Abstract
Analysis of Listeria monocytogenes ptsH, hprK, and ccpA mutants defective in carbon catabolite repression (CCR) control revealed significant alterations in the expression of PrfA-dependent genes. The hprK mutant showed high up-regulation of PrfA-dependent virulence genes upon growth in glucose-containing medium whereas expression of these genes was even slightly down-regulated in the ccpA mutant compared to the wild-type strain. The ptsH mutant could only grow in a rich culture medium, and here the PrfA-dependent genes were up-regulated as in the hprK mutant. As expected, HPr-Ser-P was not produced in the hprK and ptsH mutants and synthesized at a similar level in the ccpA mutant as in the wild-type strain. However, no direct correlation was found between the level of HPr-Ser-P or HPr-His-P and PrfA activity when L. monocytogenes was grown in minimal medium with different phosphotransferase system (PTS) carbohydrates. Comparison of the transcript profiles of the hprK and ccpA mutants with that of the wild-type strain indicates that the up-regulation of the PrfA-dependent virulence genes in the hprK mutant correlates with the down-regulation of genes known to be controlled by the efficiency of PTS-mediated glucose transport. Furthermore, growth in the presence of the non-PTS substrate glycerol results in high PrfA activity. These data suggest that it is not the component(s) of the CCR or the common PTS pathway but, rather, the component(s) of subsequent steps that seem to be involved in the modulation of PrfA activity.
Listeria monocytogenes, a gram-positive, facultative intracellular human pathogen, escapes from the primary phagosome, replicates efficiently in the host cell's cytosol, and spreads from cell to cell. These processes, which are of major importance for pathogenesis of an L. monocytogenes infection, require several, well-characterized virulence factors (for recent reviews, see references 17, 32 and 63), like internalins (InlA, InlB, and InlC) for active internalization into nonprofessional phagocytic cells, listeriolysin O (LLO) for the release from the phagosomal compartment into the cytosol, or ActA for intra- and intercellular mobility. The genes that encode these and other virulence factors are either clustered or dispersed on the listerial chromosome (31, 63).
Most listerial virulence genes are regulated by the central transcription factor PrfA (8, 36, 41). This regulator belongs to the Crp/Fnr family of transcription activators; it recognizes as a dimer a conserved 14-bp sequence of dyad symmetry (“PrfA box”) located about −40 bp 5′ upstream of the transcription start site of PrfA-regulated promoters (22, 34, 40, 55). The synthesis of PrfA is autoregulated by a PrfA-dependent promoter in front of a bicistronic operon comprising the genes plcA and prfA (11, 21, 41). In addition, two promoters, P1 and P2, in front of prfA itself constitutively transcribe this gene at a low level (21, 22, 51, 64). This small amount of PrfA is essential for initiating transcription at the autoregulated plcA promoter. As recently shown (29), the nontranscribed 5′ region of the mRNA starting at P1 can form a secondary structure which acts as a thermoswitch for the translation of this prfA transcript, resulting in PrfA synthesis only above 30°C.
Furthermore, there is evidence for the interaction of PrfA with other listerial factors that modulate its activity (6, 7, 16, 19). Mutations within specific regions of PrfA have been obtained (45, 52, 56, 64, 65, 68) which lead to a permanently active state of PrfA; e.g., the addition of activated charcoal to brain heart infusion (BHI) medium or incubation in minimal essential medium (MEM) does not cause further activation, and cellobiose no longer inhibits the activity of these mutant PrfA proteins (termed PrfA*).
Readily metabolizable sugars which are taken up by phosphoenolpyruvate (PEP)-dependent phosphotransferase systems (PTS), like glucose, mannose, and especially cellobiose, strongly inhibit the activity of PrfA (23, 42, 46, 51), suggesting that component(s) involved in these pathways or the connected carbon catabolite repression (CCR) system interfere with PrfA, thereby modulating its activity. CCR control in gram-positive bacteria of low G+C content depends on several components. The catabolite control protein A (CcpA), the activity of which is modulated by different cofactors, regulates gene expression by different modes (5, 26, 44). A major cofactor of CcpA is HPr phosphorylated at Ser46. The HPr protein (encoded by ptsH) is a central component of all PTS pathways and possesses two phosphorylation sites. During active PTS-mediated sugar uptake, HPr is phosphorylated (by PEP) at His15. This HPr phosphorylation is catalyzed by enzyme I, encoded by ptsI which forms an operon structure with ptsH (25, 66). The phosphate group of HPr-His-P is then transferred to the sugar-specific transport component EIIA and further to EIIB. The membrane-associated EIIB activates the sugar translocation via EIIC. EIIA is also involved in other regulatory functions (9). The second phosphorylation site of HPr is Ser46. This phosphorylation is catalyzed by a specific ATP-dependent HPr kinase/phosphorylase (HPrK/P) (48, 49) which is stimulated by intermediates of the glycolytic pathway, in particular, by fructose 1,6-bisphosphate. Glucose starvation, increased concentration of inorganic phosphate, and low concentrations of glycolytic intermediates trigger phosphorylase activity of HPrK/P, leading to dephosphorylation of HPr-Ser-P (20). HPr-Ser-P binds to CcpA, and this complex interacts with the catabolite responsive element (Cre-box) located in or near promoter regions (1, 28) of many genes, especially those involved in catabolic pathways (44, 61). In most cases this interaction leads to repression of gene expression. Consequently, these CcpA/HPr-Ser-P-controlled genes are up-regulated in ccpA- and hprK-deficient mutants. These components, which were studied in great detail particularly in Bacillus subtilis (59), have also been identified in L. monocytogenes (2, 3, 10, 12, 24), and the same arrangement of ptsH and ptsI in an operon structure as in B. subtilis was reported (12, 13).
A direct influence of the central regulatory CCR protein CcpA on PrfA activity has been ruled out (3), but a recent report describing activation and inhibition of PrfA in appropriate B. subtilis mutants suggests that the second key player of CCR control, HPr-Ser-P (14), might be responsible for the modulation of the PrfA activity.
To analyze the interference of components of CCR and PTS with PrfA activity, we characterized ptsH, hprK, and ccpA mutations in L. monocytogenes and studied the effect of these mutations on gene expression with emphasis on PrfA-dependent virulence gene expression. Additionally, the phosphorylation status of HPr of L. monocytogenes wild-type after growth with different carbon sources was examined. The results suggest that HPr-Ser-P does not directly modulate PrfA activity but, rather, that other component(s) involved in uptake of carbohydrates by PTS permeases may interfere with PrfA.
MATERIALS AND METHODS
General techniques.
PCR amplifications, cloning procedures, isolation of chromosomal DNA, and DNA manipulations were carried out according to standard procedures (54). Cycle sequencing was performed using a CEQ Dye Terminator Cycle Sequencing Quick Start Kit (Beckman Coulter), and sequencing reactions were run on an XL2000 Beckman Coulter Sequencer. The Listeria home page of the Institut Pasteur (http://www.genolist.pasteur.fr/ListiList/) was used for sequence comparison. All oligonucleotides used in this study were synthesized by Sigma-Genosys.
Bacterial strains and growth conditions.
The Escherichia coli strain XL2-Blue was used for cloning and construction of the mutagenesis vectors. L. monocytogenes EGD-e strains and L. monocytogenes P14-A (52) were grown under aerobic conditions in brain heart infusion (BHI) medium (Difco), in Luria-Bertani medium (LB), or in chemically defined minimal medium (MM) (50) supplemented with different sugars for L. monocytogenes at 37°C or 42°C with antibiotics if required. Erythromycin was used at a concentration of 5 μg/ml for L. monocytogenes and at 300 μg/ml for E. coli. Fresh stock solutions of carbohydrates (glucose, cellobiose, mannose, and glycerol) were filter sterilized and added to the culture medium at a final concentration of 50 mM. The linear extrapolation method was used to calculate the growth rate, reported as generation time.
Disruption of the ccpA, hprK (lmo2483), and ptsH (lmo1002) gene in L. monocytogenes.
The insertion mutants were constructed by using L. monocytogenes Sv1/2a EGD-e as the parental strain. Insertion mutants were obtained by homologous recombination using constructs derived from the pLSV101 mutagenesis vector (30, 69). Plasmid pLSV101 carries an erythromycin resistance gene, a gram-negative ori, and a gram-positive temperature-sensitive origin of replication.
Internal (N-terminal) fragments of 307 bp (ccpA), 306 bp (hprK), and 146 bp (ptsH) were PCR amplified from chromosomal DNA derived from L. monocytogenes EGD-e by using the following primer pairs: ccpA-BamHI (TGTTGCACGGGATCCGAACG), ccpA-EcoRI (AAGTACTTGGAATTCTTTATCCTC), hprK-BamHI (ATGACAAAATCGGATCCGGTAAAG), hprK-EcoRI (TGCTGCTACGAATTCTTTCGG), ptsH-BamHI(2) (ATGTAAATTATGGAT CCAGCAAG), and ptsH-SalI (GATTTAAGGTCGACTTTTTTACC) (sites for the restriction endonucleases are underlined; boldface indicates deviation from original sequence). The purified PCR products were digested with the corresponding restriction enzymes and cloned via the restriction sites into pLSV101 to yield the mutagenesis plasmids pSM1 (ccpA), pSM2 (hprK), and pSM3 (ptsH). These plasmids were transformed by electroporation into L. monocytogenes EGD-e, and insertion mutants were obtained by selection on erythromycin at 42°C. Integration of the vectors in the genes ccpA, hprK, and ptsH were confirmed by PCR and sequencing using the oligonucleotides listed in Table 1. Revertants were obtained by subcultivating the insertion mutants at 30°C without erythromycin. Precise excision of the single plasmid insertion was confirmed by PCR analysis and sequencing.
TABLE 1.
Oligonucleotides used to confirm integration of the vectors pSM1-3 in the genes ccpA, hprK, and ptsH
| Oligonucleotide | Sequence (5′→3′) | Target DNA |
|---|---|---|
| ccpA-1 | TACAATGGTAGGTAGAGCC | Upstream region of ccpA |
| ccpA-2 | TTGTTCTGAAATACGTTCGCC | ccpA |
| hprK-1 | CTAAATAATATTTTGTAGCGAACAG | Upstream region of hprK |
| hprK-2 | AAGTCTGCTTTCTAAGTAATTCG | hprK |
| ptsH-1 | TCAATAATGTCTCCAACATGTGC | Upstream region of ptsH |
| ptsH-2 | AGCAGATAAGCTTTCGCAATG | Downstream region of ptsH |
| pLSV101BamHI | AATAAGCTTGGCTGCAGGTC | Vector pLSV101 |
| pLSV101EcoRI | GTTTTCCCAGTCACGACGTT | Vector pLSV101 |
Preparation of supernatant and cellular and surface-associated proteins of L. monocytogenes strains.
Overnight cultures of L. monocytogenes were diluted 1:10 into fresh BHI medium or defined MM and grown to an optical density of 0.6 and 1.0 at 600 nm (OD600), respectively. Each culture was then centrifuged for 10 min at 6,000 rpm at 4°C.
The supernatant (containing LLO and ActA) was precipitated on ice with 10% trichloroacetic acid, pelleted by centrifugation at 6,000 rpm for 30 min at 4°C, and washed twice in acetone. After a washing step, the pellet was resuspended in urea buffer (7 M urea, 2 M thiourea, and dithiothreitol) and CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propansulfonate).
For extraction of cell wall proteins (containing LLO), the bacteria pellet was washed twice in 1× phosphate-buffered saline (PBS) and resuspended in 1.8 ml of 1× PBS with 1% sodium dodecyl sulfate (SDS). Subsequently, the sample was mixed gently for 20 min at room temperature and then centrifuged at 6,000 rpm for 5 min at room temperature. The supernatant was precipitated with trichloroacetic acid as described previously (see above).
For preparation of cellular proteins (containing PrfA and ActA), the pellet was washed twice in 1× PBS, resuspended in cold lysis buffer (1× PBS with additional protease inhibitor [Roche]), and transferred into a 2-ml BLUE TUBE (Q-Biogene) filled with silica sand. The tube was shaken six times for 30 s each at speed 6.5 in a bead beater (FP120 Fast Prep cell disrupter; Savant Instruments, Inc.). The cell debris was removed by centrifugation at 14,000 rpm for 30 min at 4°C. Total protein concentrations were determined using a Bio-Rad-Protein Microassay (Bio-Rad).
SDS-PAGE and immunoblotting.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed according to standard protocols (33). After SDS-PAGE cytoplasmatic cell surface proteins and proteins from the culture supernatant were subjected to Western blotting onto nitrocellulose membranes, and L. monocytogenes proteins were immunodetected using the following Listeria-specific rabbit polyclonal antibodies and dilutions: anti-LLO (1:1,000; J. Kreft), anti-ActA (1:1,000 [47]), anti-PrfA (1:750; see below) and anti-HPr (1:3,000; see below).
Cloning and expression of HPr and PrfA and generation of antisera against both proteins.
A PCR fragment encoding the open reading frame (ORF) of ptsH was amplified with Pfu DNA polymerase (Promega) using L. monocytogenes EGD-e wild-type (WT) as a template and the primers ptsH-NdeI (5′-GGAGAATGTAACATATGGAACAAG CAAG-3′, introducing an NdeI site [underlined]) and ptsH-BamHI (5′-TGCTGCGGATCCTTTCAACTCTTT-3′, introducing a BamHI site [underlined]) and subsequently cloned into the pET3c expression vector (Novagen), yielding the plasmid pET3c-HPr. The nucleotide sequence of the amplified sequence was verified by automated sequencing. The pET3c-HPr vector was transformed into BL21(DE3) delta(ptsHIcrr)/pLysS (W. Hillen, University of Erlangen-Nürnberg), and protein purification was carried out following the manufacturer's instructions (pET expression system; Novagen) with the following modifications: HPr was purified according to its molecular weight using a Superdex75 HiLoad 16/60 gel filtration column applied to an ÄKTAprime protein purification system (Amersham Biosciences). Gel filtration was achieved in 20 mM Tris-HCl (pH 7.5) plus 50 mM NaCl at a flow rate of 1 ml/min. Following elution, protein fractions were analyzed by Coomassie staining of SDS-polyacrylamide gels according to the method of Laemmli (33). HPr-containing fractions were pooled, and native HPr protein was finally stored in 20 mM Tris (pH 7.5) and 50 mM NaCl at −80°C.
The N-terminal His-tagged PrfA protein was isolated from a recombinant E. coli strain (7). The purification of recombinant PrfA was performed with an ÄKTAprime protein purification system (Amersham Biosciences) and HiTrap Chelating HP columns (Amersham Biosciences) as recommended by the manufacturer. Following elution, protein fractions were analyzed by Coomassie staining of SDS-polyacrylamide gels according to the method of Laemmli (33). PrfA-containing fractions were pooled, and recombinant PrfA protein was finally stored in 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 2.5 mM CaCl2, 1 mM dithiothreitol, and 20% (vol/vol) glycerol at −80°C.
Anti-HPr and anti-PrfA sera were raised in rabbits by using purified HPr and His-tagged PrfA, respectively, as antigens. Aliquots of 150 to 200 μg of the particular protein per injection were mixed with Freud's adjuvant (Difco Laboratories), and after a total of four injections at 2-week intervals, the antisera were obtained. The immunization was performed by Charles River, Kisslegg, Germany.
Determination of HPr and HPr-Ser46/His15-P in the L. monocytogenes strains.
The strains of L. monocytogenes were grown in defined MM supplemented with 50 mM glucose, 50 mM cellobiose, 50 mM mannose, or 50 mM glycerol to OD600 of 0.6 and 1.0. Each culture was then centrifuged for 10 min at 6,000 rpm at 4°C.
For the preparation of cellular proteins, the pellet was washed twice with 50 mM Tris-HCl, pH 7.5, and 50 mM EDTA, pH 8.0, buffer and resuspended in the same buffer with additional protease inhibitor (Roche). The cells were disrupted by a FP120 Fast Prep bead beater (see the paragraph on preparation of cellular proteins above), and subsequently total protein concentrations were determined using a Bio-Rad-Protein Microassay. Cell extracts untreated or incubated at 70°C for 10 min to hydrolyze the heat-labile HPr-His15-P were separated on a 15% nondenaturing polyacrylamide gel and immunoblotted using specific rabbit polyclonal antibodies against HPr.
Determination of hemolytic activity.
L. monocytogenes strains were grown in different media at 37°C to an OD600 of 0.6 or 1.0. The hemolytic activity in the supernatants was determined as described previously (53). Briefly, 50 μl of culture supernatant was incubated in 1 ml of 4% sheep erythrocyte suspension for 30 min at 37°C. After incubation the tubes were centrifuged at 2,500 rpm for 5 min at room temperature. Hemolytic activity was estimated at 543 nm using an Ultrospec 2100 Pro photometer (Amersham).
RNA isolation.
RNA from the L. monocytogenes strains grown to a later growth phase (corresponding to an OD600 of 1.0) was extracted using an RNeasy Mini Kit (QIAGEN) according to the manufacturer's protocol with some modifications to lyse the bacteria. Cell pellets were suspended in lysis buffer and placed in a 2-ml BLUE TUBE filled with silica sand (Q-Biogene). The tube was shaken three times for 45 s each, with a 1-min interval on ice in between each shaking, at a speed setting of 6.5 in a FP120 FastPrep bead beater (Savant Instruments, Inc.). Residual DNA was removed on a column with QIAGEN RNase-free DNase.
Microarray hybridization and data analysis.
Transcriptome analyses were performed using whole-genome DNA microarrays that contained synthetic 70-mer oligodeoxyribonucleotides covering all ORFs of the L. monocytogenes genome. The oligonucleotides (sequences available at http://www.operon.com/arrays/oligosets_listeria.php) were spotted on epoxy-coated glass slides from Quantifoil according to the manufacturer's instructions by T. Chakraborty (Institut für Medizinische Mikrobiologie, Giessen, Germany). Each oligonucleotide was spotted twice on a slide to generate two replicates for each oligonucleotide. A total of two RNA samples was prepared for cDNA labeling and hybridization for each combination. Briefly, equal amounts (40 μg) of the RNAs were used to synthesize cDNA differentially labeled with Cy3-dCTP and Cy5-dCTP (Amersham Pharmacia) during a first-strand reverse transcription reaction with Superscript II RNase H− reverse transcriptase and 9 μg of random primers (Life Technologies) with dye swap. The two cDNA samples were combined, diluted in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) SDS, hybridized to the microarray, and incubated at 65°C for 16 h. After a washing step, the slides were scanned using ScanArray HT and analyzed using ScanArray express software (Perkin Elmer). Spots were flagged and eliminated from analysis when the signal-to-noise ratio was less than 3 or in obvious instances of high background or stray fluorescent signals. The LOWESS method of normalization (70) was performed for the background corrected median intensity of the spots. We had a total of four slides per experiment from the two biological replicates. The eight normalized ratios per spot were further analyzed with Microsoft Excel and Significance Analysis of Microarrays software for statistical significance (62). To determine the significance of differential expression, RNA was isolated from L. monocytogenes wild-type strain grown in BHI medium, and 30 to 40 μg of this RNA was labeled either with Cy3-dCTP or with Cy5-dCTP. The two cDNA probes generated were hybridized onto the same slide, and the data were analyzed as described above. Since the values ranged from 0.6 to 1.6, we considered values of >1.8 and <0.55 to be significant in our experiments (data not shown).
Real-time (RT-PCR).
Real-time reverse transcription-PCR (RT-PCR) was conducted on total RNA samples independently isolated as for transcriptome analysis experiments. Before RT-PCR was performed, the absence of DNA from RNA samples was verified by PCR amplification of the genes to be assayed with 1 μg of RNA as template. cDNA synthesis was performed as described above from 5 μg of total RNA. Instead of the labeled nucleotides, equal amounts of each (20 mM) dATP, dCTP, dGTP, and dTTP were used. RT-PCR was done in a final volume of 25 μl. Protocol and cycling conditions were carried out according to the manufacturer's protocol of the qPCRCore Kit for SYBR Green-I (Eurogentec). The relative expression levels of the genes studied were normalized to the housekeeping gene rpoB (43, 60). All primers used for real-time RT-PCR are listed in Table 2.
TABLE 2.
Oligonucleotides used in real-time RT-PCR
| Oligonucleotide | Sequence (5′→3′) | Target DNA |
|---|---|---|
| actA-F | AGCAGATGAGTCTTCACCACA | actA |
| actA-R | CCCTGCACTTTTATCAACAATC | actA |
| hly-F | ATGCAATTTCGAGCCTAACCT | hly |
| hly-R | TTATTGTCTTGATTAGTCATAC | hly |
| hpt-F | AAGCGCTAGGATGGAGCACAA | uhpt |
| hpt-R | CAACTGCAATAATCGAGCAAAG | uhpt |
| inlA-F | ATATTAGTATTTGGCAGCGG | inlA |
| inlA-R | TTTTTCCTAAGACCGTCTTC | inlA |
| inlB-F | TTTCTATCAGCCAGTCACTATTGGA | inlB |
| inlB-R | CGCGTCCCTGCTTCTACTTTTGT | inlB |
| inlC-F | CAAATACAGGTGGACTAACTAGA | inlC |
| inlC-R | GATATCCATCTTCCATCTGGGT | inlC |
| lgt-F | CGAGAAGACTGCCATTGCCTATAA | lgt |
| lgt-R | CCCAACACGATACTCCGAAAGTAA | lgt |
| lmo1001-F | GAATTTAAGGATGGCCTTACAGGAA | lmo1001 |
| lmo1001-R | GGTAATTCTTGTGGTTAACCACTGT | lmo1001 |
| lmo1003-F | GGGGAAGCAGTAGGACTTTATCGTA | ptsI |
| lmo1003-R | CACAGATTTTCCGTCCATTCCGGAT | ptsI |
| lmo2480-F | GCGAGAAGACTGCCATTGCCTATAA | lmo2480 |
| lmo2480-R | CCCCAACACGATACTCCGAAGTAA | lmo2480 |
| lmo2481-F | GTATGATACGATTATGCGAGGTCTT | lmo2481 |
| lmo2481-R | AGAAAGAGCCATCTCAATACCTTCA | lmo2481 |
| lmo2484-F | CGACAGCGCTACTAGCGAGTTTTA | lmo2484 |
| lmo2484-R | GCATAATCGCATTGACAACGAAGGT | lmo2484 |
| mpl-F | TTGCTCCAGAGGCCACTACATGT | mpl |
| mpl-R | GATACCACTTTCCCAAACGAAGTG | mpl |
| plcA-F | AATGCATCACTTTCAGGTGTATTAGA | plcA |
| plcA-R | GTTGATTAGTGGTTGGATCCGATAA | plcA |
| plcB-F | TCAAGGAATATATGATGCGGATCAT | plcB |
| plcB-R | CTTTGCTCCTGTTATTTTCGCATTA | plcB |
| prfA-F | CAGGCTACCGCATACGTTATCAAA | prfA |
| prfA-R | AGCCAAGCTTCCCGTTAATCGAAA | prfA |
| rpoB-F | AAGTAACTGGCGGAATCGATA | rpoB |
| rpoB-R | GGAATCCATAGATGGACCGTT | rpoB |
| tyrS-F | TGCCGTTTGCAAATTGGTGGTAGT | tyrS |
| tyrS-R | GATTTCCCAAATTTCGTTCCATCAG | tyrS |
Glucose transport assay.
Different L. monocytogenes strains were grown in LB medium with 50 mM glucose to an OD600 of 1.0. Each culture was then harvested by centrifugation at 5,000 rpm for 3 min at 4°C. The pellet was washed three times in transport buffer (50 mM Tris-HCl [pH 7.2] and 20 mM MgCl2) and resuspended in the same buffer. Labeled d-[U-14C]glucose (2 μCi/ml; Amersham Pharmacia) was mixed with unlabeled sugar d-[U-12C]glucose and added to the cells (final concentration, 2 mM) and incubated at 37°C. Aliquots (50 μl) were taken out at different time points (0 s, 15 s, 30 s, 60 s, 90 s, and 120 s) and filtered rapidly under vacuum through a 0.45-μm-pore-size cellulose nitrate filter (Sartorius). The filters were washed three times with 3 ml of cold saline (0.9% NaCl) and dried for 20 min at 42°C. Radioactivity was determined using a liquid scintillation counter (1214 Rackbeta; PerkinElmer). Additionally, the number of CFU of each sample was determined, and the glucose uptake was calculated for each strain.
Microarray data accession number.
The data obtained in this study have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under GEO Series accession number GSE6028.
RESULTS
Construction and characterization of ccpA, hprK (lmo2483), and ptsH mutants of L. monocytogenes EGD-e.
Mutations were introduced in the genes ccpA, hprK (lmo2483), and ptsH by insertion of the plasmids pSM1 to pSM3 (pSM1-3) (Fig. 1), each carrying a specific fragment from the 5′ part of the gene. After electroporation of the plasmids into L. monocytogenes EGD-e at 30°C, insertion mutants were selected on erythromycin-containing BHI agar plates by incubation at 42°C, as described earlier (69). The insertions were confirmed by PCR and sequence analysis (results not shown). No Emr colonies were obtained with the EGD-e strain carrying the vector plasmid alone after shift to 42°C. Southern blot analyses confirmed that only a single (specific) insertion had occurred in all three mutants. The absence of CcpA and HPr in the corresponding insertion mutants was shown by immunoblotting with polyclonal antibodies against these proteins. The loss of functional HPrK/P was determined by the absence of HPr phosphorylation at Ser46 by Western blot analysis (data not shown).
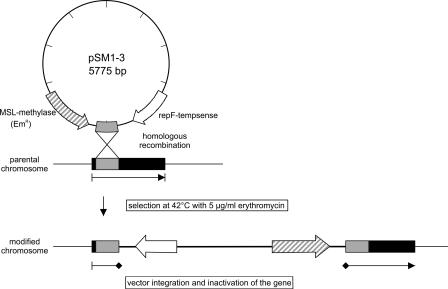
FIG. 1.
Schematic presentation of vector integration and inactivation of the genes ccpA, hprK, and ptsH. A fragment of L. monocytogenes EGD-e chromosomal DNA was cloned into the multiple cloning site of pLSV101 resulting in the plasmids pSM1 (ccpA), pSM2 (hprK), and pSM3 (ptsH). Replication of pSM1-3 in Listeria is stopped by increasing the temperature to 42°C. This selects for events in which pSM1-3 has integrated into the host chromosome by homologous recombination at the point of homology provided by the cloned DNA.
All attempts to obtain in-frame deletion mutants of these genes failed although we applied all well-established methods.
The three insertion mutants were stable upon growth in all growth media tested at 37°C while a >95% loss of Emr, indicative of plasmid excision, was observed after growth at 30°C for 10 generations. The erythromycin-sensitive colonies seem to represent mostly precise revertants, as judged by the growth kinetics in culture medium and the wild-type sequences of the corresponding genes in the colonies tested (see below).
To rule out the effect of erythromycin on the growth rate or protein expression of the insertion mutants, we compared the erythromycin-sensitive L. monocytogenes parental strain (WT) with an erythromycin-resistant strain (WT Emr) bearing pLSV101 inserted in the int gene (putative integrase of bacteriophage A118). Insertion in the int gene has been shown previously to have no influence on growth and gene expression of Listeria (A. Frentzen, unpublished data). Expression levels of PrfA and PrfA-regulated genes in these two strains were similar after growth with (WT Emr) or without (WT and WT Emr) erythromycin (data not shown).
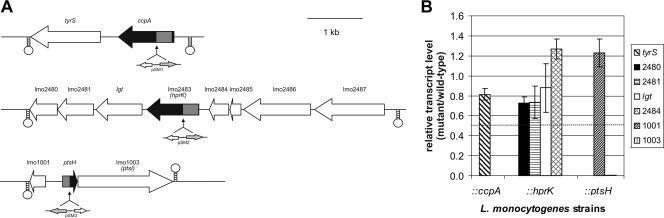
Since the three mutated genes are part of operons (Fig. 2A), we determined the transcript levels of the flanking genes of each of the three mutants by real-time RT-PCR and compared them to those of the corresponding wild-type genes. As shown in Fig. 2B, the insertions in ccpA and hprK did not significantly reduce the transcript levels of the genes located downstream (tyrS in the case of ccpA and lmo2480, lmo2481, and lgt in the case of hprK) or of the gene lmo2484 located upstream of hprK. The insertion in ptsH appears to completely abolish the transcription of ptsI (lmo1003), which is expected as ptsI is located downstream of ptsH in the ptsHI operon (12). However, this strong reduction of ptsI transcription in the ptsH mutant should not affect the phenotype caused by the ptsH mutation since the gene product of pstI is enzyme I, which needs HPr (the gene product of ptsH) as a substrate. No polar effect was detected on gene expression of lmo1001 located upstream of ptsH.
FIG. 2.
(A) Map of genetic loci disrupted during the present study with surrounding regions. All genes are drawn approximately to scale by using the L. monocytogenes EGD-e genome sequence data (http://genolist.pasteur.fr/ListiList/). The genes disrupted in L. monocytogenes EGD-e are indicated in black; adjacent ORFs are shown in white. The sites of pSM1-3 insertion into the corresponding genes are indicated by black arrows. Shaded areas within black arrows depict the regions of the genes cloned into pSM1-3. Stem-loop structures are used to illustrate putative terminator regions. Descriptions of the genes are as follows: tyrS, tyrosyl-tRNA synthetase; ccpA, catabolite control protein A; lmo2480, similar to acetyltransferase; lmo2481, similar to B. subtilis HPr-Ser-P phosphatase; lgt, highly similar to prolipoprotein diacylglyceryl transferase; lmo2483, HPr-Ser-P kinase/phosphatase; lmo2484, similar to B. subtilis YvlD protein; lmo2485, similar to B. subtilis YvlC protein; lmo2486, unknown; lmo2487, similar to B. subtilis YvlB protein; lmo1001, similar to B. subtilis protein YkvS; ptsH, PTS phosphocarrier protein HPr; lmo1003, phosphotransferase system enzyme I. (B) Transcriptional analysis with real-time RT-PCR to study the polar effect of pSM1-3 insertion on the transcription of genes located up- and downstream in the ccpA (tyrS), hprK (lmo2480, lmo2481, lgt, and lmo2484), and ptsH (lmo1001 and lmo1003) insertion mutants (indicated as ::ccpA, ::hprK, and ::ptsH, respectively) (see panel A). The strains were grown in BHI medium to an OD600 of 1.0, where the RNAs were prepared. The relative increases in the expression of the neighboring genes in the mutants compared to the wild-type strain (relative transcript level of mutant/wild-type) are depicted here. The relative expression levels of the genes studied was normalized to the housekeeping gene rpoB as described elsewhere (43, 60). The RT-PCR was performed with three independently isolated RNAs from the various strains in duplicate. The values represented here are means of the six obtained values, and the error bars indicate the standard deviations from the means. Relative expression levels of >1.8 or <0.55 (marked by dotted line) were considered to be differentially regulated based on microarray data (for details, see Material and Methods).
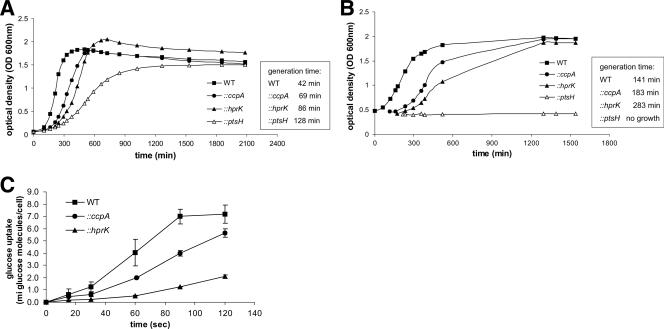
Growth rates of the three mutants were determined in BHI medium (this nutrient-rich medium contains, in addition to glucose, several other poorly defined carbon sources) and in a defined minimal medium (MM) with 50 mM glucose. All three mutants showed a delay in growth in BHI medium (Fig. 3A) which—as expected—was most pronounced for the ptsH mutant, which can no longer take up carbohydrates via the PTS pathway. In the glucose-containing minimal medium, the ccpA mutant also grew at a slower growth rate than the wild-type strain while the ptsH mutant did not grow at all in this medium (Fig. 3B). The hprK mutant exhibited an even slower growth rate than the ccpA mutant (Fig. 3B), suggesting that the uptake of glucose [or other essential nutrient(s)] might be impaired in this mutant. This assumption is strongly supported by the reduced rate of d-[U-14C]glucose uptake in the hprK mutant and (to a lesser extent) in the ccpA mutant compared to the wild-type strain (Fig. 3C).
FIG. 3.
Growth of L. monocytogenes EGD-e wild-type, ccpA, hprK, and ptsH insertion mutants. The generation times of WT and insertion mutants are shown to the right. (A) The strains were grown in BHI medium. (B) Strains were shifted from BHI medium to MM with 50 mM glucose. The cells were grown first in BHI medium to an OD600 of 0.5. After centrifugation the cells were washed twice in MM supplemented with 50 mM glucose and then resuspended in MM with 50 mM glucose where growth was determined. The growth curves are representative of four replicates. (C) Glucose uptake in L. monocytogenes EGD-e wild-type, the ccpA, and hprK insertion mutants. To measure the uptake of radioactively labeled d-[U-14C]glucose the strains were grown in LB medium supplemented with 50 mM glucose to an OD600 of 1.0. The y axis indicates the number (106) of molecules of glucose taken up per bacterial cell. The glucose uptake measurements were performed in triplicate, and the error bars indicate standard deviations of the means for the three measurements.
Revertants from each of the three mutants behaved like the wild-type strain with respect to growth and glucose uptake (data not shown).
Expression of PrfA-dependent virulence genes is enhanced in hprK and ptsH mutants but not in the ccpA mutant.
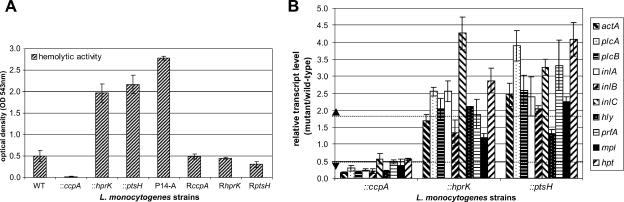
When the three mutants and the wild-type strain were grown on blood agar plates, we noticed much stronger hemolytic zones around the colonies of the hprK and ptsH mutants than around those of the wild-type strain and the ccpA mutant (data not shown), suggesting enhanced synthesis of LLO in the hprK and ptsH mutants. Measurement of the hemolytic activity in the supernatants of BHI-grown cultures of these strains (arising from secreted LLO), indicated, indeed, an enhanced activity of the PrfA-dependent virulence factor in the hprK and ptsH mutants compared to the wild-type strain and the ccpA mutant (Fig. 4A). Real-time RT-PCR performed with RNA isolated from the three mutants and the wild-type strain (harvested in the exponential growth phase in BHI medium) showed three- to fivefold increases in transcript levels of not only the hly gene but also the other PrfA-dependent virulence genes of L. monocytogenes (Fig. 4B) in the hprK and the ptsH mutants compared to the wild-type strain, while transcription of these genes appeared to be slightly reduced in the ccpA mutant.
FIG. 4.
(A) Hemolytic activity of WT EGD-e, insertion mutants (::ccpA, ::hprK, and ::ptsH), revertants (RccpA, RhprK, and RptsH), and the P14-A strain (expressing constitutively active PrfA* due to a G145S exchange [52]) grown in BHI medium to an OD600 of 1.0. The hemolytic activity was determined in three independently performed experiments; the error bars indicate standard deviations of the means for the three experiments. (B) Transcriptional analysis of the virulence genes in the insertion mutants ccpA, hprK, and ptsH. The strains were grown in BHI medium to an OD600 of 1.0, and RNAs were prepared. The relative changes in the expression of the virulence genes in the mutants compared to the wild-type strain (relative transcript level of mutant/wild-type) are depicted here. The relative expression of the genes studied was normalized to the housekeeping gene rpoB as described elsewhere (43, 60). RT-PCR was performed with three independently isolated RNAs from the various strains in duplicate. The values represented here are the means of the six obtained values, and the error bars indicate the standard deviations from the means. Relative expression levels of >1.8 or <0.55 (marked by dotted line) were considered to be differentially regulated based on microarray data (for details, see Material and Methods).
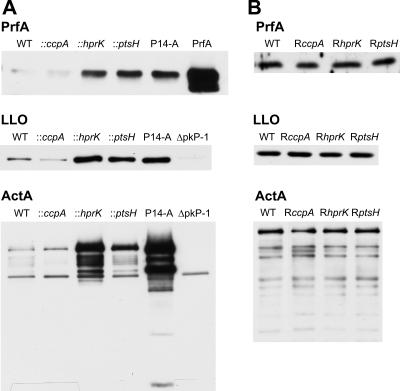
As shown in Fig. 5A, the amounts of PrfA, LLO, and ActA in the hprK and ptsH mutants measured by immunoblotting also reached higher levels than the wild-type strain and the ccpA mutant, and these levels—especially in the case of ActA—were even higher than expected from the increased transcript levels, which may be due to posttranscriptional control of ActA expression (67).
FIG. 5.
Western blot analysis of PrfA and PrfA-regulated virulence proteins LLO and ActA. WT EGD-e (A and B), insertion mutants (::ccpA, ::hprK, and ::ptsH) and strain P14-A (A), and revertants (RccpA, RhprK, and RptsH) (B) were grown in BHI medium. At an OD600 of 1.0 (later growth phase) proteins were prepared, equal amounts of proteins were separated by SDS-PAGE, and equivalent loading of the gels was controlled by Coomassie-staining (data not shown). Loading controls: PrfA, purified PrfA protein; ΔpkP-1, EGD with a deletion in the virulence gene cluster (18).
The increased amounts of the PrfA-dependent gene products were similar to results in L. monocytogenes P14-A, which expresses a constitutively active PrfA* protein (due to a G145S exchange in PrfA [52]). The parental P14 strain expresses the same amount of all studied PrfA-dependent proteins as the EGD-e wild-type strain (data not shown).
Revertants of all three mutants which had lost the insertions again showed similar hemolytic activity as the wild-type strain (Fig. 4A) and produced wild-type levels of PrfA, LLO, and ActA (Fig. 5B).
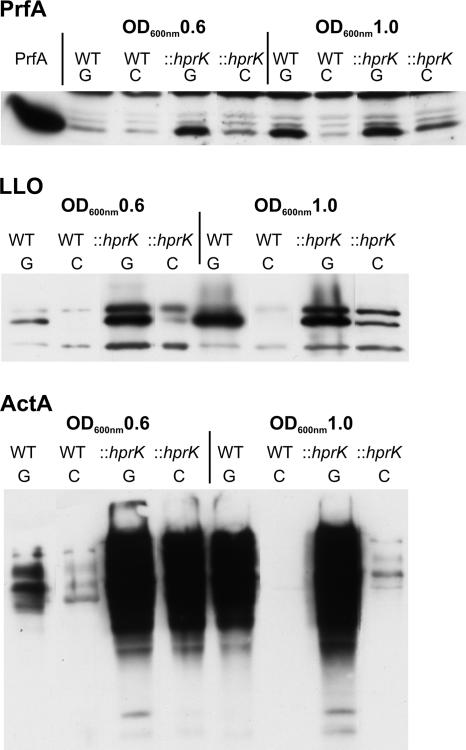
In the glucose-containing minimal medium, production of the tested PrfA-dependent virulence factors was rather low in the L. monocytogenes wild-type strain during the early exponential growth phase (OD600 of 0.6), when balanced growth is expected, but highly enhanced in the late growth phase (OD600 of 1.0). In the presence of cellobiose (25 or 50 mM), synthesis of these proteins (Fig. 6) was strongly inhibited after growth of the L. monocytogenes strains in this minimal medium.
FIG. 6.
Western blot analysis of PrfA and PrfA-regulated virulence proteins LLO and ActA. WT EGD-e and the hprK insertion mutant (::hprK) were grown in MM supplemented with 50 mM glucose (G) or 50 mM cellobiose (C). At OD600 values of 0.6 (early growth phase) and 1.0 (later growth phase) proteins were prepared, equal amounts of proteins were separated by SDS-PAGE, and equivalent loading of the gels was controlled by Coomassie-staining (data not shown). PrfA, purified PrfA protein (loading control).
In the glucose-containing minimal medium, the hprK mutant showed enhanced synthesis of PrfA and PrfA-dependent proteins (compared to the wild-type strain) in both growth phases. PrfA activity in this mutant was still inhibited by cellobiose (Fig. 6).
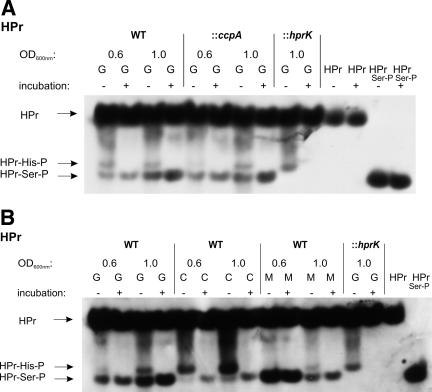
PrfA activity does not correlate with the level of HPr-Ser-P when L. monocytogenes is cultured in the presence of different PTS sugars.
As expected, the hprK mutant was unable to phosphorylate HPr at Ser46 (Fig. 7A). The ptsH mutant cannot produce HPr and, hence, also not HPr-Ser-P. In both mutants up-regulation of the PrfA-dependent virulence genes was observed (Fig. 4B). The level of HPr-Ser-P in the ccpA mutant was similar to that of the wild-type strain (Fig. 7A). These data seem to be in line with a recent report (14) which suggests that HPr-Ser-P may be directly or indirectly involved in the negative modulation of PrfA activity.
FIG. 7.
Western blot analysis of the HPr and its phosphorylated forms (HPr-His15-P or HPr-Ser46-P) to determine the amount of HPr-Ser46-P in L. monocytogenes strains. Equal amounts of cell extracts untreated (−) or incubated at 70°C for 10 min (+) to hydrolyze the heat-labile HPr-His15-P were separated on a 15% nondenaturing polyacrylamide gel and immunoblotted using specific rabbit polyclonal antibodies against HPr. The positions of HPr, HPr-Ser46-P, and HPr-His15-P are indicated. Equivalent loading of the gels was controlled by Coomassie-staining (data not shown). (A) Detection of HPr and its phosphorylated forms (HPr-His15-P or HPr-Ser46-P) in WT EGD-e and the ccpA mutant grown in MM supplemented with 50 mM glucose to OD600s of 0.6 and 1.0. The hprK mutant (control; only able to phosphorylate HPr at His15; grown in MM supplemented with 50 mM glucose to an OD600 of 1.0) and purified HPr and HPr-Ser46-P with and without incubation were separated on the nondenaturing polyacrylamide gel to show that the incubation at 70°C for 10 min has no effect on HPr or HPr-Ser46-P. (B) Determination of HPr and its phosphorylated forms (HPr-His15-P or HPr-Ser46-P) in WT EGD-e grown in MM supplemented with 50 mM glucose (G), 50 mM cellobiose (C), or 50 mM mannose (M) to OD600s of 0.6 and 1.0. The hprK mutant (control; see panel A) and purified HPr and HPr-Ser46-P were separated on the gel to indicate the different HPr forms. ::ccpA and ::hprK, insertion mutants of ccpA and hprK, respectively.
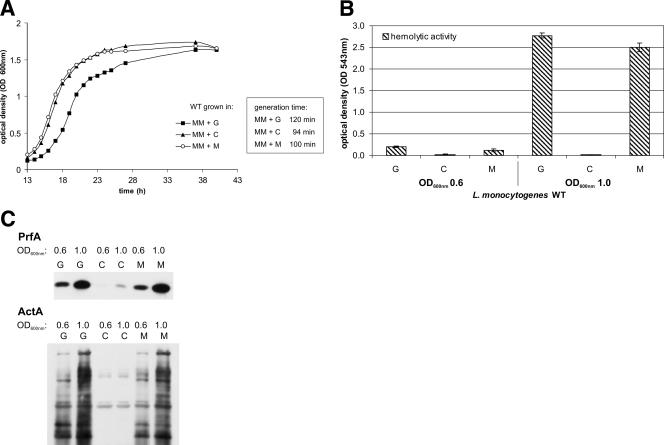
The presence of β-glucoside cellobiose, a PTS sugar which, similar to mannose, can serve as an efficient carbon source for L. monocytogenes, is known to strongly inhibit PrfA activity (46). We therefore compared the level of HPr-Ser-P in L. monocytogenes grown in minimal medium with glucose, cellobiose, or mannose (Fig. 7B). L. monocytogenes EGD-e grew with these three PTS sugars as carbon sources with generation times of 120 min (glucose), 94 min (cellobiose), and 100 min (mannose) in the minimal medium (Fig. 8A). As expected, hemolytic activity and expression of prfA and the PrfA-dependent actA gene were much lower in the presence of cellobiose than in the presence of glucose or mannose (Fig. 8B and C). The level of HPr-Ser-P in L. monocytogenes grown in minimal medium with cellobiose was, however, even lower than that in L. monocytogenes grown in the presence of glucose or mannose, both in the early (OD600 of 0.6) and later (OD600 of 1.0) growth phases (Fig. 7B). There was no significant difference whether 25 or 50 mM cellobiose was added to the medium.
FIG. 8.
(A) Growth of L. monocytogenes WT EGD-e in MM supplemented with 50 mM glucose (MM+G), cellobiose (MM+C), or mannose (MM+M), respectively. The generation times in the different media are shown to the right. The growth curves are representative of three replicates. (B) Hemolytic activity of WT EGD-e grown in MM supplemented with either 50 mM glucose (G), 50 mM cellobiose (C), or 50 mM mannose (M) to OD600s of 0.6 and 1.0. Hemolytic activity was determined in three independently performed experiments; error bars indicate standard deviations of the means for the three experiments. (C) Western blot analysis of PrfA and the PrfA-regulated virulence protein ActA in WT EGD-e grown in MM supplemented with either 50 mM glucose (G), 50 mM cellobiose (C), or 50 mM mannose (M) to OD600s of 0.6 and 1.0. Equal amounts of proteins were separated by SDS-PAGE, and equivalent loading of the gels was controlled by Coomassie-staining (data not shown).
These data thus do not show an inverse correlation between the level of HPr-Ser-P and PrfA activity in L. monocytogenes, as suggested by recent studies (14, 27). The data shown in Fig. 8B and 9B also do not demonstrate a correlation between the level of HPr-His-P and PrfA activity, suggesting that this phosphorylated HPr component is not a direct modulator of PrfA activity either.
FIG. 9.
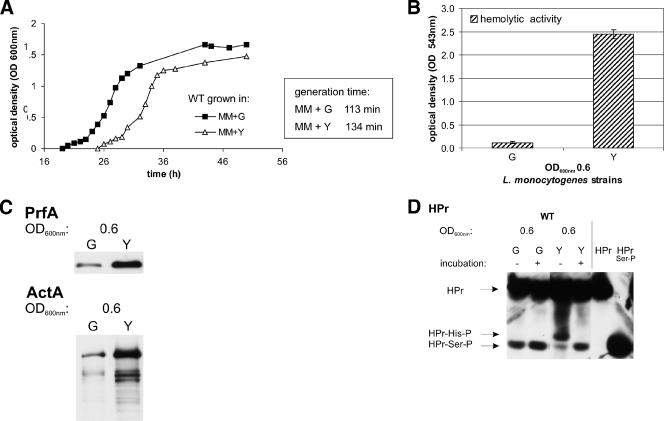
(A) Growth of L. monocytogenes WT EGD-e in MM supplemented with 50 mM glucose (MM+G) or glycerol (MM+Y). The generation times in the different media are shown at right. The growth curves are representative of three replicates. (B) Hemolytic activity of WT EGD-e grown in MM supplemented with either 50 mM glucose (G) or 50 mM glycerol (Y) to an OD600 of 0.6. Hemolytic activity was determined in three independently performed experiments; error bars indicate standard deviations of the means for the three experiments. (C) Western blot analysis of the PrfA and PrfA-regulated virulence protein ActA in WT EGD-e grown in MM supplemented with either 50 mM glucose (G) or 50 mM glycerol (Y) to an OD600 of 0.6. Equal amounts of proteins were separated by SDS-PAGE, and equivalent loading of the gels was controlled by Coomassie-staining (data not shown). (D) Detection of HPr and its phosphorylated forms (HPr-His15-P or HPr-Ser46-P) in WT EGD-e grown in MM supplemented with 50 mM glucose (G) or 50 mM glycerol (Y) to an OD600 of 0.6 (see legend of Fig. 7). As a control, purified HPr and HPr-Ser46-P were separated on the gel to indicate the different HPr forms.
Comparative transcript patterns of hprK versus WT and ccpA versus WT using whole L. monocytogenes genome microarrays.
As shown in Fig. 3B the growth rate of the hprK mutant in glucose-containing minimal medium is strongly impaired compared to the wild-type strain and the ccpA mutant (generation time of 283 min versus 141 min). This observation correlates with the reduced rate of d-[U-14C]glucose uptake in the hprK mutant (Fig. 3C). Similar to the ccpA mutant, the hprK mutant is expected to derepress genes and operons which are under CCR control. However, the PrfA-regulated genes are up-regulated in the hprK mutant but not in the ccpA mutant (Fig. 4B), suggesting that genes other than the typical genes controlled by CCR are differently regulated in the two mutants. The information on these genes was expected to provide some idea of how PrfA may be activated in the hprK mutant.
To obtain a more general view on the L. monocytogenes genes that are differently regulated in the hprK and the ccpA mutant, we compared the transcription profiles of these two mutants with that of the wild-type strain using oligonucleotide-based whole-genome microarrays. All strains were cultured in BHI medium. In this complex growth medium, most genes subject to CCR were expected to be repressed in the wild-type strain.
Three groups of differently regulated genes were obtained. In group I are genes that were up- or down-regulated in both the ccpA and the hprK mutants. These genes are presumably repressed or activated by CcpA/HPr-Ser-P in the presence of glucose.
In group II are genes that were up- or down-regulated in the ccpA mutant but not in the hprK mutant (with few exceptions). This group of genes is expected to be positively or negatively regulated by CcpA alone, i.e., without HPr-Ser-P as a cofactor.
In group III are genes that were up- or down-regulated in the hprK mutant but not in the ccpA mutant. We assume that this group of genes is either directly regulated by the level of HPr-Ser-P or by the ratio of HPr-His-P/HPr-Ser-P and hence by the efficiency of glucose uptake. Since the PrfA-regulated genes are also specifically up-regulated in the hprK mutant, we expected to observe a correlation between the regulation of the PrfA-dependent genes and other genes that are specifically up- or down-regulated in this mutant.
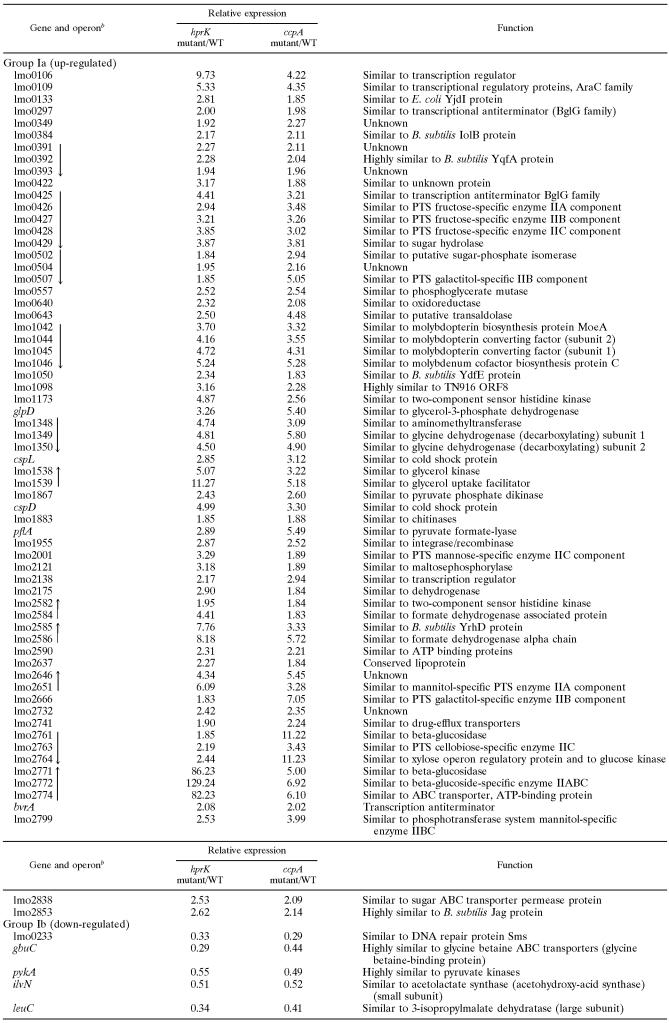
Group I (Table 3) contains a large number of up-regulated genes or operons (Ia) involved in sugar transport (mainly PTS) and metabolism, ABC transporters, coenzyme metabolism, and synthesis of regulation factors. Many genes of this group are similar or even identical to those previously identified in B. subtilis as being subject to repression by CcpA/HPr-Ser-P. There are very few down-regulated genes in this group (Ib), suggesting that only a small number of genes in L. monocytogenes are activated by CcpA/HPr-Ser-P.
TABLE 3.
Expression of group I genes in hprK and ccpA insertion mutants relative to L. monocytogenes wild typea
Genes up-regulated (group Ia) and down-regulated (group Ib) in hprK and ccpA insertion mutants relative to (their isogenic) WT Listeria monocytogenes EGD-e (grown in BHI medium) were identified by microarray analysis.
Black arrows indicate genes which are probably organized in an operon structure and the direction of transcription based on the L. monocytogenes EGD-e genome sequence data (http://genolist.pasteur.fr/ListiList/).
Among the group II genes being up-regulated in the absence of CcpA only (Table 4, IIa) are notably some genes (operons) which encode several PTS and transcription factors other than those of group Ia. These genes are apparently repressed by CcpA without HPr-Ser-P. A similar number of genes are down-regulated (greater than twofold) in the ccpA mutant (IIb) and hence activated in L. monocytogenes by CcpA in the presence of glucose; these genes encode mainly metabolic enzymes. Interestingly inlA and inlB which, like the other PrfA-dependent virulence genes, are up-regulated in the hprK mutant (see below) also belong to this set of genes, suggesting that expression of the inlAB operon which is under the control of several promoters, including a PrfA-dependent one (37, 57), is also affected directly or indirectly by CcpA.
TABLE 4.
Group II genes only up- or down-regulated in a ccpA insertion mutant relative to L. monocytogenes wild-typea
Genes identified by microarray analysis as up-regulated (group IIa) or down-regulated (group IIb) in the ccpA insertion mutant relative to (its isogenic) wild-type Listeria monocytogenes EGD-e (grown in BHI medium) are shown.
Black arrows indicate genes which are probably organized in an operon structure and the direction of transcription based on the L. monocytogenes EGD-e genome sequence data (http://genolist.pasteur.fr/ListiList/).
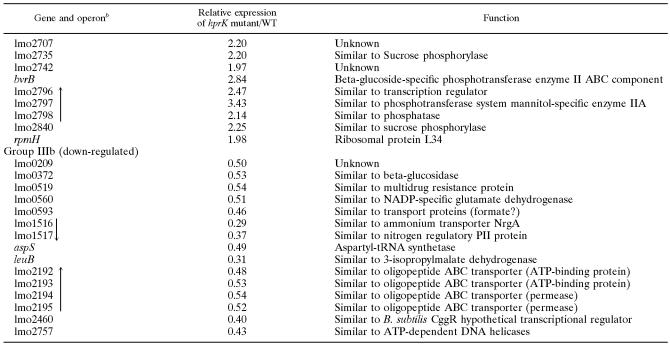
The number of up-regulated genes (up-regulation greater than twofold) in group III is rather large (Table 5; IIIa) and interestingly contains the PrfA-regulated virulence genes, which accords with the data described above. In addition, this group again comprises genes involved in sugar transport via PTS, carbohydrate metabolism, and transcription factors. Several phage-specific genes belonging to the A118 prophage are also found in group IIIa.
TABLE 5.
Group III genes only up- or down-regulated in an hprK insertion mutant relative to L. monocytogenes wild-typea
Genes identified by microarray analysis as up-regulated (group IIIa) or down-regulated (group IIIb) in an hprK insertion mutant relative to (its isogenic) wild-type Listeria monocytogenes EGD-e (grown in BHI medium) are shown.
Black arrows indicate genes which are probably organized in an operon structure and the direction of transcription based the L. monocytogenes EGD-e genome sequence data (http://genolist.pasteur.fr/ListiList/).
The genes actA and prfA are missing from this group because of the nonfunctioning of the oligonucleotides, but the transcriptional up-regulation of these genes was confirmed by real-time RT-PCR.
The down-regulated genes (greater than twofold) of group III (IIIb) are predominantly genes involved in the biosynthesis of branched amino acids and pyrimidine and in nitrogen metabolism. Interestingly, several of these genes were previously shown to be down-regulated by impaired glucose uptake in B. subtilis (38, 44).
PrfA activity is high when L. monocytogenes grows in minimal medium with the non-PTS substrate glycerol as the carbon source.
The above data show that PrfA activity is low when L. monocytogenes wild-type grows in BHI medium and also in minimal medium in the presence of PTS sugars, particularly in the early growth phase when the glucose uptake is highest (Fig. 5A, 6, and 8). On the other hand, expression of PrfA-regulated genes appears to be induced when glucose uptake is impaired, and expression of genes controlled by the efficiency of PTS-mediated glucose transport is down-regulated (see group III of the hprK mutant). Based on the described data the two major components of CCR, CcpA or HPr-Ser-P, do not act as direct modulators of the PrfA activity, although the data do not strictly rule out the possibility that additional factors may modify HPr-Ser-P activity, which could then act as a modulator of PrfA activity.
However, in our opinion the results suggest, rather, that components of the specific PTS-mediated sugar transport may affect PrfA activity. We therefore decided to study PrfA activity in the presence of the non-PTS carbon source glycerol.
As shown in Fig. 9A, L. monocytogenes grew in minimal medium containing 50 mM glycerol—after a longer lag phase—at a similar rate as in the presence of 50 mM glucose. Under these growth conditions, the expression of the genes encoding a glycerol uptake facilitator (lmo1539), glycerol kinase (lmo1538), and glycerol-phosphate dehydrogenase (glpD) as well as the genes for gluconeogenesis were highly up-regulated (B. Joseph, personal communication) compared to growth in the presence of glucose or cellobiose.
The amount of PrfA and PrfA activity measured by the hemolytic activity and the amount of ActA (Fig. 9B and C) in L. monocytogenes grown to the early (balanced) growth phase (OD600 of 0.6) were significantly higher in the presence of the non-PTS sugar glycerol compared to the PTS sugar glucose. In the later growth phase (OD600 of 1.0) the enhancement of the PrfA activity was less pronounced in glycerol-containing medium compared to glucose-containing medium (data not shown). However, in the later growth phase PrfA activity is already rather high even in glucose-containing medium.
The amount of HPr-Ser-P and HPr-His-P in L. monocytogenes grown in glycerol-containing minimal medium was rather similar to that observed in the presence of cellobiose (Fig. 7B and 9D) although PrfA activity was much higher in the glycerol-containing medium (Fig. 8B and C and 9B and C).
These data again argue against the idea that HPr-Ser-P and HPr-His-P are direct modulators of PrfA activity and rather favor component(s) of subsequent steps (i.e., PTS permeases or others phosphorylated by HPr-His-P) as a negative effector of PrfA activity.
DISCUSSION
In this study the influence of components involved in the CCR control system and (directly or indirectly) in PTS-mediated sugar transport on the activity of the major virulence regulator PrfA of L. monocytogenes was studied. Specifically, PrfA-dependent gene expression was determined in L. monocytogenes insertion mutants defective in ccpA (encoding the central catabolite control protein CcpA), hprK (encoding the ATP-dependent HPr kinase-phosphorylase, the central regulator of CCR), and ptsH (encoding HPr) when these listerial strains were grown either in nutrient-rich medium (BHI) or in a defined glucose-containing minimal medium.
We could not obtain in-frame deletions in these genes, possibly due to a massive outgrowth of the wild-type strain relative to these mutants even when grown in BHI medium. Nevertheless, the effects on PrfA activity observed with the insertion mutants seem to be a direct consequence of the absence of functional CcpA, HPr, and HPrK/P and not due to polar effects caused by the insertion on the adjacent genes. In contrast to the insertions in ccpA and hprK, which showed no significant polar effects on the expression of the upstream or downstream located genes, no transcription of ptsI (located downstream of ptsH in the ptsHI operon [12]) was observed apparently as a result of the insertion in ptsH. But since enzyme I (the gene product of ptsI) acts only on HPr, it is unlikely that this polar effect on ptsI expression affects the PrfA activity differently than the ptsH mutation alone. Revertants of all three mutants behaved in all tested properties similar to the wild-type strain, thus excluding the possibility that additional, unrecognized mutations might affect the phenotype of the mutants.
In agreement with previous findings (3), the ccpA mutation did not enhance PrfA activity. Our studies showed, rather, even a slightly reduced expression of the PrfA-dependent virulence genes compared to the EGD-e wild-type strain, which is in accord with the recent findings of Herro et al. (27).
Highly significant up-regulation of all tested PrfA-dependent virulence genes was observed in the hprK and ptsH mutants upon growth in glucose-containing medium. The degree of enhanced transcription of most of these genes including prfA itself in the two mutants was, however, not as high as that observed in the L. monocytogenes prfA* mutant (used as standard for a constitutively active PrfA), suggesting that PrfA is not as active in the hprK and ptsH mutants as in the prfA* mutant. Besides, in minimal medium cellobiose could still inhibit—albeit at a reduced rate compared to the wild type strain—PrfA-dependent gene expression in the hprK mutant but at a lower rate than in the prfA* mutant (52, 53).
A major difference between the ptsH and hprK mutants, on one hand, and the ccpA mutant, on the other, lies in the capability of these mutants to phosphorylate HPr. Obviously, no phosphorylated HPr can be produced in the ptsH mutant since HPr is not synthesized. The loss of HPrK/P activity in the hprK mutant restricts HPr phosphorylation to His15 (essential for PTS-mediated sugar transport) while the ccpA mutant can still phosphorylate HPr at both positions, His15 and Ser46. The level of HPr-Ser-P in the ccpA mutant appears to be similar or even slightly higher than in the wild-type strain and virtually zero in the hprK mutant. Increased levels of HPr-Ser-P were also observed in ccpA mutants in other gram-positive bacteria, e.g., Enterococcus faecalis (35) and B. subtilis (39).
Thus, there seems to be an inverse correlation between PrfA activity and the cellular HPr-Ser-P concentration in L. monocytogenes when cultured in glucose-containing medium. This observation would therefore be in accord with a recent study on PrfA modulation in B. subtilis (27). However, when L. monocytogenes was grown in minimal medium in the presence of the PTS sugar cellobiose instead of glucose (both sugars in equimolar quantities), the growth rates were similar in both cases, but the level of HPr-Ser-P was even lower in the presence of cellobiose than in the presence of glucose although the PrfA activity was much higher in the latter case. The reason for this reproducible result is presently unclear. Possibly the activity of HPrK, catalyzing synthesis of HPr-Ser-P, is lower in presence of cellobiose than with glucose due to lower levels of the HPrK-activating glycolysis intermediates, such as fructose-1,6-diphosphate or PEP.
These data do not support the notion that HPr-Ser-P acts as a direct negative modulator of PrfA activity. This conclusion is also in line with our recent studies aimed at demonstrating direct binding of purified PrfA with HPr-Ser-P in Biacore assays or inhibition of PrfA-mediated in vitro transcription initiated at several PrfA-dependent promoters by HPr-Ser-P, which were all negative (S. Müller-Altrock, personal communication). However, the data do not rule out the possibility that PrfA activity is inhibited by HPr-Ser-P in combination with an additional factor that may modify HPr-Ser-P activity. Synthesis of this factor could be enhanced when L. monocytogenes grows in the presence of cellobiose.
The lack of correlation between the amount of HPr-Ser-P or HPr-His-P and PrfA activity also does not favor the assumption that HPr-His-P may directly phosphorylate PrfA, as shown for the activation of several transcription regulators that contain a conserved PTS regulation domain (58). This is not surprising as a PTS regulation domain (PRD) could not be identified in PrfA.
The comparative transcript profiling carried out with RNAs from the ccpA and the hprK mutants versus the wild-type strain shows that a rather large number of genes (operons) are up-regulated in both the ccpA and the hprK mutants compared to the wild-type strain when strains are cultured in BHI medium, suggesting that they were repressed upon growth of L. monocytogenes in BHI medium. Many of these genes and operons encode proteins involved in transport and metabolism of various carbohydrates and some transcription factors, and they are likely to be under negative control of the CcpA/HPr-Ser-P complex, as in B. subtilis (44). This assumption is further supported by the observation that most of these genes carry typical cre sites (CcpA/HPr-Ser-P binding sites) in their upstream regulatory regions. Clearly, the PrfA-regulated virulence genes do not belong to this group of genes, which is in accord with previous findings (3).
There is, however, another set of genes which are up- or down-regulated in the hprK but not in the ccpA mutant compared to the wild-type strain. All PrfA-controlled genes are found among the up-regulated genes of this group. Among the down-regulated genes are mainly those that were previously termed class II CCR-controlled genes (39, 44). Several of these down-regulated genes are involved in nitrogen metabolism, like nrgAB (encoding an ammonium transporter and a PII-like protein) and in biosynthesis of branched-chain amino acids. Their expression was shown to be reduced when glucose uptake was impaired (38), which is also the case in the hprK (and ptsH) mutant.
Thus, there seems to be an inverse correlation between the regulation of PrfA-dependent genes and class II CCR-controlled genes; i.e., PrfA-controlled genes are up-regulated (suggesting increased PrfA activity) when PTS-mediated sugar uptake is impaired, whereas the class II CCR-controlled genes are down-regulated under these conditions.
The notion that PTS-mediated sugar transport may modulate PrfA activity can also be deduced from the observation that PrfA and its regulated genes are highly active when L. monocytogenes is grown in a minimal medium containing glycerol as the carbon source in comparison to growth in glucose- or cellobiose-containing medium (B. Joseph, unpublished data). Uptake of glycerol is not mediated by PTS, but probably, as in B. subtilis (4), by the glycerol uptake facilitator (encoded by lmo1539) which is highly up-regulated in L. monocytogenes grown in glycerol-containing medium. Metabolism of glycerol requires glycerol kinase and glycerol-phosphate dehydrogenase (the genes encoding these two enzymes are also highly up-regulated in glycerol-containing minimal medium), and glycerol kinase activation requires phosphorylation by HPr-His-P (15). The amounts of HPr-His-P and HPr-Ser-P are rather similar in the presence of glycerol and cellobiose, despite the much higher expression of PrfA-controlled genes in the presence of glycerol, and thus shows again that there is no correlation between the amount of HPr-Ser-P and PrfA activity (see above).
Glucose, mannose, and especially cellobiose are mediated by several PTS permeases (2, 10), which seem to functionally overlap in part and are either constitutively expressed or substrate induced (R. Ecke, personal communication). It is reasonable to assume that the EIIA and EIIB components of the PEP-PTS mediating transport of glucose, mannose, or cellobiose will be in a mainly nonphosphorylated state during active sugar transport, while these components are expected to be either not expressed or in a phosphorylated state in the presence of glycerol as carbon source. Under the first condition PrfA activity is low while it is high under the latter condition. We therefore propose that it is not the cellular level of HPr-Ser-P, HPr-His-P, or other components of the common PTS pathway that negatively affects PrfA activity but, rather, the phosphorylation status of the component(s) that is phosphorylated by HPr-His-P (i.e., specific PTS permeases or others). We are currently in the process of testing this hypothesis further.
Acknowledgments
We thank Jürgen Kreft, University of Würzburg, Germany, for providing the anti-LLO antibody and Nico Marr for purification and phosphorylation of HPr protein.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB473 and SFB479-B1) and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Aung-Hilbrich, L. M., G. Seidel, A. Wagner, and W. Hillen. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J. Mol. Biol. 319:77-85. [DOI] [PubMed] [Google Scholar]
- 2.Barabote, R. D., and M. H. Saier, Jr. 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69:608-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behari, J., and P. Youngman. 1998. A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J. Bacteriol. 180:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beijer, L., R. P. Nilsson, C. Holmberg, and L. Rutberg. 1993. The glpP and glpF genes of the glycerol regulon in Bacillus subtilis. J. Gen. Microbiol. 139:349-359. [DOI] [PubMed] [Google Scholar]
- 5.Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stülke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133-149. [DOI] [PubMed] [Google Scholar]
- 6.Böckmann, R., C. Dickneite, W. Goebel, and J. Bohne. 2000. PrfA mediates specific binding of RNA polymerase of Listeria monocytogenes to PrfA-dependent virulence gene promoters resulting in a transcriptionally active complex. Mol. Microbiol. 36:487-497. [DOI] [PubMed] [Google Scholar]
- 7.Böckmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. [DOI] [PubMed] [Google Scholar]
- 8.Bohne, J., H. Kestler, C. Uebele, Z. Sokolovic, and W. Goebel. 1996. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol. Microbiol. 20:1189-1198. [DOI] [PubMed] [Google Scholar]
- 9.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 10.Buchrieser, C., C. Rusniok, F. Kunst, P. Cossart, and P. Glaser. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35:207-213. [DOI] [PubMed] [Google Scholar]
- 11.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1998. Cloning and expression of the Listeria monocytogenes Scott A ptsH and ptsI genes, coding for HPr and enzyme I, respectively, of the phosphotransferase system. Appl. Environ. Microbiol. 64:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1999. Mutational analysis of the role of HPr in Listeria monocytogenes. Appl. Environ. Microbiol. 65:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutscher, J., R. Herro, A. Bourand, I. Mijakovic, and S. Poncet. 2005. P-Ser-HPr—a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim. Biophys. Acta 1754:118-125. [DOI] [PubMed] [Google Scholar]
- 15.Deutscher, J., and H. Sauerwald. 1986. Stimulation of dihydroxyacetone and glycerol kinase activity in Streptococcus faecalis by phosphoenolpyruvate-dependent phosphorylation catalyzed by enzyme I and HPr of the phosphotransferase system. J. Bacteriol. 166:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickneite, C., R. Böckmann, A. Spory, W. Goebel, and Z. Sokolovic. 1998. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol. Microbiol. 27:915-928. [DOI] [PubMed] [Google Scholar]
- 17.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 18.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 19.Ermolaeva, S., S. Novella, Y. Vega, M. T. Ripio, M. Scortti, and J. A. Vázquez-Boland. 2004. Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor. Mol. Microbiol. 52:601-611. [DOI] [PubMed] [Google Scholar]
- 20.Fieulaine, S., S. Morera, S. Poncet, I. Mijakovic, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2002. X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr. Proc. Natl. Acad. Sci. USA 99:13437-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 22.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbreth, S. E., A. K. Benson, and R. W. Hutkins. 2004. Catabolite repression and virulence gene expression in Listeria monocytogenes. Curr. Microbiol. 49:95-98. [DOI] [PubMed] [Google Scholar]
- 24.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 25.Gonzy-Tréboul, G., M. Zagorec, M. C. Rain-Guion, and M. Steinmetz. 1989. Phosphoenolpyruvate:sugar phosphotransferase system of Bacillus subtilis: nucleotide sequence of ptsX, ptsH and the 5′-end of ptsI and evidence for a ptsHI operon. Mol. Microbiol. 3:103-112. [DOI] [PubMed] [Google Scholar]
- 26.Gösseringer, R., E. Küster, A. Galinier, J. Deutscher, and W. Hillen. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 266:665-676. [DOI] [PubMed] [Google Scholar]
- 27.Herro, R., S. Poncet, P. Cossart, C. Buchrieser, E. Gouin, P. Glaser, and J. Deutscher. 2005. How seryl-phosphorylated HPr inhibits PrfA, a transcription activator of Listeria monocytogenes virulence genes. J. Mol. Microbiol. Biotechnol. 9:224-234. [DOI] [PubMed] [Google Scholar]
- 28.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15:395-401. [DOI] [PubMed] [Google Scholar]
- 29.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 30.Joseph, B., K. Przybilla, C. Stühler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreft, J., J. A. Vázquez-Boland, S. Altrock, G. Domínguez-Bernal, and W. Goebel. 2002. Pathogenicity islands and other virulence elements in Listeria. Curr. Top. Microbiol. Immunol. 264:109-125. [PubMed] [Google Scholar]
- 32.Kuhn, M., and W. Goebel. 1995. Molecular studies on the virulence of Listeria monocytogenes. Genet. Eng. (NY) 17:31-51. [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Lampidis, R., R. Gross, Z. Sokolovic, W. Goebel, and J. Kreft. 1994. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol. Microbiol. 13:141-151. [DOI] [PubMed] [Google Scholar]
- 35.Leboeuf, C., L. Leblanc, Y. Auffray, and A. Hartke. 2000. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by ccpA. J. Bacteriol. 182:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig, H., C. Meinken, A. Matin, and J. Stülke. 2002. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J. Bacteriol. 184:5174-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludwig, H., N. Rebhan, H. M. Blencke, M. Merzbacher, and J. Stülke. 2002. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol. Microbiol. 45:543-553. [DOI] [PubMed] [Google Scholar]
- 40.Luo, Q., M. Rauch, A. K. Marr, S. Müller-Altrock, and W. Goebel. 2004. In vitro transcription of the Listeria monocytogenes virulence genes inlC and mpl reveals overlapping PrfA-dependent and -independent promoters that are differentially activated by GTP. Mol. Microbiol. 52:39-52. [DOI] [PubMed] [Google Scholar]
- 41.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vázquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 42.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 43.Milohanic, E., P. Glaser, J. Y. Coppée, L. Frangeul, Y. Vega, J. A. Vázquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 44.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 45.Mueller, K. J., and N. E. Freitag. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect. Immun. 73:1917-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, S. F., and R. G. Kroll. 1993. Expression of listeriolysin and phosphatidylinositol-specific phospholipase C is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol. Microbiol. 8:653-661. [DOI] [PubMed] [Google Scholar]
- 47.Pfeuffer, T., W. Goebel, J. Laubinger, M. Bachmann, and M. Kuhn. 2000. LaXp180, a mammalian ActA-binding protein, identified with the yeast two-hybrid system, co-localizes with intracellular Listeria monocytogenes. Cell Microbiol. 2:101-114. [DOI] [PubMed] [Google Scholar]
- 48.Pompeo, F., Y. Granet, J. P. Lavergne, C. Grangeasse, S. Nessler, J. M. Jault, and A. Galinier. 2003. Regulation and mutational analysis of the HPr kinase/phosphorylase from Bacillus subtilis. Biochemistry 42:6762-6771. [DOI] [PubMed] [Google Scholar]
- 49.Poncet, S., I. Mijakovic, S. Nessler, V. Gueguen-Chaignon, V. Chaptal, A. Galinier, G. Boel, A. Mazé, and J. Deutscher. 2004. HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in gram-positive bacteria. Biochim. Biophys. Acta 1697:123-135. [DOI] [PubMed] [Google Scholar]
- 50.Premaratne, R. J., W. J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renzoni, A., A. Klarsfeld, S. Dramsi, and P. Cossart. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect. Immun. 65:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripio, M. T., G. Domínguez-Bernal, M. Lara, M. Suárez, and J. A. Vázquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ripio, M. T., G. Domínguez-Bernal, M. Suárez, K. Brehm, P. Berche, and J. A. Vázquez-Boland. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res. Microbiol. 147:371-384. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Sheehan, B., A. Klarsfeld, R. Ebright, and P. Cossart. 1996. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol. Microbiol. 20:785-797. [DOI] [PubMed] [Google Scholar]
- 56.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48:1537-1551. [DOI] [PubMed] [Google Scholar]
- 57.Stritzker, J., C. Schoen, and W. Goebel. 2005. Enhanced synthesis of internalin A in aro mutants of Listeria monocytogenes indicates posttranscriptional control of the inlAB mRNA. J. Bacteriol. 187:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stülke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD-a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 59.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 60.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. sigmaB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 61.Tobisch, S., D. Zuhlke, J. Bernhardt, J. Stülke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vega, Y., C. Dickneite, M. T. Ripio, R. Böckmann, B. Gonzalez-Zorn, S. Novella, G. Domínguez-Bernal, W. Goebel, and J. A. Vázquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vega, Y., M. Rauch, M. J. Banfield, S. Ermolaeva, M. Scortti, W. Goebel, and J. A. Vázquez-Boland. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol. Microbiol. 52:1553-1565. [DOI] [PubMed] [Google Scholar]
- 66.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong, K. K., H. G. Bouwer, and N. E. Freitag. 2004. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell Microbiol. 6:155-166. [DOI] [PubMed] [Google Scholar]
- 68.Wong, K. K., and N. E. Freitag. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186:6265-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wuenscher, M. D., S. Köhler, W. Goebel, and T. Chakraborty. 1991. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol. Gen. Genet. 228:177-182. [DOI] [PubMed] [Google Scholar]
- 70.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]