteristics of TIGR4Δrr09.
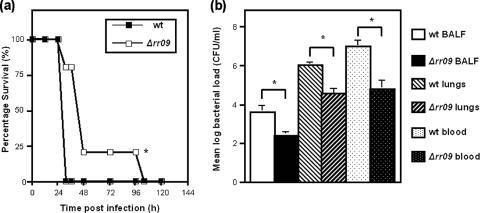
Previous studies indicated that RR09 has a strain-specific role in pneumococcal virulence, as D39 with deletion of the response regulator rr09 was found to be avirulent in all murine models tested, while 0100993Δrr09 was found to be attenuated only upon intranasal infection (3). Here, we extended these observations by analyzing a TIGR4Δrr09 mutant in a murine pneumonia model of infection. Mice infected intranasally with 106 CFU of TIGR4Δrr09 were found to have significantly longer survival times than mice infected with the parental strain (Fig. 1a). None of the mice infected with the wild-type strain survived longer than 30 h after inoculation. The level of survival of mice infected with Δrr09 dropped to 20% after 48 h, and none of these mice survived longer than 100 h after inoculation. Furthermore, the TIGR4Δrr09 bacterial counts in BALF, lungs, and blood showed that there were significant reductions compared to the wild-type strain (P < 0.01) (Fig. 1b). This indicated that like 0100993Δrr09, the TIGR4Δrr09 mutant was attenuated upon intranasal infection but was not avirulent like D39Δrr09.
FIG. 1.
(a) Survival of mice after intranasal infection with 106 CFU TIGR4. The asterisk indicates that the P value is <0.01 for a comparison of the survival times of TIGR4Δrr09 and wild-type strain TIGR4. (b) Bacterial loads in BALF, homogenized lungs, and blood 24 h after intranasal infection with 106 CFU. An asterisk indicates that the P value is <0.01 for a comparison of the bacterial loads recovered from different sites. The bars indicate geometric means, and the error bars indicate standard errors of the means. wt, wild type.
Microarray analysis of pneumococcal mutants.
To identify genes controlled by RR09, we compared the transcriptional profiles of S. pneumoniae wild-type strains D39 and TIGR4 and their isogenic mutant Δrr09 derivatives at various stages of growth. During this analysis, we used two different amplicon-based microarrays, both designed to provide nonredundant representation of the sequenced R6 and TIGR4 genomes. Importantly, no differences in in vitro growth between the wild-type strains and the Δrr09 mutants were detected, indicating that the observed differential gene expression was not a result of altered growth rates.
Comparison of D39 and TIGR4.
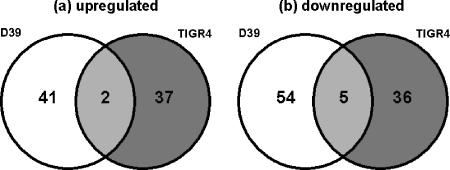
Little overlap was observed in the sets of genes controlled by RR09 in D39 and TIGR4, with a few notable exceptions (Fig. 2a and b and Table 1). The levels of transcription of two genes, encoding a putative lactose phosphotransferase system (PTS) repressor (sp0875) and a 1-phosphofructokinase (sp0876), were increased in both D39 and TIGR4 Δrr09 mutants in BHI broth at an OD600 of 0.6 (Fig. 2a and Table 1). Five genes were downregulated in both Δrr09 derivatives (Fig. 2b and Table 1). Five genes appeared to be oppositely regulated by RR09 in D39 and TIGR4 when they were grown in THY broth; these genes included glnR and pyrR, which were upregulated in D39Δrr09 and downregulated in TIGR4 when the organisms were grown in THY broth (Table 1).
FIG. 2.
Distribution of genes regulated by RR09 in D39 and TIGR. (a) Upregulation in the Δrr09 mutants; (b) downregulation in the Δrr09 mutants.
TABLE 1.
Genes regulated by RR09 in both D39 and TIGR4a
| Gene | Annotation | Ratiosb
|
|||||
|---|---|---|---|---|---|---|---|
| D39
|
TIGR4
|
||||||
| OD600 = 0.1 | OD600 = 0.2 | OD600 = 0.6 | OD600 = 0.1 | OD600 = 0.2 | OD600 = 0.6 | ||
| Commonly regulated | |||||||
| sp0875 | Lactose phosphotransferase system repressor, lacR | −0.06 | −0.51 | 3.93 | −0.44 | −0.09 | 1.46 |
| sp0876 | 1-Phosphofructokinase, putative | −0.12 | −0.33 | 3.74 | −0.40 | −0.08 | 1.37 |
| sp0647 | PTS, galactitol-specific IIC, putative | −2.40 | −2.48 | NAc | 0.11 | −0.17 | −1.11 |
| sp0648 | Beta-galactosidase, bgaA | −3.26 | −3.47 | NA | −0.02 | 0.82 | −1.80 |
| sp1804 | General stress protein 24, putative | −1.05 | −0.66 | NA | −1.39 | −0.55 | −1.04 |
| sp1883 | Dextran glucosidase dexS, putative | −1.55 | −2.47 | NA | −0.26 | 0.28 | −1.45 |
| sp1884 | Trehalose PTS, IIABC components | −1.19 | −1.50 | NA | 0.03 | 0.11 | −1.87 |
| Oppositely regulated | |||||||
| sp0090 | ABC transporter, permease protein | NA | 0.36 | −2.01 | 2.38 | 0.21 | NA |
| sp0501 | Transcriptional regulator, glnR | NA | NA | 1.51 | −0.10 | −1.06 | NA |
| sp0964 | Dihydroorotate dehydrogenase B, pyrD | 0.82 | 1.36 | NA | −1.00 | −1.05 | NA |
| sp1278 | Pyrimidine operon regulatory protein, pyrR | 1.45 | 1.50 | NA | −2.93 | −1.22 | NA |
| sp2141 | Glycosyl hydrolase-related protein | −0.05 | 0.01 | −3.62 | 1.96 | 0.17 | NA |
Commonly regulated genes showed the same expression pattern in D39 and TIGR4, while oppositely regulated genes were found to be upregulated in the rr09 mutant in one strain and downregulated in the other.
The values are log2(expression in Δrr09 mutant/expression in wild type) values. The values in bold type are the values for genes that are considered to be differentially expressed.
NA, no valid data were acquired.
Gene regulation by RR09: D39.
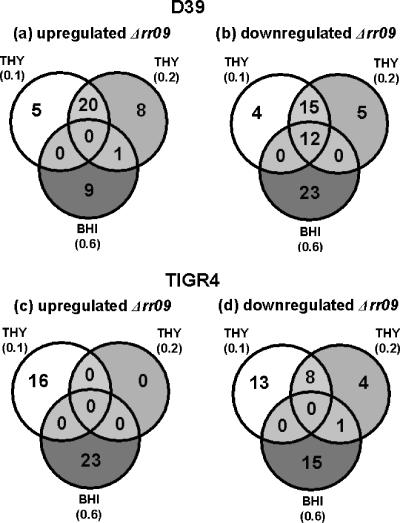
A total of 102 genes were found to be differentially expressed in wild-type strain D39 and the Δrr09 mutant. Forty-three of these genes could be considered to be directly or indirectly repressed by RR09 as they were upregulated in the D39 Δrr09 derivative compared to the expression in the wild type (Fig. 3a; see Tables S1 to S3 in the supplemental material).
FIG. 3.
Distribution of genes regulated by RR09 at OD600 of 0.1 and 0.2 in THY broth and at an OD600 of 0.6 in BHI broth. (a) Genes upregulated in D39Δrr09; (b) genes downregulated in D39Δrr09; (c) genes upregulated in TIGR4Δrr09; (d) genes downregulated in TIGR4Δrr09.
The transcription of genes involved in genetic competence, such as comAB, comDE, cinA, comX, and cglABCD, increased significantly in D39Δrr09 in the early and mid-log phases of growth in THY broth. Upregulation of comDE induces competence, which in turn induces transcription of comX. Peterson et al. described genes induced by competence-stimulating peptide and described three groups of responding genes: early, late, and delayed genes (24). In this study, upregulation of early genes (regulated by ComE) and late genes (cglABCD and cinA) may have been the result of premature development of competence caused by the lack of RR09.
A similar pattern of increased expression in the rr09 mutant was observed for genes encoding products predicted to be involved in pyrimidine and purine synthesis (e.g., sp1277 and sp1278), riboflavin kinase (sp1110), and a polypeptide deformylase (sp1549). Interestingly, the polypeptide deformylase has been suggested to be a broad-range antimicrobial target (5). In BHI broth at an OD600 of 0.6, the notable genes showing increased expression in the Δrr09 mutant included glnAR, encoding glutamate synthetase and its repressor, and a putative operon encoding a lactose phosphotransferase system repressor, 1-phosphofructokinase, and a fructose-specific PTS (sp0875 to sp0877). Finally, expression of a putative operon containing the pneumolysin gene, as well as several hypothetical ORFs (sp1923 to sp1926), was found to be upregulated in D39Δrr09 only at an OD600 of 0.6.
The levels of transcription of 59 genes were significantly decreased in D39Δrr09, meaning that these genes are directly or indirectly activated by RR09 (Fig. 3b; see Tables S1 to S3 in the supplemental material). These genes include a putative operon (sp2141 to sp2144) predicted to be involved in N-glycan degradation and the serine protease htrA gene (10, 11, 25), which are downregulated at an OD600 of 0.6 in BHI broth.
The set of genes downregulated in D39Δrr09 was found to be enriched for genes predicted to encode proteins involved in carbohydrate metabolism, particularly genes encoding enzyme II of nine sugar-specific phosphotransferase systems. These membrane-associated proteins facilitate the uptake of carbohydrates such as mannose and fructose. For instance, several genes encoding the putative mannose-specific PTS IIABC (sp0061 to sp0064), the cellobiose-specific PTS IIABC (sp0305 to sp0310), and the galactitol-specific PTS IIABC (sp0645 to sp0647) were in this group. However, genes encoding other components of these phosphotransferase systems, including enzyme I, HPr, and CcpA (global regulator of carbon metabolism) (for a review, see reference 30), did not appear to be regulated by RR09 in D39. Expression of these genes may be controlled by a different transcriptional regulator.
Gene regulation by RR09: TIGR4.
In TIGR4, there were significant differences in expression of 80 genes, and there was hardly any overlap between the growth phases sampled, underscoring the importance of sampling at multiple times. Thirty-nine genes were upregulated in TIGR4Δrr09 (Fig. 3c; see Tables S4 to S6 in the supplemental material), including genes encoding the iron compound ABC transporter (piuD [sp1871]) and five hypothetical proteins expressed early during growth (OD600 = 0.1). At the highest cell density, increased expression of a whole cluster of genes predicted to be involved in purine metabolism (sp0044 to sp0056) was observed in the mutant.
Several of the 41 genes found to be repressed in TIGRΔrr09 are predicted to encode products involved in metabolism of (amino) sugars and purines or pyrimidines (e.g., sp0266 and sp1278), particularly during early growth in THY broth (Fig. 3d; see Tables S4 to S6 in the supplemental material). Later during growth in BHI broth, the downregulated genes included the gene encoding pneumococcal surface protein A (pspA), clpL, coding for the ATP-dependent ClpL protease, and various stress response genes (hrcA, grpE, dnaK, and dnaJ), as well as genes in an ABC transporter operon (sp1895 to sp1897).
An interesting pattern of regulation was observed for genes located on the rlrA pathogenicity islet: the genes were strongly downregulated in THY broth but strongly upregulated in BHI broth. This 12-kb islet, which is not present in the D39 genome (2, 23, 28), encodes the transcriptional activator RlrA, three surface proteins (RrgA, RrgB, and RrgC), and three putative sortases (SrtB, SrtC, and SrtD). RlrA has been shown to positively regulate the expression of these seven genes on the pathogenicity islet, including itself (7). While repression of the pathogenicity islet by another regulator, MgrA, has been reported (8), no differential expression of mgrA was observed in this study, possibly due to a low level of expression and hence low signals. The pathogenicity islet has been shown to contribute to virulence (2, 6, 7), and thus, unbalanced regulation in TIGR4Δrr09 may have resulted in the attenuation that we observed in the murine model (Fig. 1). Although the regulatory mechanism of this pattern of expression remains unclear, the differences in regulation between the two broth media used suggest that there is indirect regulation, which might involve a number of downstream regulators controlled by RR09 in a growth-phase-dependent manner.
Real-time PCR analysis.
To validate our microarray data, relative transcript levels were determined by quantitative real-time PCR for a selection of genes for all growth phases sampled. Overall, the expression ratios obtained by microarray and real-time PCR analyses were found to be strongly positively correlated for both D39 (R2 = 0.86; n = 24) and TIGR4 (R2 = 0.90; n = 31). For example, expression of comA was confirmed to be strongly upregulated in D39Δrr09, and the comA expression levels in TIGR4Δrr09 and the wild-type strain were found to be similar by both methods (not shown).
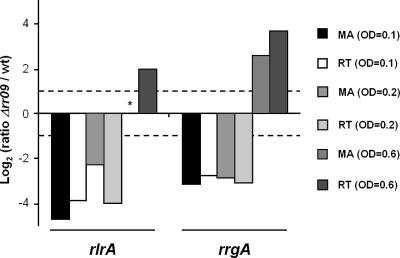
An interesting target of RR09 identified by our microarray analysis was the rlrA pathogenicity islet of TIGR4, which had an unusual, growth-phase-dependent pattern of expression. Real-time PCR analysis of two genes of this locus, rrgA and rlrA, confirmed this transcriptional profile: there was strong downregulation at the lower optical densities, and there was strong upregulation later (Fig. 4).
FIG. 4.
Expression of rlrA and rrgA in TIGR4. Log2(Δrr09/wild type) was determined by microarray analysis (MA) or real-time PCR (RT). The dashed lines indicate the twofold change microarray cutoff values for differential expression. The asterisk indicates that no microarray data were obtained for rlrA at an OD600 of 0.6. wt, wild type.
In addition, we validated the levels of expression of other genes (htrA, spoJ, pspA, sp1896, and sp2141 to sp2144) identified as differentially expressed by microarray analysis and found that there was agreement between the microarray and real-time PCR results (not shown).
Expression of pneumococcal genes during experimental virulence.
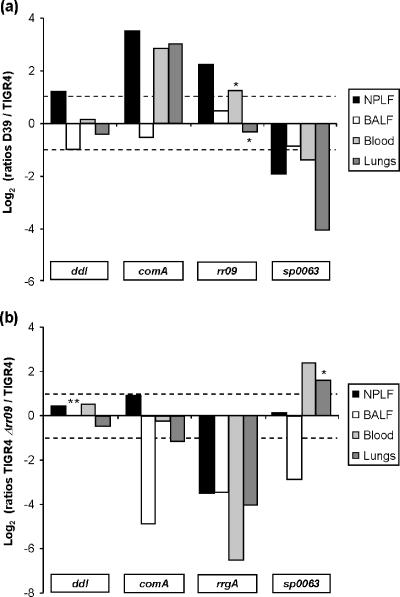
To determine to what extent the observed in vitro gene regulation by RR09 reflected the in vivo situation, we examined expression of rr09 and three putative targets during experimental infection of mice with D39, TIGR4, and TIGR4Δrr09. Due to its avirulent phenotype, analysis of gene expression in D39Δrr09 during infection was not possible. To correct for potential differences in the bacterial load or RNA yield, the levels of expression of sampled genes were normalized to the level of expression of gyrA. The levels of expression of the housekeeping gene ddl (sp1671) did not vary much in all of the strains in the compartments sampled (Fig. 5). The rr09 transcript could be detected in mice infected with both wild-type D39 and TIGR4, indicating that rr09 is indeed expressed in vivo. With the exception of the nasopharynx, the levels of rr09 expression were comparable for the two strains in all compartments sampled (Fig. 5a). Strikingly, the level of rr09 expression was higher in the lumen of the lungs (BALF) than in the other compartments sampled (not shown). The in vivo expression levels of putative RR09 targets appeared to be different in wild-type D39 and TIGR4. For example, the levels comA of expression were higher in D39 than in TIGR4 (Fig. 5a). Differential in vivo gene expression in wild-type strain TIGR4 and TIGR4Δrr09 was also observed. The level of expression of comA was lower in TIGR4Δrr09 than in wild-type strain TIGR4 in BALF, in contrast to the in vitro expression data (in THY broth), which indicated that there was no regulation of this gene by RR09 in TIGR4.
FIG. 5.
Expression of pneumococcal genes during experimental infection measured by real-time PCR. Log2 ratios of wild-type strain D39/wild-type strain TIGR4 (a) and TIGR4Δrr09/wild-type strain TIGR4 (b) are the averages of expression measured in three individual mice; the only exceptions were when two mice were used (indicated by one asterisk) and when no valid data were acquired (indicated by two asterisks). NPLF, nasopharyngeal lavage fluid.
The levels of expression of the rrgA gene, encoding a structural unit of the pilus encoded by the rlrA pathogenity islet (2, 16), were also determined during experimental virulence, and this gene found to be expressed in all compartments sampled in the TIGR4 wild-type strain. The levels of expression of rrgA in TIGR4Δrr09 were found to be 10- to 90-fold lower than the levels of expression in the TIGR4 wild-type strain (Fig. 5b). This finding correlated well with our in vitro microarray and real-time PCR data, supporting the hypothesis that RR09 has a role in the regulation of this pathogenicity islet.
Strain-dependent regulation of sp0063 does not explain the observed difference in virulence.
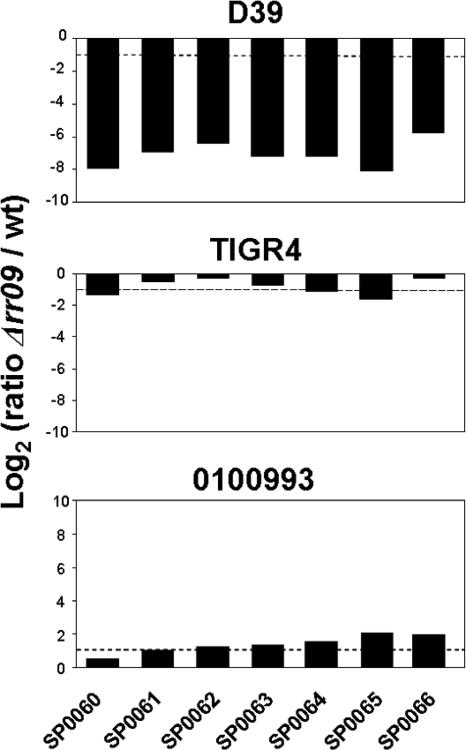
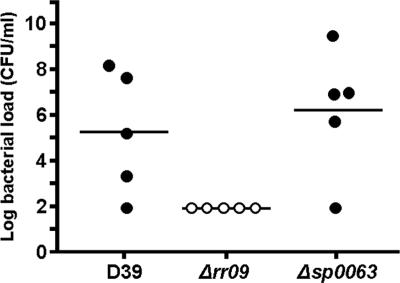
While our analysis indicated that RR09 has a prominent role in regulation of sugar metabolism in D39, only three putative PTSs were found to be regulated by RR09 in TIGR4. It seems unlikely that there is a difference in the sugar diets of D39 and TIGR4, as both genomes basically contain the same putative PTSs (20 PTSs common to both strains and one PTS specific for TIGR4) (9, 28). A clear example of a PTS locus differentially regulated by RR09 in D39 and TIGR4 was the locus comprising sp0060 to sp0066, and therefore we also examined expression of this locus by real-time PCR in a third strain, 0100993. This and previous studies showed that RR09 mutants of the three strains used here all have different phenotypes in murine models of infection (2). While strong downregulation of sp0060 to sp0066 was confirmed to occur in D39Δrr09, no RR09-dependent regulation was observed in either TIGR4 or 0100993 (Fig. 6). The observed downregulation of this locus in D39Δrr09 may result in reduced sugar uptake and thus a smaller supply of carbohydrates for the polysaccharide capsule, which in turn might explain the avirulent phenotype of D39Δrr09. To test this hypothesis, an sp0063 mutant was constructed and tested in a murine model, and it was compared with the D39 wild-type strain and the rr09 mutant. There was no significant difference in the survival rates between wild-type strain D39- and sp0063 mutant-infected mice; all five wild-type strain-infected mice died, as did four of the five Δsp0063-infected mice. Likewise, the blood counts at 24 h postinfection were similar (Fig. 7). In agreement with our previous work (3), the rr09 mutant was essentially avirulent; all infected mice survived, and the blood counts at 24 h were below the detection limit (∼83 CFU/ml). Indeed, the mice showed no overt clinical signs of infection. Thus, the gene product of sp0063 alone does not significantly contribute to virulence in D39 in this model and hence does not explain the dramatic phenotype seen after deletion of rr09 in D39. The Δrr09 derivatives of TIGR4 and 0100993 conferred a less severe in vivo phenotype. In vivo expression of sp0063 could be demonstrated for both D39 and TIGR, and the levels in TIGR4 were higher (almost fourfold) (Fig. 5a). In TIGR4, sp0063 appears to be regulated by RR09 in vivo (Fig. 5b) but not in vitro (Fig. 6), highlighting the importance of measuring gene expression in vivo. Moreover, this finding provides further evidence that the differential regulation of this operon observed in vitro does not explain the difference between D39 and TIGR4 in vivo. Also, there appears to be a redundancy of PTS in the pneumococcal genome, which could complement the lower level of expression of the PTS. This is underscored by our preliminary data, which indicate that the rr09 and sp0063 mutants are able to ferment mannose (not shown).
FIG. 6.
Expression of sp0060 to sp0066 in wild-type and Δrr09 mutant D39, TIGR4, and 0100993 as determined by real-time PCR. The dashed line indicates the twofold change microarray cutoff value for differential expression. wt, wild type.
FIG. 7.
Bacterial loads in blood of mice 24 h after intraperitoneal infection with 106 CFU. No differences between D39Δsp0063 and the wild-type strain were observed; in contrast, in the Δrr09 strain the bacterial loads were below the detection limit (log10 1.92) and were assigned a value of 1.90.
In conclusion, we identified several targets of RR09 in D39 and TIGR4 that could account for the phenotypes of their mutants, and we demonstrated that rr09 and three of its targets are expressed in vivo. These targets include both common and strain-specific targets of RR09. During in vitro growth, some genes were found to be controlled by RR09 at all phases of growth examined, while other genes appeared to be regulated in a growth-phase-dependent manner. Although this differential expression could have been the result of the different growth conditions and/or of the use of two different microarrays, this explanation appears less likely considering the considerable overlap between the sets of genes regulated at the different growth phases (e.g., sp0060 to sp0066, sp0645 to sp0648, and the rlrA pathogenicity island). The expression of pneumococcal genes in different broth media has been shown to differ substantially (21), but our results show that apparent putative targets can be identified with different experimental setups. The targets identified can be either directly or indirectly regulated by RR09. Further experiments, such as DNA binding assays, are necessary to distinguish between these two possibilities. DNase footprinting can be used to identify the RR09 DNA-binding sequence, and in order to obtain an initial view of the RR09 regulon in other strains, the consensus binding sequence could subsequently be used for in silico screening. Although the exact roles of most of the identified targets in pneumococcal virulence still have to be investigated, a few targets have already been studied extensively. Furthermore, predictions of the functions of many of the novel RR09 targets identified here are still based on sequence homology alone, and further studies are required to identify the exact roles in pneumococcal virulence. Pneumococci are likely to have different nutritional needs during the various stages of infection. Possibly, RR09 plays a role in this process by regulating genes required for nutrient uptake in response to various conditions characteristic of different sites in the host. The signal that triggers RR09 might be present at one site and not at another, and particular strains might respond differently to these signals, potentially explaining the phenotype differences observed in animal studies.
Supplementary Material
Acknowledgments
This work was supported by grant SSWO 356 from the Sophia Foundation for Medical Research (Rotterdam, The Netherlands), by the Wellcome Trust, and by the Foundation of Science and Technology (Portugal).
We thank the Pathogen Functional Genomics Resource Centre for providing microarrays (array 2 in this study) and Silvia Estevão for technical assistance.
Footnotes
Published ahead of print on 3 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Appleby, J. L., J. S. Parkinson, and R. B. Bourret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86:845-848. [DOI] [PubMed] [Google Scholar]
- 2.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 103:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blue, C. E., and T. J. Mitchell. 2003. Contribution of a response regulator to the virulence of Streptococcus pneumoniae is strain dependent. Infect. Immun. 71:4405-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia de la Nava, J., S. van Hijum, and O. Trelles. 2003. PreP: gene expression data pre-processing. Bioinformatics 19:2328-2329. [DOI] [PubMed] [Google Scholar]
- 5.Giglione, C., M. Pierre, and T. Meinnel. 2000. Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol. Microbiol. 36:1197-1205. [DOI] [PubMed] [Google Scholar]
- 6.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 7.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemsley, C., E. Joyce, D. L. Hava, A. Kawale, and A. Camilli. 2003. MgrA, an orthologue of Mga, acts as a transcriptional repressor of the genes within the rlrA pathogenicity islet in Streptococcus pneumoniae. J. Bacteriol. 185:6640-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J. Bacteriol. 186:5258-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72:3584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadioglu, A., N. A. Gingles, K. Grattan, A. Kerr, T. J. Mitchell, and P. W. Andrew. 2000. Host cellular immune response to pneumococcal lung infection in mice. Infect. Immun. 68:492-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr, A. R., J. J. Irvine, J. J. Search, N. A. Gingles, A. Kadioglu, P. W. Andrew, W. L. McPheat, C. G. Booth, and T. J. Mitchell. 2002. Role of inflammatory mediators in resistance and susceptibility to pneumococcal infection. Infect. Immun. 70:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281:25097-25109. [DOI] [PubMed] [Google Scholar]
- 15.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 16.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 74:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 18.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 19.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. B. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCluskey, J., J. Hinds, S. Husain, A. Witney, and T. J. Mitchell. 2004. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 51:1661-1675. [DOI] [PubMed] [Google Scholar]
- 21.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045-2053. [DOI] [PubMed] [Google Scholar]
- 22.Paterson, G. K., C. E. Blue, and T. J. Mitchell. 2006. Role of two-component systems in the virulence of Streptococcus pneumoniae. J. Med. Microbiol. 55:355-363. [DOI] [PubMed] [Google Scholar]
- 23.Paterson, G. K., and T. J. Mitchell. 2006. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 8:145-153. [DOI] [PubMed] [Google Scholar]
- 24.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 25.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Standish, A. J., U. H. Stroeher, and J. C. Paton. 2005. The two-component signal transduction system RR06/HK06 regulates expression of cbpA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 102:7701-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 29.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 30.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 31.van Hijum, S. A., A. de Jong, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Hijum, S. A., J. Garcia de la Nava, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241-244. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.