Abstract
Francisella tularensis is a gram-negative coccobacillus that is capable of causing severe, fatal disease in a number of mammalian species, including humans. Little is known about the proteins that are surface exposed on the outer membrane (OM) of F. tularensis, yet identification of such proteins is potentially fundamental to understanding the initial infection process, intracellular survival, virulence, immune evasion and, ultimately, vaccine development. To facilitate the identification of putative F. tularensis outer membrane proteins (OMPs), the genomes of both the type A strain (Schu S4) and type B strain (LVS) were subjected to six bioinformatic analyses for OMP signatures. Compilation of the bioinformatic predictions highlighted 16 putative OMPs, which were cloned and expressed for the generation of polyclonal antisera. Total membranes were extracted from both Schu S4 and LVS by spheroplasting and osmotic lysis, followed by sucrose density gradient centrifugation, which separated OMs from cytoplasmic (inner) membrane and other cellular compartments. Validation of OM separation and enrichment was confirmed by probing sucrose gradient fractions with antibodies to putative OMPs and inner membrane proteins. F. tularensis OMs typically migrated in sucrose gradients between densities of 1.17 and 1.20 g/ml, which differed from densities typically observed for other gram-negative bacteria (1.21 to 1.24 g/ml). Finally, the identities of immunogenic proteins were determined by separation on two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and mass spectrometric analysis. This is the first report of a direct method for F. tularensis OM isolation that, in combination with computational predictions, offers a more comprehensive approach for the characterization of F. tularensis OMPs.
Francisella tularensis is a gram-negative, intracellular pathogen that is the causative agent of the zoonotic disease tularemia. This coccobacillus is extremely pathogenic, easily aerosolized, has a low infectious dose (<10 CFU), and causes high morbidity and mortality in a number of mammalian species, including humans (10). Given these considerations, F. tularensis long has been recognized as a potential bioweapon (8, 31) and thus has been designated as a category A select agent (39).
There are four subspecies of F. tularensis, two of which cause disease in humans (F. tularensis subsp. tularensis [type A] and F. tularensis subsp. holarctica [type B]). The type A strains are found predominantly in North America and are associated with lethal human disease. An understanding of the fundamental aspects of the type A strains has been hindered because of the requirement for manipulation of the bacterium under strict biosafety level 3 (BSL3) conditions. Schu S4, a type A human isolate, is a type strain for the tularensis subspecies. The live vaccine strain (LVS) is an attenuated type B strain that induces partial immunological protection against virulent F. tularensis, although the nature of the LVS attenuation remains unknown.
Outer membrane proteins (OMPs) often are strategic for facilitating bacterial host invasion, intracellular survival, virulence, and immune evasion (16, 18, 29, 43, 55). Their significance also is highlighted by the fact that they can serve as protective vaccines for a number of bacterial diseases (6, 25, 32, 34, 54). In the case of virulent F. tularensis, identification of OMPs, particularly those with surface-exposed domains, also could be highly advantageous for the eventual development of blocking antibodies, diagnostic reagents, or environmental detection systems. As with other intracellular pathogens, protective immunity to F. tularensis infection is presumed to rely primarily on the cellular immune response (9). However, for a number of intracellular pathogens there is substantial evidence that antibodies can block the initial infection or even enter host target cells to inhibit intracellular growth (4, 5).
Little is known about the proteins on the surface of virulent F. tularensis. Initial attempts to characterize OMPs from F. tularensis were limited in that they focused on studies with avirulent LVS and typically relied on bulk membrane extraction techniques, including sonication of cells followed by ultracentrifugation and/or detergent extraction (11, 12, 27, 41, 47). While providing an initial foundation, characterizations of these protein preparations were limited with respect to potential periplasmic and/or cytoplasmic (inner) membrane (IM) contaminants. Nonetheless, the first presumed F. tularensis OMP was a 43-kDa protein identified by probing lithium chloride extracts of bacteria with antisera collected from individuals involved in an outbreak of tularemia in Norway (2). This protein was named FopA, for Francisella outer membrane protein, because it localized with other major Escherichia coli OMPs when expressed in E. coli (27). Two separate vaccine trials demonstrated that FopA was not protective against type A F. tularensis or LVS challenge, despite its induction of antibodies (12, 13).
The second presumed F. tularensis OMP was a 17-kDa T-lymphocyte-reactive protein originally identified from an N-lauroylsarcosinate-insoluble protein preparation (41); this 17-kDa polypeptide was later named TUL4 (45). Additional studies confirmed the ability of TUL4 to stimulate lymphocyte proliferation, primarily CD4+ T cells, and noted marked production of interleukin-2 (IL-2) and gamma interferon (IFN-γ) in response to TUL4 (11, 44, 45, 47). Initial [3H]palmitate radiolabeling and detergent extraction of F. tularensis suggested that TUL4 was an integral membrane lipoprotein (LP) (46). Further studies later suggested that TUL4 most likely was in the F. tularensis OM (22). A number of other probable OMPs and LPs were identified in the aforementioned studies, yet none have been further characterized.
Current advances in proteomics and two-dimensional gel electrophoresis (2DE) have accelerated the discovery and identification of F. tularensis antigens. More than 1,500 proteins were visualized when total cellular proteins were solubilized from F. tularensis and separated by 2DE (20). However, only three of those identified by mass spectrometry were proposed to be membrane proteins, including TUL4. More recently, two separate studies have used sodium carbonate to putatively enrich for F. tularensis membrane proteins (33, 51). Whereas the former study identified roughly 200 proteins by 2DE and mass spectrometry, only 7 were predicted to be OMPs. The latter study identified a larger number of potential OMP candidates, approximately 500 in total, yet only 4 were predicted to be OMPs, including FopA. In an effort to identify immunogenic proteins from F. tularensis, total cellular proteins were separated by 2DE and probed with sera from experimentally infected mice (19). Of the 36 immunoreactive proteins identified, the vast majority were probable cytoplasmic proteins, whereas only two predicted OMPs were noted.
In the present study, we employed a two-pronged approach for a more comprehensive characterization of F. tularensis OMPs. We first utilized six independent bioinformatic analyses to putatively identify membrane proteins from the genomes of both the type A strain (Schu S4) and type B strain (LVS). A total of 16 putative OMP- and 2 IM-encoding regions were subsequently cloned and expressed in E. coli, and recombinant proteins were used for the generation of polyclonal antisera. These antibodies were deemed strategic probes for further assessing either the OM or IM compartment of F. tularensis. Computational algorithms, however, are not unequivocal for predicting OMPs. Thus, to complement theoretical predictions, we applied a variation of the sucrose density gradient ultracentrifugation “gold standard” method for the separation of F. tularensis OMs and IMs (30). F. tularensis membranes were extracted from both Schu S4 and LVS by spheroplasting and osmotic lysis, followed by sucrose density gradient centrifugation. This is the first report of a direct method for the physical separation of F. tularensis OMs and IMs. The approach allows not only for more direct OMP identification, but also for a better assessment of protein localization predictions derived from computational algorithms.
MATERIALS AND METHODS
Bacterial strains and culture.
Following all federal and institutional select agent regulations, F. tularensis type A strain Schu S4 was obtained from the Centers for Disease Control and Prevention (Fort Collins, CO). All work with Schu S4 was conducted under strict BSL3 containment conditions, including the use of liquid impervious gowns and powered air purifying respirators. F. tularensis type B LVS was obtained from Karen Elkins (Center for Biologics Evaluation and Research, Food and Drug Administration, Rockville, MD) and manipulated under BSL2 containment conditions. F. tularensis stock cultures were grown at 37°C with 5% CO2 on Mueller-Hinton agar supplemented with 2.5% (vol/vol) donor calf serum (Mediatech, Herndon, VA), 2% (vol/vol) IsoVitaleX (Becton Dickinson, Franklin Lakes, NJ), 0.1% (wt/vol) glucose, and 0.025% (wt/vol) iron pyrophosphate. Following 48 to 72 h of growth on agar, individual colonies were inoculated into modified Mueller-Hinton medium supplemented with 1.23 mM calcium chloride dihydrate, 1.03 mM magnesium chloride hexahydrate, 0.1% (wt/vol) glucose, 0.025% (wt/vol) iron pyrophosphate, and 2% (vol/vol) IsoVitaleX. Broth cultures were grown at 37°C for 12 to 16 h with shaking. Freezer stocks of F. tularensis were prepared in modified Mueller-Hinton medium with 10% (wt/vol) sucrose. Escherichia coli XL1-Blue electrocompetent cells (Stratagene, La Jolla, CA) were routinely cultivated in Luria-Bertani (LB) broth or on LB agar plates at 37°C, each supplemented with ampicillin (100 μg/ml) for selection.
Bioinformatic analyses.
A number of computational algorithms were utilized to identify signatures characteristic of OMPs and outer membrane LPs from the genomes of F. tularensis type A strain Schu S4 (accession no. NC_006570) and type B strain LVS (accession no. NC_007880): the PSORTb bacterial protein subcellular localization prediction program (http://www.psort.org/psortb/index.html), the SignalP signal peptidase cleavage site prediction program (http://www.cbs.dtu.dk/services/SignalP/), the LipoP lipoprotein and signal peptide discrimination program (http://www.cbs.dtu.dk/services/LipoP/), the PROFtmb bacterial transmembrane beta barrel prediction program (http://cubic.bioc.columbia.edu/services/proftmb/), the BOMP beta barrel integral outer membrane protein prediction program (http://www.bioinfo.no/tools/bomp), and the TMB-Hunt amino acid composition-based transmembrane beta barrel prediction program (http://www.bioinformatics.leeds.ac.uk/∼andy/betaBarrel/AACompPred/aaTMB_Hunt.cgi). These programs also were used as adjuncts for the identification of cytoplasmic proteins and IMPs, which were used as validation markers for membrane separation experiments. Additionally, similarity comparisons were made between F. tularensis open reading frames (ORFs) and known bacterial proteins by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). Compilation and comparison of the bioinformatic predictions (see Table 1, below) highlighted 16 putative OMP and LP coding regions, as well as cytoplasmic and IM markers, which were targeted for cloning and expression.
TABLE 1.
Proteins used as localization markers for analysis of sucrose density gradient fractions
| Locus | Proteina | PSORTb localizationb | PSORTb scoreb | PROFtmb scorec | BOMP scored | TMB-Hunt scoree | Predicted massf | Recombinant massg |
|---|---|---|---|---|---|---|---|---|
| FTT0842 | Pal | OM | 10.00 | 21.0 | 24.2 | |||
| FTT1095c | TolC-A | OM | 10.00 | 49.9 | 52.9 | |||
| FTT1156c | PilQ | OM | 10.00 | 10.7 | 65.0 | 64.7 | ||
| FTT1258 | SilC | OM | 10.00 | 51.9 | 54.9 | |||
| FTT1573c | FtaG | OM | 10.00 | 6.2 | 3 | 10.7 | 85.0 | 88.2 |
| FTT1724c | TolC-B | OM | 10.00 | 55.1 | 58.3 | |||
| FTT0583 | FopA | OM | 9.93 | 7.3 | 8.6 | 39.0 | 42.2 | |
| FTT1043 | Mip | OM | 9.92 | 27.1 | 29.8 | |||
| FTT0918 | YapH-Nh | OM | 9.52 | 8.1 | 3 | 7.4 | 56.8 | 27.0 |
| FTT0919 | YapH-Ch | OM | 9.49 | 7.8 | 4.2 | 50.2 | 25.7 | |
| FTL_0439 | YapH-LVSh | OM | 9.52 | 8.3 | 7.1 | 55.8 | 35.1 | |
| FTT0715 | Cht1 | Un | 2.00 | 6.2 | 81.0 | 84.1 | ||
| FTT0025c | SrfA | IM | 9.46 | 7.6 | 3 | 4.9 | 51.6 | 51.2 |
| FTT0901 | Tul4-A | Un | 2.00 | 13.8 | 16.9 | |||
| FTT0904 | Tul4-B | Un | 2.00 | 15.5 | 18.6 | |||
| FTT0223c | LamB | Cyto | 8.96 | 25.8 | 24.3 | |||
| FTT0345 | SecY | IM | 10.00 | 44.8 | 20.2 | |||
| FTT0404 | LolC | IM | 10.00 | 48.2 | 28.1 | |||
| FTT0137 | EF-Tu | Cyto | 9.97 | 43.4 | 42.8 | |||
| FTT1359c | IglA | Cyto | 8.96 | 22.3 | 24.0 |
Underlined proteins are putative lipoproteins, based upon the LipoP lipoprotein prediction program (http://www.cbs.dtu.dk/services/LipoP/).
PSORTb version 2.0.4 bacterial protein subcellular localization prediction program (http://www.psort.org/psortb/index.html) localization predicted for OM, cytoplasmic membrane (IM), cytoplasm (Cyto), or unknown (Un). Scores of >7.5 were considered significant.
PROFtmb bacterial transmembrane beta-barrel prediction program (http://cubic.bioc.columbia.edu/services/proftmb/). Only significant Z-values (>4.0) are shown.
BOMP beta-barrel integral outer membrane protein prediction program (http://www.bioinfo.no/tools/bomp). Only significant scores (>2) are shown.
TMB-Hunt transmembrane beta-barrel prediction program (http://www.bioinformatics.leeds.ac.uk/∼andy/betaBarrel/AACompPred/aaTMB_Hunt.cgi). Only significant log probability scores (>2) are shown.
For OMPs and IMPs, the predicted molecular mass (in kDa) of the protein was calculated following cleavage by signal peptidase I or II.
A portion of each coding region was cloned into the pProEX HTb six-histidine fusion vector for recombinant expression and purification in E. coli. Sequence upstream of the predicted signal sequence was excluded to aid in purification.
There are two distinct YapH ORF orthologs in Schu S4. In LVS, the YapH-LVS ORF is the result of an in-frame deletion of the C-terminal end of FTT0918 and the N-terminal end of FTT0919.
Expression and purification of recombinant proteins.
Genomic DNA was extracted from either F. tularensis Schu S4 or LVS using an Easy-DNA genomic DNA isolation kit (Invitrogen, Carlsbad, CA). Because of the high degree of sequence identity between the two strains, the majority of coding regions were amplified from LVS DNA. Only FTT0918 (YapH-N) and FTT0919 (YapH-C) were amplified from Schu S4 DNA. To aid in recombinant protein purification, all proteins generated in this study lacked their N-terminal leader peptides. Further, for YapH-N, YapH-C, YapH-LVS, and LolC, only a selected hydrophilic portion from each protein was cloned. Coding regions were PCR amplified using genomic DNA, the primers listed in Table S1 (see the supplemental material), and ExTaq Hot Start DNA polymerase (TaKaRa, Otsu, Shiga, Japan). Individual amplification products were directionally cloned into the pProEX HTb vector (Invitrogen), which created N-terminal His6 fusions. Ligations were transformed into E. coli XL1-Blue electrocompetent cells, and plasmids were purified from representative clones for PCR screening (exploiting the same primers used for PCR amplification) followed by DNA sequence confirmation. Clones containing in-frame inserts were grown in LB broth to an optical density at 600 nm (OD600) of 0.4, after which protein expression was induced for 4 h by the addition of isopropyl β-d-thiogalactopyranoside to 0.5 mM. Following induction, cells were collected by centrifugation and frozen overnight at −20°C (to aid in cell lysis). The following morning, cell pellets were thawed and lysed/solubilized in BugBuster protein extraction reagent (Novagen, Madison, WI) with Complete Mini EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics, Indianapolis, IN). The cell suspension was sonicated three times at 1-min intervals, followed by the removal of insoluble material by centrifugation at 10,000 × g for 15 min. The resulting supernatants, designated as BB, were removed and saved for affinity purification. The insoluble pellet was suspended in inclusion body reagent (Pierce, Rockford, IL) and sonicated as noted above, debris was removed by centrifugation at 3,000 × g for 20 min, and the resulting supernatant, designated as IBR, was removed and saved for affinity purification.
Affinity purification of fusion proteins was performed by applying BB and IBR supernatants to separate columns containing preequilibrated Ni-nitrilotriacetic acid-agarose (QIAGEN, Valencia, CA), followed by three column washes with wash buffer (8 M urea, 10 mM Tris, 30 mM imidizole, pH 7.5). Purified recombinant OMPs were removed from the Ni-nitrilotriacetic acid-agarose by resuspension and boiling of the agarose for 10 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Purity of the recombinant proteins was assessed by SDS-PAGE and Coomassie brilliant blue staining, the relative quantity of recombinant OMPs was compared between BB and IBR extractions, and the extraction with the highest protein quantity and purity was electroeluted from SDS-PAGE gels. Concentrations of purified recombinant proteins were determined by using the BCA protein assay kit (Pierce).
Gene building for SecY.
For the IM protein SecY, recombinant protein expression and purification were not successful due to low levels of expression (e.g., rare codon usage by E. coli), toxicity to XL1-Blue cells, or insolubility. Therefore, the portions of the secY gene that encoded hydrophilic regions were selected and assembled into a chimeric protein using MacVector sequence editing software (Accelrys, San Diego, CA). Overlapping oligonucleotide primers were designed for the piecewise assembly of the secY chimera using DNA Builder (P. Hunter and S. A. Johnston, Center for Innovations in Medicine; www.biodesign.asu.edu), a program designed for codon-optimization to enhance foreign protein expression in a cloning host (e.g., E. coli). In the first step of the chimera assembly, primer sets (see Table S2 in the supplemental material) were mixed and annealed, and gaps were filled using Ex Taq Hot Start DNA polymerase (TaKaRa). In the second step, PCR amplification was performed with the flanking 5′ and 3′ primers (SecY 1 and SecY 16) to select for the full-length product. Finally, the resulting chimera was cloned, expressed, and purified as described above.
Animals and antisera generation.
All animal procedures were approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee and the Biological and Chemical Safety Advisory Committee. Animals were housed in microisolator cages at the UT Southwestern Animal Resource Center and fed irradiated food and water ad libitum. For the generation of polyclonal antisera against putative OMPs and IMPs, 20 μg of each recombinant protein (in 200 μl of phosphate-buffered saline [PBS]) was emulsified with an equal volume of complete Freund's adjuvant (Sigma, St. Louis, MO) and injected intraperitoneally into 6-week-old, female Sprague-Dawley rats (Harlan, Indianapolis, IN). After 3 weeks, the rats were boosted with a similar amount of protein emulsified in incomplete Freund's adjuvant (Sigma). Two weeks after this boost, test bleeds were performed and, if necessary, a second boost was performed to increase antiserum affinity and titer. For antisera collection, rats were anesthetized with isoflurane and exsanguinated by cardiac puncture. Blood was allowed to coagulate for 2 h at room temperature and centrifuged at 3,000 × g for 20 min, and serum was removed and transferred to sterile tubes for storage at −20°C.
To generate immune sera reactive against both LVS and Schu S4, mice were actively infected with LVS, followed by a low-dose Schu S4 infection. Briefly, 6- to 8-week-old, female C3H/HeN mice (Charles River Laboratories, Wilmington, MA) were first intranasally inoculated with approximately 104 CFU of live LVS (in 25 μl of freezing medium). Four weeks after the initial exposure, mice were boosted with a similar inoculum. Three weeks after this boost, antisera were collected as described above or mice were transferred to the BSL3 animal facility for intranasal challenge with 100 CFU of live Schu S4 (in 25 μl of freezing medium). Mice were held for an additional 3 weeks, at which time they were exsanguinated as described above.
Immunoblotting.
Rat polyclonal antisera specificity was assessed by immunoblot analysis against whole-cell lysates of LVS and Schu S4, as well as the relevant purified recombinant proteins. F. tularensis whole-cell lysates were prepared from Mueller-Hinton broth cultures that had been grown to an OD600 of 0.4 to 0.6, which corresponded to approximately 108 CFU, suspended in SDS loading buffer, and boiled for 10 min. The relative intensities of prominent LVS and Schu S4 proteins were compared by SDS-PAGE and Coomassie brilliant blue staining, to standardize the lysates (data not shown). F. tularensis lysates and recombinant proteins were separated by SDS-PAGE and transferred to nitrocellulose. Blots were blocked overnight with blot block (0.05% [vol/vol] Tween 20 and 2% [wt/vol] bovine serum albumin in PBS) at 4°C. Antisera were diluted between 1:1,000 and 1:10,000 in blot block and incubated for 2 h at room temperature, followed by four washes for 4 min each with PBS-Tween (0.05% [vol/vol] Tween 20 in PBS), incubation for 1 h with secondary (diluted 1:20,000) goat anti-rat immunoglobulin G (IgG; H+L)-horseradish peroxidase (HRP) antibody (Jackson ImmunoResearch Laboratories, West Grove, PA), four washes for 4 min each with PBS-Tween, and development with SuperSignal West Pico chemiluminescent detection reagent (Pierce). Polyclonal antisera with weak specificity were affinity purified utilizing recombinant protein and the AminoLink Plus immobilization kit (Pierce).
Mouse and human antisera specificity against F. tularensis was assessed by immunoblot analysis against whole-cell lysates of LVS and Schu S4 as described above. For secondary antibodies, mouse immunoblot assays were performed with goat anti-mouse IgG (H+L)-HRP antibody (Jackson ImmunoResearch), whereas human immunoblot assays were performed with goat anti-human IgG (H+L)-HRP antibody (Jackson ImmunoResearch).
Spheroplasting and sucrose density gradient centrifugation.
F. tularensis cell lysis and sucrose density gradient centrifugation procedures were modified based upon previously published methods for other gram-negative bacteria (26, 28, 30). F. tularensis cultures were grown in 1-liter batches to an OD600 of 0.2 to 0.3 (∼108 CFU/ml). Cultures were centrifuged at 8,000 × g for 30 min at 10°C to collect the cells, the supernatant was removed, and centrifuge bottles were briefly tapped on absorbent material to remove excess growth medium. Within 10 min, cell pellets were suspended in 35 ml of 0.75 M sucrose (in 5 mM Tris, pH 7.5) and transferred to a sterile 250-ml flask with a stir bar. While gently mixing the cell suspension, 70 ml (two volumes) of 10 mM EDTA (in 5 mM Tris, pH 7.8) was slowly added over the course of 10 min. The 10 mM EDTA solution was added with the tip of a pipette below the cell suspension level to avoid elevated local concentrations of EDTA. After 30 min of incubation at room temperature, lysozyme was added slowly to a final concentration of 200 μg/ml (11 ml of a 2-mg/ml stock solution), and the cells were incubated for 30 min at room temperature. The cells were osmotically lysed by slowly diluting them into 520 ml (4.5 volumes in a 1-liter sterile flask with a stir bar) of Cellgro molecular-grade distilled water (Mediatech) over the course of 10 min with gentle mixing. After 30 min of room temperature incubation, the osmotic lysis solution was divided into 40-ml aliquots and centrifuged at 7,500 × g for 30 min at 10°C to remove intact cells and debris. To collect total membranes, supernatants were carefully removed and then centrifuged at 200,000 × g (44,400 rpm in a T865 ultracentrifuge rotor; Sorvall, Asheville, NC) for 2 h at 4°C. Following centrifugation, supernatants were removed, ultracentrifuge tubes were inverted on absorbent material and tapped to remove excess supernatant, and total membrane pellets were gently resuspended in 5 to 6 ml of resuspension buffer (25% [wt/wt] sucrose, 5 mM Tris, 30 mM MgCl2, 1 tablet of Complete Mini EDTA-free protease inhibitor cocktail [Roche Diagnostics], 5 U Benzonase [Novagen]). Total membrane suspensions were incubated with gentle inversion for 30 min at room temperature to degrade DNA. Protein quantitation was performed for total membranes using the DC protein assay (Bio-Rad, Hercules, CA). Total protein yield was generally between 1.0 mg/ml and 1.6 mg/ml.
Linear sucrose gradients were prepared by layering 1.8 ml each of sucrose solutions (wt/wt; prepared in 5 mM EDTA, pH 7.5) into 14- by 95-mm ultracentrifuge tubes (Beckman, Palo Alto, CA) in the following order: 55%, 50%, 45%, 40%, 35%, and 30%. Total membranes were layered on top of each sucrose gradient, with no more than 1.5 mg of protein per gradient. Sucrose gradients were centrifuged in an SW40 swinging bucket rotor (Beckman) at 256,000 × g (38,000 rpm) for 18 h at 4°C. After completion of the centrifugation, sucrose gradient tubes were removed from the rotor buckets and 500-μl fractions were collected from the bottom of each gradient by puncturing with a 21-gauge needle and allowing them to drip by gravity. The refractive index of each sucrose fraction was determined using a refractometer (Fisher Scientific, Hampton, NH) and correlated with a specific density in g/ml (37). Representative sucrose gradient fractions (density increments of 0.01; from 1.11 g/ml to 1.26 g/ml) were diluted in SDS loading buffer, boiled for 10 min, separated by SDS-PAGE, and either silver stained (Silver Quest; Invitrogen) or transferred to nitrocellulose and immunoblotted as described above.
Membrane enrichment and protein immunoreactivities.
OMP-containing (densities from 1.17 to 1.20 g/ml) and IMP-containing (densities from 1.13 to 1.14 g/ml) sucrose gradient fractions were pooled and dialyzed in 3- to 12-ml, 5,000-Da molecular weight cutoff Slide-A-Lyzer dialysis cassettes (Pierce) against 5 mM EDTA and 0.2 mM phenylmethylsulfonyl fluoride for 24 h. The refractive indices of the dialyzed solutions were determined to ensure that the amount of sucrose was less than 0.5%. Dialyzed fractions were concentrated in Amicon Ultra-4 centrifugal filter units (Millipore) followed by protein quantitation using the DC protein assay (Bio-Rad). Equal amounts of LVS and Schu S4 whole-cell lysates, IM fractions, and OM fractions were separated by SDS-PAGE and silver stained to visualize membrane separation and enrichment. Additionally, equal amounts of LVS and Schu S4 OMPs were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted (as described above) with pooled sera from F. tularensis-infected mice or individual sera from LVS-vaccinated humans (human sera kindly provided by Beverly Fogtman, U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD).
Two-dimensional gel electrophoresis.
Pooled OMP-containing fractions were dialyzed, concentrated, and quantitated as described above. Prior to isoelectric focusing, OMPs were further purified using the ReadyPrep 2-D Cleanup kit (Bio-Rad), with no more than 150 μg of OMPs per microcentrifuge tube. Purified OMPs (150 μg) were suspended in a total of 185 μl of ReadyPrep 2-D rehydration/sample buffer 1 (7 M urea, 2 M thiourea, 1% [wt/vol] ASB-14, 40 mM Tris base, 0.001% [wt/vol] bromophenol blue [Bio-Rad]), supplemented with 10 mM tributylphosphine (Bio-Rad), 4% (wt/vol) 3-[3-(cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 0.2% (vol/vol) BioLytes 3-10 (Bio-Rad), 0.1% (vol/vol) BioLytes 3-5 (Bio-Rad), and 0.1% (vol/vol) BioLytes 8-10 (Bio-Rad). The OMP suspensions were vortexed for 5 min at 15-min intervals for 2 h before being loaded onto 11-cm linear pH 3 to 10 ReadyStrip IPG strips (Bio-Rad). The OMP suspensions were passively absorbed into IPG strips overnight without the suggested mineral oil overlay. The following day, IPG strips were transferred to an 11-cm focusing tray with predampened wicks and overlaid with 3 ml of mineral oil. OMPs were focused in a Protean IEF cell (Bio-Rad) under the following conditions: 150 V for 3 h, linear ramp; 300 V for 1 h, linear ramp; 600 V for 1 h, linear ramp; 1,200 V for 1 h, linear ramp; 2,400 V for 1 h, linear ramp; 5,000 V for 1 h, linear ramp; 5,000 V for a total of 80,000 Vh, rapid ramp; 500 V hold, rapid ramp. Following focusing, IPG strips were removed from the focusing tray, excess mineral oil was blotted from the strips, and strips were transferred to a fresh tray for a 20-min wash with ReadyPrep 2-D equilibration buffer I (375 mM Tris-HCl, pH 8.8, 6 M urea, 2% [wt/vol] SDS, 2% [wt/vol] dithiothreitol [Bio-Rad]), a 20-min wash with ReadyPrep 2-D equilibration buffer II (375 mM Tris-HCl, pH 8.8, 6 M urea, 2% [wt/vol] SDS [Bio-Rad]) supplemented with 4% (wt/vol) iodoacetamide, and a brief wash in SDS running buffer. IPG strips were placed into wells of 12.5% Criterion SDS-PAGE gels (Bio-Rad), overlaid with ReadyPrep overlay agarose (0.5% [wt/vol] agarose, SDS running buffer, 0.003% [wt/vol] bromophenol blue [Bio-Rad]), and proteins were separated by running the gels at 150 V for 1.5 h. For staining of OMPs, SDS-PAGE gels were fixed for 30 min in fixing solution (10% methanol, 7% acetic acid), followed by overnight staining in SYPRO Ruby protein stain (Bio-Rad) with gentle rocking, two washes in fixing solution for 30 min, and a final rinse in distilled H2O. OMPs were visualized by excitation with a 300-nm UV light and imaging on a Gel Logic 200 image station (Kodak, New Haven, CT). Alternatively, proteins were transferred to nitrocellulose and immunoblot assays were performed as described above, probed with sera from F. tularensis-infected mice or LVS-vaccinated humans.
Protein identification by mass spectrometry.
Predominant OMPs from LVS and Schu S4 were excised from SYPRO Ruby-stained 2DE gels for identification. In-gel trypsin digestion and reversed-phase nano-high-performance liquid chromatography/ion trap mass spectrometry were performed at the UT Southwestern Protein Chemistry Core Facility using either a ThermoFinnigan LCQ Deca XP MS or LCQ Deca MS instrument. Resulting data sets were searched against the Schu S4 and LVS genomic databases noted above using Sonar database software (Genomic Solutions, Ann Arbor, MI).
RESULTS
Bioinformatic analyses.
To date, only two OMPs and/or LPs have been characterized from F. tularensis: FopA (27) and TUL4 (45). To facilitate the identification of other OMPs and LPs, we scanned the predicted ORFs from the type A strain Schu S4 (1,603 ORFs) and the type B strain LVS (1,754 ORFs) using six bioinformatic algorithms, with an emphasis on detecting OMP and LP signatures. Because of the high degree of sequence homology between putative OMPs from Schu S4 and LVS (96 to 100%) (data not shown) and nearly identical bioinformatic results obtained for both strains (data not shown), the majority of the data presented are for Schu S4 (Table 1) . Results from the PSORTb bacterial subcellular localization prediction program identified a total of 39 ORFs (data not shown) with high OM localization scores (scores > 9). These data were further screened by BLAST analysis to counterselect against probable cytoplasmic proteins (e.g., DNA/RNA enzymes, metabolic proteins), and SignalP analysis was used to identify signal peptidase cleavage sites. As a result, 11 putative OMPs were identified and named according to their orthologs as follows: Pal (FTT0842), peptidoglycan-associated lipoprotein; TolC-A (FTT1095c) and TolC-B (FTT1724c), both orthologs of the outer membrane efflux protein TolC; PilQ (FTT1156c), type IV pilin subunit; SilC (FTT1258), outer membrane efflux protein that confers resistance to silver cations; FtaG (FTT1573c), outer membrane surface antigen; FopA (FTT0583), Francisella outer membrane protein; Mip (FTT1043), macrophage infectivity potentiator; YapH-N (FTT0918), N-terminal portion orthologous to Yersinia autotransporter protein; YapH-C (FTT0919), C-terminal portion orthologous to Yersinia autotransporter; YapH-LVS (FTL_0439), LVS ortholog of the Yersinia autotransporter that is the result of an in-frame deletion of the C-terminal end of YapH-N and the N-terminal end of YapH-C.
In gram-negative bacteria, proteins with eight or more beta-strands typically form beta-barrel transmembrane structures, which play pivotal roles in molecule transport and outer membrane integrity (48). The beta-barrel prediction program PROFtmb identified a total of 18 proteins (data not shown) having more than eight predicted transmembrane beta-barrel strands and z-values of greater than 6 (40% accuracy). As noted for the PSORTb analysis, the PROFtmb results were further subjected to BLAST and SignalP analyses, resulting in a total of seven proteins that were possible beta-barrel OMPs (Table 1). Of these, five overlapped with the PSORTb analysis (FtaG, FopA, YapH-N, YapH-C, and YapH-LVS) while two were newly identified as putative OMPs. The two additional putative OMPs were named according to their orthologs as follows: Cht1 (FTT0715), chitinase family 18 protein; SrfA (FTT0025c), outer membrane surface antigen. Upon further examination, PSORTb predicted that SrfA was an IMP (score, 9.46) but was not able to predict a cellular location for Cht1. The BOMP beta-barrel prediction program selected 10 putative beta-barrel proteins from Schu S4 ORFs (scores > 2; 40% accuracy) (data not shown), with three being selected after manual BLAST and SignalP screening (Table 1): FtaG, YapH-N, and SrfA. These three BOMP-selected putative OMPs overlapped with the PROFtmb results. Additionally, TolC-A, FopA, YapH-C, and YapH-LVS were also selected by BOMP, but their scores and accuracy values were below the threshold for beta-barrel assignment (data not shown). As a final beta-barrel OMP predictor, TMB-Hunt identified 54 proteins (log probability > 2) (data not shown), 7 of which were selected after manual BLAST and SignalP screening: PilQ, FtaG, FopA, YapH-N, YapH-C, YapH-LVS, and SrfA. Taken together, FtaG, YapH-N, and SrfA were identified by all three beta-barrel prediction algorithms, with FopA, YapH-C, and YapH-LVS being identified by two of three programs.
The Schu S4 and LVS genomes were each found to contain two TolC paralogs, TolC-A and TolC-B, which were predicted to be putative OMPs by PSORTb. This apparent duplication is not unusual among gram-negative bacteria, as the E. coli genome encodes TolC and three paralogs (23). Manual sequence comparisons revealed that the F. tularensis TolC-A and TolC-B proteins share only 21% amino acid identity (data not shown). Separately, TolC-A shares 22% amino acid identity with TolC orthologs from Vibrio, Pseudomonas, and Legionella, while TolC-B shares 27% amino acid identity with TolC orthologs from Legionella, E. coli, Salmonella, and Yersinia.
Two TUL4 paralogs were identified in both the Schu S4 and the LVS genomes by manual BLAST analysis and named Tul4-A (FTT0901) and Tul4-B (FTT0904). Neither Tul4-A nor Tul4-B deduced polypeptides were determined to be an integral OMP by PSORTb or by the three beta-barrel predictors (Table 1). The TUL4 paralogs share only 32% amino acid identity, with Tul4-B likely being the 17-kDa T-lymphocyte-reactive protein (41), based upon sequence homology (45) and predicted molecular mass (Table 1). Nevertheless, both proteins were selected for further analysis based upon previous reports of outer membrane localization of TUL4 (22).
Through manual BLAST analysis, an F. tularensis lactamutilization protein B (LamB) ortholog was identified with 35% amino acid identity to its E. coli counterpart. PSORTb identified the F. tularensis LamB as a cytoplasmic protein (score, 8.96) with no predicted beta-barrel transmembrane regions (Table 1).
Lipoproteins tend to be useful reagents for serological testing, inasmuch as their covalently attached fatty acids help to induce strong antibody responses (40). The lipoprotein prediction program LipoP was utilized to identify putative lipoproteins from the 16 selected putative OMPs (Table 1). Of those, Pal, Mip, YapH-N, YapH-LVS, Tul4-A, and Tul4-B had very strong lipoprotein signatures (log-odds cleavage scores > 15) (data not shown), whereas TolC-A and SilC had weaker lipoprotein signatures (log-odds cleavage scores < 10). Manual analysis of the N-terminal amino acid sequences of TolC-A and SilC did not reveal conserved lipobox sequences (1, 21, 53), suggesting that these two putative OMPs are not lipoproteins.
The PSORTb bacterial subcellular localization prediction program was also used to putatively identity IM and cytoplasmic proteins from F. tularensis. Of those with high IM and cytoplasmic scores, manual BLAST homology searches were performed to screen for candidate proteins. Two IMPs, SecY and LolC, and two cytoplasmic proteins, EF-Tu and IglA, were selected as likely non-OMP controls (Table 1).
Antisera specificities.
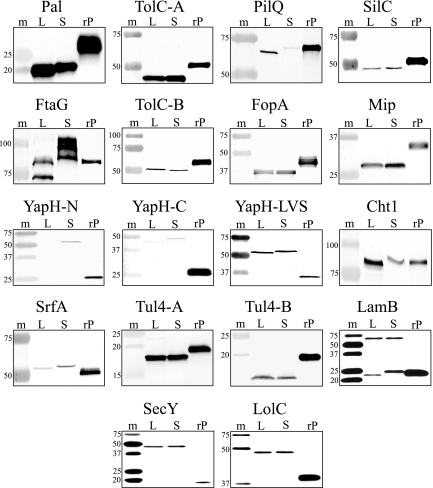
To generate antibody reagents for the evaluation of OM enrichment and separation techniques, we expressed and purified 16 putative OMPs and 2 IMPs as recombinant proteins in E. coli. Polyclonal antisera were generated against each of the purified proteins, and immunoblotting was employed to evaluate the specificities of the antisera for whole-cell lysates of LVS, Schu S4, and the respective recombinant protein (Fig. 1). Antisera reactivity to homologous proteins found in both LVS and Schu S4 lysates verified the high degree of sequence homology that was noted in bioinformatic analyses and, additionally, validated that the predicted LVS and Schu S4 coding sequences were expressed by each strain. Some minor differences were observed in the apparent molecular weights between putative OMPs from LVS and Schu S4 (e.g., Pal, Cht1, and SrfA) (Fig. 1), despite 99% protein identity. These differences are likely due to any number of well-known anomalies that can influence the mobilities of OMPs in SDS-PAGE gels (24). The majority of recombinant OMPs had higher molecular masses, compared to their LVS and Schu S4 counterparts (Fig. 1), due to the His6, spacer region, and TEV protease cleavage site that were fused to each protein. However, smaller recombinant proteins were evident for YapH-N, YapH-C, and YapH-LVS (Fig. 1), because truncated hydrophilic portions of each protein were cloned and expressed in E. coli (to simplify recombinant protein expression and purification).
FIG. 1.
Antisera generated in this study to probe for putative OMPs and IMPs. Proteins were separated by SDS-PAGE and transferred to nitrocellulose. m, prestained molecular mass standards with sizes (in kDa) noted on the left side of each immunoblot; L, whole-cell lysate of F. tularensis LVS; S, whole-cell lysate of F. tularensis Schu S4; rP, purified recombinant His6 fusion protein. Immunoblot assays were performed by probing with antisera against putative OMPs (Pal, TolC-A, PilQ, SilC, FtaG, TolC-B, FopA, Mip, YapH-N, YapH-C, YapH-LVS, Cht1, SrfA, Tul4-A, Tul4-B, and LamB) and IMPs (SecY and LolC).
The LVS and Schu S4 coding regions for PilQ share 96.8% amino acid identity, which is unexpectedly low compared to other putative F. tularensis OMPs. There are a total of 19 amino acid changes between the LVS and Schu S4 PilQ proteins, but the most significant difference is the lack of a computer-predicted signal peptidase I sequence cleavage site at the N terminus of the Schu S4 PilQ protein. As a result, the mature Schu S4 PilQ protein is predicted to be 3.3 kDa larger than its LVS homolog. This size disparity is accurately reflected by the larger Schu S4 PilQ protein weakly detected by the PilQ antiserum (Fig. 1). Despite equal protein loading for both LVS and Schu S4 lysates, the Schu S4 PilQ appears to be expressed at lower levels compared to its LVS homolog, at least under the culture conditions employed.
Immunoblot analysis demonstrated that FtaG antiserum reacted with an 88-kDa recombinant FtaG protein (Fig. 1), but two proteins were recognized in the LVS lysate and four proteins were recognized in the Schu S4 lysate. The slower-migrating polypeptide in the LVS lysate appears to be the same mass as the recombinant FtaG, while the faster-migrating protein in the Schu S4 lysate is likely the correct size. The small difference between the mobilities of the larger LVS protein and smaller Schu S4 protein is likely due to SDS-PAGE anomalies of unknown origin, inasmuch as the two proteins are 99% identical (data not shown). Antibody reactivities to the smallest (ca. 70 kDa) and largest (95 to 105 kDa) polypeptides in LVS or Schu S4, respectively, remain unexplained at this time.
YapH-N and YapH-C are adjacent Schu S4 ORFs that share a common LVS homolog, albeit with less than 50% amino acid identity among the three proteins. Manual sequence alignment and analysis suggested that the LVS homolog, YapH-LVS, is an in-frame fusion of the N-terminal portion of the Schu S4 YapH-N and the C-terminal protein of the Schu S4 YapH-C, due to an apparent deletion of the C-terminal 251 amino acids of YapH-N and the N-terminal 227 amino acids of YapH-C. Antisera generated against the unique regions of YapH-N and YapH-C reacted specifically, albeit weakly, with proteins in Schu S4, but not in LVS lysates (Fig. 1). Antiserum against YapH-LVS, however, recognized a single protein in both LVS and Schu S4 lysates. The size of the reactive protein in Schu S4 lysates (ca. 55 kDa) suggests that YapH-LVS antiserum only reacts with YapH-N and not YapH-C, likely due to the intended exclusion of the C-terminal hydrophobic region of YapH-LVS during cloning.
Both monomeric (23-kDa) and trimeric (69-kDa) forms of LamB were observed in LVS and Schu S4 lysates (Fig. 1), suggesting that the F. tularensis LamB might form trimers that were difficult to denature or separate by SDS-PAGE (Fig. 1).
Antisera were also generated against the putative F. tularensis IMPs SecY and LolC as additional markers to evaluate cell fractionation experiments (below). Because the complete coding sequences for both SecY and LolC were difficult to express and purify as recombinant proteins in E. coli (data not shown), alternative cloning approaches were employed. For LolC, a truncated hydrophilic region was cloned, expressed, and purified for antiserum generation (Fig. 1). For SecY, hydrophilic epitopes (8 to 12 amino acids) were selected and assembled into a chimeric protein by gene building, and the chimeric protein was expressed in E. coli and purified for antiserum production (Fig. 1).
Enrichment and isolation of OMPs.
In gram-negative bacteria, the OM can be separated from the IM based on differences in buoyant densities, believed to be predicated largely on the presence of lipopolysaccharide (LPS) in the OM (24, 28). The unusual lipid A structure of the F. tularensis LPS (52), however, raised doubts as to whether a sucrose gradient centrifugation approach could be applied for the separation of F. tularensis OMs and IMs. Typically, OMs are found within densities between 1.21 and 1.24 g/ml, whereas IMs display densities from 1.14 to 1.16 g/ml (28, 30). Separation of relatively pure OM and IM fractions is highly dependent on the integrity of spheroplasting/lysis procedures, which usually need to be determined empirically. We evaluated a number of methods to generate spheroplasts from F. tularensis, including sonication, high-pressure lysis using EmulsiFlex-C5 (Avestin, Ottawa, Canada), and osmotic lysis. Sonication and high-pressure lysis resulted in poor or incomplete separation of F. tularensis OMs and IMs (data not shown), likely due to the well-documented formation of mixed-membrane fusions using these procedures (28, 49). By comparison, osmotic lysis resulted in optimal F. tularensis spheroplast generation, as indicated by the near-complete separation of putative OMPs and IMPs by sucrose density gradient centrifugation (Fig. 2 and 3). Although our modified osmotic lysis procedure (see Materials and Methods) reproducibly generated relatively pure OMs, a number of parameters were found to be sensitive to experimental variation. (i) F. tularensis broth cultures were lightly inoculated for overnight incubation, and cells were harvested only when densities were less than an OD600 of 0.3, as densities greater than an OD600 of 0.4 yielded mixed-membrane fusions. (ii) Gentle mixing of the cell suspension was necessary during osmotic lysis to avoid local concentrations of solutions; vigorous stirring or frothing at any step resulted in mixed-membrane fusions. (iii) In contrast to the classical osmotic lysis protocol (30), we obtained higher total membrane yields and less mixed membranes when sucrose-suspended F. tularensis was treated with EDTA before the addition of lysozyme. (iv) Concentrations of lysozyme less than 200 μg/ml resulted in lower total membrane yields. (v) Washing or vigorous suspension of total membrane pellets after ultracentrifugation resulted in mixed-membrane fusions. (vi) The addition of nucleases and MgCl2 to the ultracentrifugation suspension buffer was necessary to degrade large amounts of genomic DNA that copelleted with the total membranes. DNA contamination caused total membrane suspensions to be extremely viscous, which disrupted sucrose gradient buoyancy. (vii) The addition of EDTA-free protease inhibitors to the ultracentrifugation suspension buffer was also necessary, as a dramatic loss in protein yield occurred without them, likely due to protease activation by MgCl2.
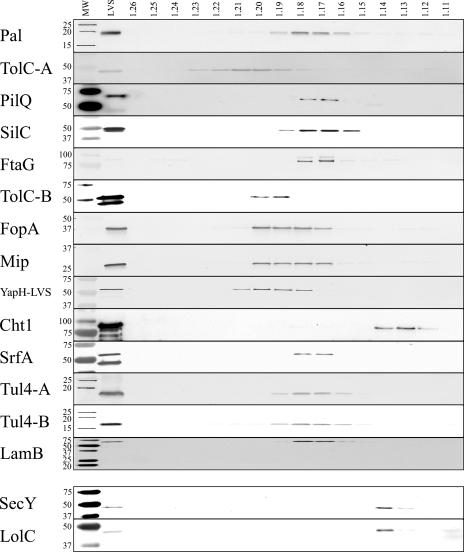
FIG. 2.
Immunoblotting of F. tularensis LVS sucrose density gradients. Sequential fractions were collected from gradients, and densities (in g/ml) were calculated based upon refractive indices. Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted to detect OMP and IMP fractionation. MW, prestained molecular mass standards with sizes (in kDa) noted on the left side of each blot; LVS, whole-cell lysates of F. tularensis LVS. The corresponding sucrose gradient fraction densities are noted above their respective lanes. OMPs and IMPs, as noted in the left margin, were detected with polyclonal, monospecific antisera.
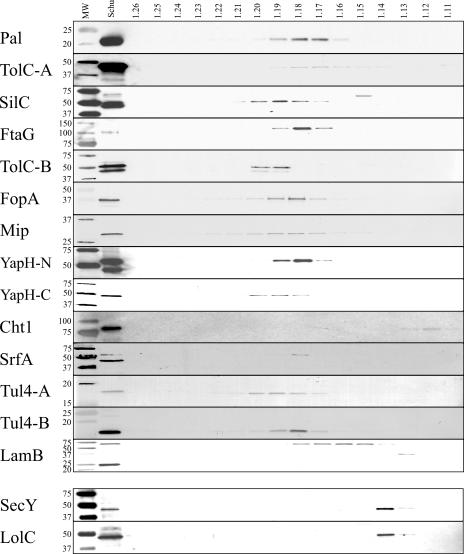
FIG. 3.
Immunoblotting of F. tularensis Schu S4 sucrose density gradients. Sequential fractions were collected from gradients, and densities (in g/ml) were calculated based upon refractive indices. Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted to detect OMP and IMP fractionation. MW, prestained molecular mass standards with sizes (in kDa) noted on the left side of each blot; Schu, whole-cell lysates of F. tularensis Schu S4. The corresponding sucrose gradient fraction densities are noted above their respective lanes. OMPs and IMPs, as noted in the left margin, were detected with polyclonal, monospecific antisera.
Separation of F. tularensis OMs from IMs was accomplished by osmotic lysis and sucrose density gradient centrifugation. Proteins from gradient fractions (densities from 1.11 g/ml to 1.26 g/ml, at 0.01 increments) were separated by SDS-PAGE and visualized by silver stain or transferred to nitrocellulose for immunoblotting with specific antisera. Silver stain visualization of individual sucrose gradient fractions confirmed that different protein subsets were separated and enriched across sucrose gradients (data not shown). The immunoblot results presented here (Fig. 2 and 3) are representative data from a series of LVS and Schu S4 sucrose gradients. The majority of putative OMPs were detected between densities of 1.17 and 1.20 g/ml for both LVS (Fig. 2) and Schu S4 (Fig. 3), while putative IMPs were detected between densities of 1.13 and 1.14 g/ml for both LVS and Schu S4 (Fig. 2 and 3). Whereas OMPs and IMPs separate in sucrose density gradients based upon the densities of their associated membranes, cytoplasmic (water-soluble) proteins are not membrane tethered and, thus, diffuse equally in soluble form across the gradients. Thus, as expected, cytoplasmic proteins EF-Tu and IglA were detected equally across all gradient fractions (data not shown). Taken together, the immunoblot data demonstrate that Pal, TolC-A, SilC, FtaG, TolC-B, FopA, Mip, SrfA, Tul4-A, Tul4-B, and LamB are OMPs in both LVS and Schu S4. Additionally, the three YapH proteins (LVS YapH-LVS [Fig. 2] and Schu S4 YapH-N and YapH-C [Fig. 3]) colocalized with other OMPs from their respective F. tularensis subspecies.
The LVS TolC-A was detected at densities from 1.18 to 1.23 g/ml (Fig. 2), whereas the Schu S4 TolC-A was detected at densities from 1.16 to 1.19 g/ml (Fig. 3). While both density ranges overlapped with other F. tularensis OMPs, we cannot account for the observed difference in TolC-A membrane localization between LVS and Schu S4. It is worth noting, however, that of the 16 putative OMPs and 2 IMPs examined in this study, TolC-A densities were the most difficult to reproduce among experiments (data not shown). The LVS PilQ colocalized with other F. tularensis OMPs (densities between 1.17 and 1.18 g/ml Fig. 2), indicating that it likely is an OMP. The Schu S4 PilQ, however, was not detectable in Schu S4 sucrose gradient fractions (data not shown). The lack of PilQ detection from Schu S4 sucrose gradient fractions was likely due to low expression levels, as noted earlier (Fig. 1). Only a trimeric (69 kDa) form of LamB was detected in LVS and Schu S4 sucrose gradient fractions (Fig. 2 and 3). While LamB was detected at OM densities of 1.17 to 1.18 g/ml for both LVS and Schu S4, the Schu S4 LamB also migrated to densities between 1.15 and 1.16 g/ml (Fig. 3). At this time, we cannot account for this difference in the Schu S4 LamB. Finally, immunoblots against Cht1 detected the 81-kDa protein at densities between 1.12 and 1.14 in both LVS (Fig. 2) and Schu S4 (Fig. 3). When compared to the localization of IMPs SecY and LolC (densities between 1.13 and 1.14 g/ml) (Fig. 2 and 3), membrane separation results suggest that Cht1 is an IMP.
OMP antigenicity and identification.
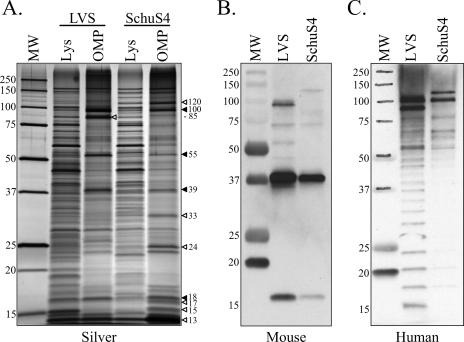
F. tularensis membrane separation and enrichment was directly visualized by comparing whole-cell lysates with pooled sucrose gradient fractions containing IMPs (1.13 to 1.14 g/ml) and OMPs (1.17 to 1.20 g/ml) from both LVS and Schu S4. Distinct protein subsets were observed when IM and OM fractions were compared (data not shown), which further demonstrated the efficacy of sucrose density gradients for the separation of the two membrane compartments and their associated proteins. A more comprehensive analysis of sucrose gradient membrane separation and enrichment was performed by comparing bacterial lysates with OMPs from both LVS and Schu S4 (Fig. 4A). Most noteworthy was the parallel enrichment of 18-, 39-, 55-, and 100-kDa proteins in both LVS and Schu S4 OM fractions, which emphasized the utility of this technique in both Francisella subspecies (Fig. 4A). Differences in LVS and Schu S4 OMP enrichment were also observed, including greater enrichment of 13-, 15-, 17-, 24-, 33-, and 120-kDa OMPs from Schu S4 (Fig. 4A) compared to enhanced amounts of an 85-kDa OMP from LVS (Fig. 4A). When OMPs from LVS and Schu S4 were probed with sera from F. tularensis-infected mice, similar immunoreactive proteins were observed, including highly reactive 17-kDa and 39-kDa proteins (Fig. 4B). Reactivity was also observed, although to a lesser extent, against an 85-kDa LVS OMP, but no 85-kDa protein was detected in Schu S4 OMPs (Fig. 4B). The mouse antisera reactivities observed at 39 kDa (LVS and Schu S4 OMPs) and 85 kDa (LVS OMPs alone) (Fig. 4B) correlated with the OMP enrichment observed by silver staining (Fig. 4A). Unlike mouse sera, sera from LVS-vaccinated humans displayed a ladder-like pattern of immunoreactivity against both LVS and Schu S4 OMPs (representative human serum immunoblot displayed in Fig. 4C). This pattern is typical of antibody reactivity to LPS, which has been previously noted for hyperimmune sera to Francisella (7). Indeed, each human serum examined in this study gave similar ladder-like reactivity (data not shown). Nevertheless, three immunodominant proteins were commonly detected by human antisera, including 85- and 100-kDa proteins in LVS OMPs and 100- and 120-kDa proteins in Schu S4 OMPs (Fig. 4C). In addition to being highly immunogenic, these high-molecular-mass proteins were substantially enriched in OM fractions (Fig. 4A).
FIG. 4.
SDS-PAGE and immunoblot analysis of F. tularensis OMPs. (A) Silver stain comparison of LVS and Schu S4 whole-cell lysates (Lys) with sucrose gradient-enriched OMPs. MW, molecular mass standard with sizes (in kDa) noted on the left side of the gel. OMPs that were significantly enriched in both LVS and Schu S4 are indicated by solid arrowheads. OMPs that were enriched in only one subspecies are indicated by hollow arrowheads. The estimated molecular mass (in kDa) of each enriched OMP is indicated to the right of the arrowhead. (B) Immunoreactivity of LVS and Schu S4 OMPs with pooled sera from C3H/HeN mice sequentially infected with F. tularensis LVS and then Schu S4. (C) Representative immunoreactivity of LVS and Schu S4 OMPs with serum from an F. tularensis LVS-vaccinated human.
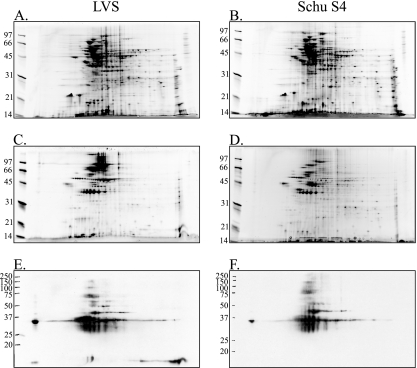
Because conventional SDS-PAGE is limited in protein separation capabilities, we utilized 2DE to more completely characterize and define the repertoire of enriched OMPs from LVS and Schu S4. The enrichment of F. tularensis OMs by sucrose density gradient centrifugation was directly visualized by comparing whole-cell lysates of LVS (Fig. 5A) and Schu S4 (Fig. 5B) with enriched OMPs from LVS (Fig. 5C) and Schu S4 (Fig. 5D) by 2DE. As similarly noted for the conventional SDS-PAGE visualizations (Fig. 4A), a subset of proteins was enriched in OMP-containing fractions (Fig. 5C and D), further indicating OM separation and enrichment.
FIG. 5.
2DE of F. tularensis proteins. Proteins were focused in 11-cm IPG strips and separated on 12.5% SDS-PAGE gels. Molecular mass sizes (in kDa) are noted on the left side of each gel. (A to D) Gels were stained with SYPRO Ruby, and proteins were visualized by excitation at 300 nm. (A and B) Whole-cell lysates of LVS (A) and Schu S4 (B). (C and D) Sucrose gradient-enriched OMPs from LVS (C) and Schu S4 (D). (E and F) Immunoreactivity of sucrose gradient-enriched OMPs from LVS (E) and Schu S4 (F) using pooled sera from C3H/HeN mice sequentially infected with F. tularensis LVS and then Schu S4.
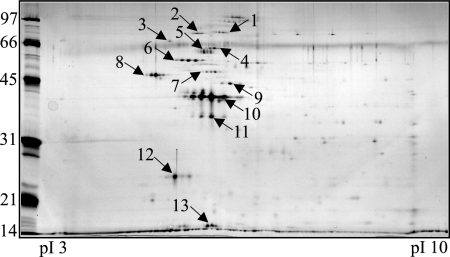
A group of proteins was excised from LVS OMP 2DE gels (Fig. 6) and identified by mass spectrometry (Table 2). Mass spectrometric protein identifications were also confirmed from Schu S4 2DE gels (data not shown). Of the 13 proteins identified by mass spectrometry, 5 were predicted to be OMPs (KatG, PilQ, YapH-LVS, FopA, and Pal), 2 were predicted to be cytoplasmic proteins (pyruvate dehydrogenase and GroEL), 1 was predicted to be a periplasmic protein (DnaK), and 4 were of unknown cellular localization (ATP synthase, lipoprotein FTT1103, OmpA, and Tul4-A) by PSORTb analysis (Table 2). A comparison of 2DE-identified F. tularensis proteins (Fig. 6 and Table 2) with 2DE immunoblot analysis using pooled mouse sera (Fig. 5E and F) indicated that FopA, OmpA, ATP synthase, PilQ, GroEL, KatG, and Tul4-A are immunogenic during mouse infections.
FIG. 6.
Representative 2DE of pooled F. tularensis LVS OMPs from sucrose gradient fractions. Gels were stained with SYPRO Ruby for visualization, and select protein spots (numbered) were excised for mass spectrometry identification. Protein identities are listed in Table 2.
TABLE 2.
F. tularensis LVS proteins identified by 2DE and mass spectrometry
| Spot no.a | Massb | Protein name | Locusc | PSORTb localizationd | PSORTb score |
|---|---|---|---|---|---|
| 1 | 100.2 | Pyruvate dehydrogenase | FTT1485c | Cyto | 8.96 |
| 2 | 82.4 | Catalase-peroxidase KatG | FTT0721c | OM | 9.52 |
| 3 | 69.2 | Chaperone protein DnaK | FTT1269c | Peri | 9.83 |
| 4 | 64.9 | Type IV pilin subunit PilQ | FTT1156c | OM | 10.00 |
| 5 | 57.4 | Heat shock protein GroEL | FTT1696 | Cyto | 9.26 |
| 6 | 55.8 | YapH-LVS | FTL_0439 | OM | 9.52 |
| 7 | 49.8 | ATP synthase beta chain | FTT0064 | Un | 2.00 |
| 8 | 38.7 | Conserved hypothetical lipoprotein | FTT1103 | Un | 2.00 |
| 9 | 46.7 | OmpA family protein | FTT0831c | Un | 2.00 |
| 10 | 41.4 | FopA | FTT0583 | OM | 9.93 |
| 11 | 41.4 | FopA | FTT0583 | OM | 9.93 |
| 12 | 23.2 | Peptidoglycan-associated lipoprotein Pal | FTT0842 | OM | 10.00 |
| 13 | 15.8 | Tul4-A | FTT0901 | Un | 2.00 |
Protein spots were excised from 2DE gels and submitted for mass spectrometric protein identification. A representative 2DE image is shown in Fig. 6.
Mass spectrometry-calculated molecular mass (in kDa).
Locus numbers are listed for the Schu S4 homologs, except for YapH-LVS.
The PSORTb bacterial sublocalization prediction program was utilized to predict protein localization for cytoplasm (Cyto), periplasm (Peri), outer membrane (OM), or unknown (Un).
DISCUSSION
To date, only two F. tularensis OMPs have been described: FopA and TUL4 (22, 27). This study describes, for the first time, the physical separation and enrichment of F. tularensis OMs from other cellular components for the identification of additional OMPs and LPs from both LVS and Schu S4. Of the 16 putative OMP and LP coding regions preliminarily identified by bioinformatic analyses (Table 1), 15 were ultimately confirmed as OMPs by sucrose density gradient centrifugation (Fig. 2 and 3). Additionally, SecY and LolC were confirmed as IM-localized proteins (Fig. 2 and 3). PSORTb was the most accurate computational program examined in this study, with 11 of 11 predicted OMPs (Table 1) localizing in sucrose gradient-derived OM fractions from both LVS and Schu S4 (Fig. 2 and 3). However, PSORTb failed to select Tul4-A or Tul4-B as OMPs and, further, designated LamB as a cytoplasmic protein. As demonstrated by immunoblot analysis of sucrose gradient fractions, all three of these proteins colocalized with other F. tularensis OMPs (Fig. 2 and 3), emphasizing that computational algorithms are not unequivocal for predicting OMPs. On average, putative beta-barrel OMPs were identified by two of three beta-barrel prediction programs (Table 1), all of which overlapped with PSORTb OMP predictions. Cht1, which received a significant beta-barrel score by PROFtmb, but for which localization could not be predicted by PSORTb, did not fractionate with other F. tularensis OMPs (Fig. 2 and 3). This suggests that the three beta-barrel prediction algorithms utilized here are not as well developed as PSORTb for identifying OMPs. Further, while these results demonstrate a general utility of computational prediction programs, such programs do not substitute for direct experimental evidence. Given the lack of available information about the F. tularensis OM, our two-pronged, complementary approach was strategic for initially identifying candidate F. tularensis OMPs. This led to the generation of antibody reagents to assess membrane separations and, thus, to further evaluate bioinformatic prediction programs.
The data presented in this study clearly demonstrate the utility of osmotic lysis and sucrose density gradient centrifugation for the separation of OMs from IMs in F. tularensis (Fig. 2 and 3). While our optimized method was rigorous and subject to some experimental variability (see Results), our results appear to have much greater utility than those garnered from the use of detergents, lithium chloride, or sodium carbonate (2, 20, 33, 41, 46, 51). We identified 15 bona fide F. tularensis OMPs, which colocalized at densities between 1.17 and 1.20 g/ml. However, these OM densities were slightly lower than the OM densities of 1.21 to 1.24 g/ml noted for other gram-negative bacteria (28, 30). Because buoyant densities of OMs in sucrose solutions are believed to be influenced by LPS, the observed shift of F. tularensis OM densities, relative to those of other gram-negative bacteria, was likely due, at least in part, to the unusual nature of the F. tularensis LPS (35, 52). Despite the lighter densities of F. tularensis OMs, their separation and colocalization from 1.17 to 1.20 g/ml (Fig. 2 and 3) is distinct from that of F. tularensis IMs, which had densities of 1.13 to 1.14 g/ml (Fig. 2 and 3).
Lipoproteins, in addition to being highly immunogenic, are believed to play crucial roles in gram-negative bacterial pathogenesis and immune evasion (55). Of the 15 OMPs characterized in this study, the LipoP prediction program identified 6 as likely lipoproteins (Table 1). Whereas we did not provide direct experimental evidence for lipid modification of this protein subset, BLAST homology analysis indicated that F. tularensis contains a complete Lol system, including inner membrane ABC transport proteins LolC (FTT0404) and D (FTT0405), lipid modification enzymes Lgt (FTT1228), LspA (FTT0914c), and Lnt (FTT0614c), periplasmic chaperone LolA (FTT1636), and outer membrane-associated LolB (FTT0270). Given the previously reported lipidation of Tul4-B (46) and the confirmed IM localization of LolC (Fig. 2 and 3), it is plausible that the Lol system is fully functional in F. tularensis. Of course, the presence of a functional Lol system to shuttle lipoproteins across the periplasm does not provide insights into whether any lipoproteins are subsequently translocated to the outer leaflet of the F. tularensis OM. Future studies are planned not only to verify lipoprotein identities (via [3H]palmitate radiolabeling experiments), but also to discern their possible surface topologies.
Little is known about the function of the majority of F. tularensis OMPs identified in this study. However, a recently published report demonstrated the involvement of both F. tularensis TolC paralogs in LVS multidrug resistance (15). Although deletion mutants of tolC-A and tolC-B were still able to replicate in macrophages, the tolC-B mutant was attenuated for virulence in mice and thus has been implicated as a virulence factor. Based upon sequence homology and the findings noted above, that study proposed that the TolC paralogs may be located in the OM. Our data provide direct experimental evidence that both TolC paralogs are OMPs (Fig. 2 and 3). In light of these findings, it will be of interest to determine whether the Schu S4 TolC paralogs perform similar antibiotic resistance and virulence-associated functions.
A type 4 pilus gene cluster, including a gene encoding PilQ, was reported for both type A and type B Francisella (14). Expression of multiple LVS pilin subunits was confirmed by reverse transcription-PCR and electron microscopic visualization of long, polar fibers extending from the surface of LVS. More recently, an alternative function for PilQ was proposed, as pilin fibers could not be visualized from Francisella novicida, and an F. novicida pilQ mutant was unable to secrete proteins into culture supernatants (17). That study proposed that, at least for F. novicida, PilQ forms part of an OM secretion system. Given these apparent differences in F. novicida and LVS pilin function, the roles of the LVS and Schu S4 PilQ proteins need to be more thoroughly explored. Indeed, this study demonstrated that Schu S4 PilQ expression was dramatically reduced, compared to LVS (Fig. 1). Given our results, it is thus possible that PilQ functions in different capacities in LVS and Schu S4.
A recently described spontaneous Schu S4 mutant was examined by 2DE and found to be deficient in expression of eight proteins, including YapH-N (FTT0918) and YapH-C (FTT0919), compared to wild-type Schu S4 (50). When site-directed mutants were generated in that study, YapH-C deletion mutants remained virulent in a mouse infection model, whereas YapH-N deletion mutants were highly attenuated. Given that attenuated LVS encodes a hybrid protein that consists of the N-terminal portion of YapH-N and the C-terminal portion of YapH-C, it is likely that the C-terminal portion of YapH-N, or the complete YapH-N protein, contributes to virulence. Whereas that study suggested that YapH-N and YapH-C were possibly membrane-associated proteins, our results provided direct evidence of the OM localization of YapH-N and YapH-C from Schu S4 and YapH-LVS from LVS. Despite our suggested YapH nomenclature for this group of proteins, their classification is tentative and should be interpreted with caution. Indeed, no close homologs exist for either YapH-N or YapH-C, and thus the actual functions of these proteins remain to be determined.
Although we have not yet performed an exhaustive analysis of all the OMPs in sucrose gradient-enriched fractions, 2DE and mass spectrometry allowed us to identify the following dominant OMPs from F. tularensis LVS: PilQ, YapH-LVS, FopA, Pal, and Tul4-A (Table 2 and Fig. 6). Additionally, YapH-N and YapH-C were identified as bona fide OMPs from Schu S4 OM fractions (data not shown). While KatG (FTT0721c) had a high PSORTb OM localization score during our initial bioinformatic analysis (data not shown), we did not target it for our initial study. Given the identification of KatG from pooled OM fractions (Table 2) and its high PSORTb OM localization score, it is likely that KatG is an OMP. Our results are consistent with the previous identification of KatG as a putative OMP from F. tularensis extracts (33). Antibodies are currently being developed against KatG to verify its membrane localization in sucrose gradient fractions. A putative lipoprotein (FTT1103) and OmpA family protein (FTT0831c) were also identified from 2DE gels (Table 2 and Fig. 6), but bioinformatic analyses do not support their OM localization. Previous 2DE studies that used sodium carbonate to extract membrane proteins from F. tularensis also identified FTT1103 and FTT0831c (33, 51), and we plan to generate antibodies to confirm their OM localization. The presence of certain cytoplasmic (water-soluble) proteins, including pyruvate dehydrogenase (FTT1485c) and GroEL (FTT1696), was not surprising in that other cytoplasmic proteins were uniformly distributed across the sucrose gradients (data not shown) and thus were difficult to exclude from OM fractions. However, given the relative number of cytoplasmic contaminants compared to OMPs (Table 2), the utility of osmotic lysis and sucrose density gradient centrifugation is evident. Previous membrane enrichment procedures extracted hundreds of proteins, but relatively few were proposed to be OMPs (33, 51).
A comparison of 2DE immunoblots with mass spectrometry identification revealed that KatG, PilQ, GroEL, ATP synthase, OmpA, FopA, and Tul4-A induce antibody production during the infection process in C3H/HeN mice (Fig. 5 and 6; Table 2). By comparison, the majority of antibody induced in humans vaccinated with LVS was generated against LPS (Fig. 4C). These divergent immune responses are difficult to discern, as C3H/HeN mice are capable of generating an immune response against LPS (36). Our results confirm similar reactivities previously reported for mouse sera against KatG, GroEL, and FopA (19). Currently, we are in the process of optimizing our 2DE of OMPs from both LVS and Schu S4 for continued mass spectrometry identification of the complete repertoire of F. tularensis OMPs. Additionally, we are generating OMP deletion mutants in F. tularensis to assist in evaluating their surface exposure and contribution to cell entry and intracellular survival, as well as to assess their potential roles in virulence.
Supplementary Material
Acknowledgments
Funding for this work was provided by grant P01-AI055637 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank Richard Scheuermann and Shubhada Godbole for assistance with bioinformatic analyses and Rachel Blackwell for technical assistance. We are indebted to Karen Elkins and Francis Nano for advice on Francisella sp. strains and cultivation and to J. Wayne Conlan for advice on SchuS4 and for providing draft standard operating procedures for BSL3 activities.
Footnotes
Published ahead of print on 17 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Babu, M. M., M. L. Priya, A. T. Selvan, M. Madera, J. Gough, L. Aravind, and K. Sankaran. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188:2761-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevanger, L., J. A. Maeland, and A. I. Naess. 1988. Agglutinins and antibodies to Francisella tularensis outer membrane antigens in the early diagnosis of disease during an outbreak of tularemia. J. Clin. Microbiol. 26:433-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Casadevall, A. 1998. Antibody-mediated protection against intracellular pathogens. Trends Microbiol. 6:102-107. [DOI] [PubMed] [Google Scholar]
- 5.Casadevall, A. 2003. Antibody-mediated immunity against intracellular pathogens: two-dimensional thinking comes full circle. Infect. Immun. 71:4225-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassataro, J., S. M. Estein, K. A. Pasquevich, C. A. Velikovsky, S. de la Barrera, R. Bowden, C. A. Fossati, and G. H. Giambartolomei. 2005. Vaccination with the recombinant Brucella outer membrane protein 31 or a derived 27-amino-acid synthetic peptide elicits a CD4+ T helper 1 response that protects against Brucella melitensis infection. Infect. Immun. 73:8079-8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlan, J. W., H. Shen, A. Webb, and M. B. Perry. 2002. Mice vaccinated with the O-antigen of Francisella tularensis LVS lipopolysaccharide conjugated to bovine serum albumin develop varying degrees of protective immunity against systemic or aerosol challenge with virulent type A and type B strains of the pathogen. Vaccine 20:3465-3471. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDad, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russel, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 9.Elkins, K. L., S. C. Cowley, and C. M. Bosio. 2003. Innate and adaptive immune responses to an intracellular bacterium, Francisella tularensis live vaccine strain. Microbes Infect. 5:135-142. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ericsson, M., G. Sandstrom, A. Sjostedt, and A. Tarnvik. 1994. Persistence of cell-mediated immunity and decline of humoral immunity to the intracellular bacterium Francisella tularensis 25 years after natural infection. J. Infect. Dis. 170:110-114. [DOI] [PubMed] [Google Scholar]
- 12.Fulop, M., R. Manchee, and R. Titball. 1995. Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine 13:1220-1225. [DOI] [PubMed] [Google Scholar]
- 13.Fulop, M., R. Manchee, and R. Titball. 1996. Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS Immunol. Med. Microbiol. 13:245-247. [DOI] [PubMed] [Google Scholar]
- 14.Gil, H., J. L. Benach, and D. G. Thanassi. 2004. Presence of pili on the surface of Francisella tularensis. Infect. Immun. 72:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil, H., G. J. Platz, C. A. Forestal, M. Monfett, C. S. Bakshi, T. J. Sellati, M. B. Furie, J. L. Benach, and D. G. Thanassi. 2006. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. USA 103:12897-12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hager, A. J., D. L. Bolton, M. R. Pelletier, M. J. Brittnacher, L. A. Gallagher, R. Kaul, S. J. Skerrett, S. I. Miller, and T. Guina. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227-237. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, R. E., R. Siehnel, and N. Martin. 1990. Outer membrane proteins of Pseudomonas. Mol. Microbiol. 4:1069-1075. [DOI] [PubMed] [Google Scholar]
- 19.Havlasova, J., L. Hernychova, M. Brychta, M. Hubalek, J. Lenco, P. Larsson, M. Lundqvist, M. Forsman, Z. Krocova, J. Stulik, and A. Macela. 2005. Proteomic analysis of anti-Francisella tularensis LVS antibody response in murine model of tularemia. Proteomics 5:2090-2103. [DOI] [PubMed] [Google Scholar]
- 20.Hubálek, M., L Hernychová, J. Havlasová, I. Kasalová, V. Neubauerová, J. Stulík, A. Macela, M. Lundqvist, and P. Larsson. 2003. Towards proteome database of Francisella tularensis. J. Chromatogr. B 787:149-177. [DOI] [PubMed] [Google Scholar]
- 21.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khlebnikov, V. S., I. R. Golovliov, D. P. Kulevatsky, N. V. Tokhtamysheva, S. F. Averin, V. E. Zhemchugov, S. Y. Pchelintsev, S. S. Afanasiev, and G. Y. Shcherbakov. 1996. Outer membranes of a lipopolysaccharide-protein complex (LPS-17 kDa protein) as chemical tularemia vaccines. FEMS Immunol. Med. Microbiol. 13:227-233. [DOI] [PubMed] [Google Scholar]
- 23.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467-489. [DOI] [PubMed] [Google Scholar]
- 24.Lugtenberg, B., and L. Van Alphen. 1983. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim. Biophys. Acta 737:51-115. [DOI] [PubMed] [Google Scholar]
- 25.McSorley, S. J., and M. K. Jenkins. 2000. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar Typhimurium. Infect. Immun. 68:3344-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuno, T., and M. Kageyama. 1978. Separation and characterization of the outer membrane of Pseudomonas aeruginosa. J. Biochem. 84:179-191. [DOI] [PubMed] [Google Scholar]
- 27.Nano, F. E. 1988. Identification of a heat-modifiable protein of Francisella tularensis and molecular cloning of the encoding gene. Microb. Pathog. 5:109-119. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido, H. 1994. Isolation of outer membranes. Methods Enzymol. 235:225-234. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 30.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 31.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 212:967-978. [DOI] [PubMed] [Google Scholar]
- 32.Pal, S., E. M. Peterson, and L. M. de la Maza. 2005. Vaccination with the Chlamydia trachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteria. Infect. Immun. 73:8153-8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pávkova, I., M. Hubálek, J. Zechovská, J. Lenco, and J. Stulík. 2005. Francisella tularensis live vaccine strain: proteomic analysis of membrane proteins enriched fraction. Proteomics 5:2460-2467. [DOI] [PubMed] [Google Scholar]
- 34.Philipovskiy, A. V., C. Cowan, C. R. Wulff-Strobel, S. H. Burnett, E. J. Kerschen, D. A. Cohen, A. M. Kaplan, and S. C. Straley. 2005. Antibody against V antigen prevents Yop-dependent growth of Yersinia pestis. Infect. Immun. 73:1532-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, N. J., B. Schilling, M. K. McLendon, M. A. Apicella, and B. W. Gibson. 2004. Novel modification of lipid A of Francisella tularensis. Infect. Immun. 72:5340-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 37.Price, C. A. 1982. Centrifugation in density gradients, p. 335-343. Academic Press, New York, N.Y.
- 38.Reference deleted.
- 39.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez, P. J., G. H. McCracken, Jr., G. D. Wendel, K. Olsen, N. Threlkeld, and M. V. Norgard. 1989. Molecular analysis of the fetal IgM response to Treponema pallidum antigens: implications for improved serodiagnosis of congenital syphilis. J. Infect. Dis. 159:508-517. [DOI] [PubMed] [Google Scholar]
- 41.Sandström, G., A. Tärnvik, and H. Wolfe-Watz. 1987. Immunospecific T-lymphocyte stimulation by membrane proteins from Francisella tularensis. J. Clin. Microbiol. 25:641-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Schuch, R., and A. T. Maurelli. 1999. The mxi-Spa type III secretory pathway of Shigella flexneri requires an outer membrane lipoprotein, MxiM, for invasin translocation. Infect. Immun. 67:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjöstedt, A., G. Sandström, and A. Tärnvik. 1990. Several membrane polypeptides of the live vaccine strain Francisella tularensis LVS stimulate T cells from naturally infected individuals. J. Clin. Microbiol. 28:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sjöstedt, A., G. Sandström, A. Tärnvik, and B. Jaurin. 1990. Nucleotide sequence and T cell epitopes of a membrane protein of Francisella tularensis. J. Immunol. 145:311-317. [PubMed] [Google Scholar]
- 46.Sjöstedt, A., A. Tärnvik, and G. Sandström. 1991. The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect. Immun. 59:3163-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surcel, H.-M, M. Sarvas, I. M. Halander, and E. Herva. 1989. Membrane proteins of Francisella tularensis LVS differ in ability to induce proliferation of lymphocytes from tularemia-vaccinated individuals. Microb. Pathog. 7:411-419. [DOI] [PubMed] [Google Scholar]
- 48.Tamm, L. K., H. Hong, and B. Liang. 2004. Folding and assembly of beta-barrel membrane proteins. Biochim. Biophys. Acta 1666:250-263. [DOI] [PubMed] [Google Scholar]
- 49.Tsukagoshi, N., and C. F. Fox. 1971. Hybridization of membranes by sonic irradiation. Biochemistry 10:3309-3313. [DOI] [PubMed] [Google Scholar]
- 50.Twine, S., M. Bystrom, W. Chen, M. Forsman, I. Golovliov, A. Johansson, J. Kelly, H. Lindgren, K. Svensson, C. Zingmark, W. Conlan, and A. Sjostedt. 2005. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73:8345-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twine, S. M., N. C. S. Mykytczuk, M. Petit, T.-L. Tremblay, J. W. Conlan, and J. F. Kelly. 2005. Francisella tularensis proteome: low levels of ASB-14 facilitate the visualization of membrane proteins in total protein extracts. J. Proteome Res. 4:1848-1854. [DOI] [PubMed] [Google Scholar]
- 52.Vinogradov, E., M. B. Perry, and J. W. Conlan. 2002. Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 269:6112-6118. [DOI] [PubMed] [Google Scholar]
- 53.von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2:531-534. [DOI] [PubMed] [Google Scholar]
- 54.Winslow, G. M., E. Yager, K. Shilo, E. Volk, A. Reilly, and F. K. Chu. 2000. Antibody-mediated elimination of the obligate intracellular bacterial pathogen Ehrlichia chaffeensis during active infection. Infect. Immun. 68:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, H., D. W. Niesel, J. W. Peterson, and G. R. Klimpel. 1998. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 66:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.