Abstract
The 3′ cis-acting element for mouse hepatitis virus (MHV) RNA synthesis resides entirely within the 301-nucleotide 3′ untranslated region (3′ UTR) of the viral genome and consists of three regions. Encompassing the upstream end of the 3′ UTR are a bulged stem-loop and an overlapping RNA pseudoknot, both of which are essential to MHV and common to all group 2 coronaviruses. At the downstream end of the genome is the minimal signal for initiation of negative-strand RNA synthesis. Between these two ends is a hypervariable region (HVR) that is only poorly conserved between MHV and other group 2 coronaviruses. Paradoxically, buried within the HVR is an octanucleotide motif (oct), 5′-GGAAGAGC-3′, which is almost universally conserved in coronaviruses and is therefore assumed to have a critical biological function. We conducted an extensive mutational analysis of the HVR. Surprisingly, this region tolerated numerous deletions, rearrangements, and point mutations. Most striking, a mutant deleted of the entire HVR was only minimally impaired in tissue culture relative to the wild type. By contrast, the HVR deletion mutant was highly attenuated in mice, causing no signs of clinical disease and minimal weight loss compared to wild-type virus. Correspondingly, replication of the HVR deletion mutant in the brains of mice was greatly reduced compared to that of the wild type. Our results show that neither the HVR nor oct is essential for the basic mechanism of MHV RNA synthesis in tissue culture. However, the HVR appears to play a significant role in viral pathogenesis.
Coronaviruses are large, positive-strand RNA viruses with a reproductive cycle that involves both replication of genomic RNA (gRNA) and transcription of a 3′ nested set of subgenomic mRNAs (sgRNAs) (34). RNA sequences and structures that participate in these functions are embedded at various loci in the genome and are thought to interact with viral and host cellular components (2). The cis-acting elements that are required for coronavirus replication have been studied principally through the use of defective interfering (DI) RNAs, which are extensively deleted genomic remnants that parasitize the RNA synthetic machinery of a helper virus. Most DI RNA studies have been performed with mouse hepatitis virus (MHV) and bovine coronavirus (BCoV), closely related members of the second of the three phylogenetic groups into which coronaviruses are classified.
The 3′ end of the genome is the site of initiation of synthesis of negative-strand gRNA and sgRNA. Independent deletion analyses of MHV DI RNAs concluded that the minimal amount of the genomic 3′ terminus that can support RNA synthesis ranges between 378 and 462 nucleotides (nt) (23, 28, 32). This suggested that the 3′ genomic cis-acting element of MHV must encompass the entire 301-nt 3′ untranslated region (3′ UTR) as well as a portion of the adjacent nucleocapsid (N) gene. However, genetic manipulations of the intact viral genome have demonstrated that the 3′ UTR and the 3′ cis-acting signal for RNA synthesis are one and the same. The MHV N gene can be moved to a distant genomic site without affecting viral replication (10, 11). Additionally, the distal portion of the N gene tolerates considerable modification by point mutations, substitutions, and deletions (16, 19, 25, 42), and it is thus highly unlikely to harbor elements essential to RNA synthesis.
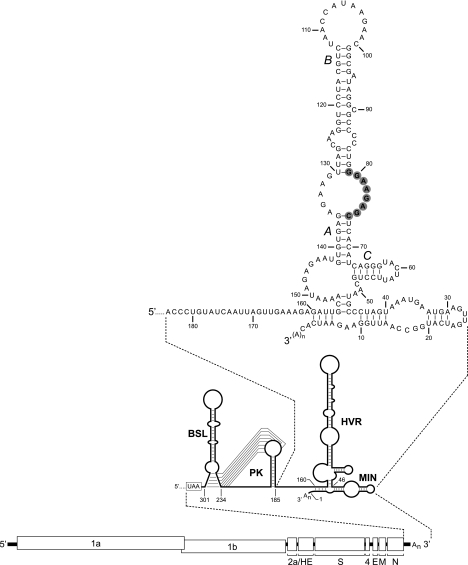
Work from a number of separate investigations has led to the overall picture of the MHV 3′ UTR shown in Fig. 1. At the upstream end of the 3′ UTR, immediately following the N gene stop codon, are two RNA secondary structures, a bulged stem-loop (16) and a classical hairpin-type pseudoknot (55), the latter of which was first identified in BCoV. Both of these structures have been confirmed by extensive genetic analysis, as well as by chemical and enzymatic probing, and both have been shown to be functionally essential in DI RNAs and in the intact viral genome (11, 15, 16, 55). Notably, the bulged stem-loop and the pseudoknot overlap, and they therefore cannot form simultaneously. This has led to the proposal that the two structures are part of a molecular switch regulating a transition between different steps of the viral replication scheme (11, 15). The presence of both structures and their overlap are conserved among all group 2 coronaviruses (58), including the severe acute respiratory syndrome coronavirus and human coronavirus HKU1 (12, 57). Moreover, the bulged stem-loops and pseudoknots of different group 2 coronavirus species are functionally equivalent, given that the entire 3′ UTRs of BCoV and the severe acute respiratory syndrome coronavirus were able to replace the corresponding region of the MHV genome (12, 16). Additionally, any of a number of group 2 helper viruses closely related to MHV and BCoV were able support replication of a BCoV DI RNA (58).
FIG. 1.
Landscape of the MHV 3′ UTR. At the bottom is shown the organization of the 31.3-kb MHV genome. Above this is an expanded view of the secondary structure of the 301-nt 3′ UTR. The 3′ UTR comprises the bulged stem-loop (BSL) (nt 234 through 301) (11, 15, 16) and the overlapping pseudoknot (PK) (nt 185 through 238) (11, 55), the HVR (nt 46 through 156) (30), and the minimal element for the initiation of negative-strand RNA synthesis (MIN) (nt 1 through 45) (29). At the top is a detailed view of the structure of the downstream end of the 3′ UTR, as reported by Liu and coworkers (30), beginning at the nucleotide following the pseudoknot. The oct motif, 5′-GGAAGAGC-3′, is highlighted in gray. Stem segments A, B, and C of the HVR are those designated previously (30). Nucleotides are numbered from the first base at the 3′ end of the genome, excluding poly(A), and the N gene stop codon is boxed.
At the opposite end of the 3′ UTR is the minimal region, as defined by DI RNA analysis, necessary to support the initiation of MHV negative-strand RNA synthesis (29). This element has been mapped to fall within the 3′-most 45 to 55 nt of the genome, and it must act in concert with a polyadenylate tract of at least 5 to 10 residues (50).
Falling between the upstream and downstream ends of the 3′ UTR is a more complex RNA secondary structural element, running from nt 46 through nt 156 from the 3′ end of the genome (Fig. 1, top) (30). Neither the sequence nor the structure of this segment of the 3′ UTR is well conserved, either within or across coronavirus groups or even between the highly homologous MHV and BCoV 3′ UTRs, which are 60% divergent in this interval (16). We refer to it here as the hypervariable region (HVR). (It must be noted that in this report, the designation HVR refers to a different region than the previously described HVR at the 5′ end of the infectious bronchitis virus [IBV] 3′ UTR [8, 54]. The latter HVR now appears to be one or more degenerate accessory open reading frames [22]).
Paradoxically, buried within the HVR is a conserved octanucleotide, 5′-GGAAGAGC-3′, designated oct. This motif, which is always situated some 70 to 80 nt from the 3′ end of the genome, was noted first in a comparison of IBV and MHV (1) and shortly thereafter in BCoV (27) and human coronavirus 229E (49). It is now clear that oct is a characteristic signature for coronaviruses. To date, among GenBank sequences containing the 3′ UTR regions from over 300 coronavirus strains and isolates spanning all three groups, there are only three deviations from absolute conservation of oct: GGGAGAGC in IBV isolate UK/919/68 (8) (accession no. AJ278334), GGAGGAGC in pigeon coronavirus (22) (accession no. AJ871022), and GGAAGGGC in ferret enteric coronavirus (56) (accession no. DQ340562). Such stringent conservation strongly implies that the oct motif has a critical function in the biology of coronaviruses.
The structure of the MHV HVR has previously been confirmed by enzymatic probing, and mutagenesis of DI RNAs suggested that stem segments A and B (Fig. 1), but not stem segment C, play a critical role in RNA synthesis (30). A complex of host proteins has been shown to bind to the region that includes stem A and the internal loop opposite oct (31, 60). In addition, pyrimidine tract-binding protein (PTB) (17) and heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) (18) were found to bind to the negative and positive strands of the HVR, respectively. Small deletions generated in the oct motif or in the loop opposite oct reduced binding by PTB and hnRNP A1 and had profound effects on DI RNA transcription and replication (17, 18).
To further investigate the functional importance of the HVR and oct, we carried out a detailed genetic analysis of this region of the 3′ UTR in the intact MHV genome. Our results show that the HVR, including oct, is dispensable for the basic mechanism of MHV replication in tissue culture. By contrast, removal of the HVR resulted in a dramatic attenuation of virulence in the mouse host.
MATERIALS AND METHODS
Cells and viruses.
Wild-type MHV-A59 and MHV mutants were propagated in mouse 17 clone 1 (17Cl1) cells; plaque assays and plaque purifications were carried out in mouse L2 cells. The interspecies chimeric virus fMHV.v2 (11) was grown in feline FCWF cells.
Transcription template construction.
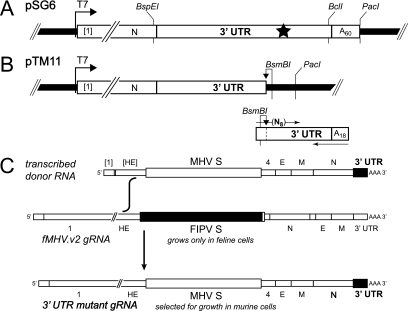
Templates for donor RNAs were derived from pSG6, which contains a cDNA segment of the 5′-most 0.5 kb of the MHV genome linked to the 3′-most 8.6 kb of the MHV genome (11). pSG6 was generated from the previously described pMH54 (24) by creation of a coding-silent unique BspEI site near the end of the N gene, changing codons 444 through 446 from GTGCCAGAT to GTTCCGGAT (the BspEI site is underlined). In most cases, defined mutations were constructed in pSG6 by splicing overlap extension-PCR (14), followed by cloning of the amplified product between the unique BspEI and BclI sites located near the ends of the N gene and the 3′ UTR, respectively (Fig. 2A). For the plasmid corresponding to the octG1C mutation, the mutation was initially generated in a subclone, pB85, and was then transferred to pMH54 by exchange of the NheI-SacI fragment that encompasses most of the N gene and the 3′ UTR, as described previously for other mutations (15). For the plasmid corresponding to the Δ(3′ UTR) mutation, a junction linking the N gene stop codon to a tract of 66 A residues was created by replacement of the BstBI-PacI fragment of pA122. pA122 (10) has a rearranged gene order and contains a coding-silent unique BstBI site near the end of the N gene, changing codons 451 through 453 from GACTCTAAT to GATTCGAAT (the BstBI site is underlined). The wild-type order of the S, 4, E, M, and N genes in the resulting clone was then restored through replacement of the SbfI-NheI fragment with that of pSG6.
FIG. 2.
Selection of MHV 3′ UTR mutations by targeted RNA recombination. (A) Transcription vector pSG6 (1), the precursor of the plasmids used for synthesis of donor RNA for most recombinants, was derived from pMH54 (24) and contains a 5′ segment of the MHV genome (denoted by [1]) fused to a partial HE gene and all genes downstream of HE. The region between the 5′ segment and the N gene is not shown. The T7 RNA polymerase start site is indicated by an arrow. Particular 3′ UTR mutations, represented by a star, were constructed by splicing overlap extension-PCR and transferred into pSG6 by exchange of the BspEI-BclI restriction fragment. Donor RNAs were in vitro transcribed from PacI-linearized vectors. (B) For random mutagenesis of the oct motif, pTM11 was constructed from pSG6 to contain a fragment of the 3′ UTR preceding a site for BsmBI, which cuts at a distance from its recognition sequence. A PCR product was also generated to contain the remainder of the 3′ UTR, including a complementary BsmBI site in the opposite orientation and a randomized oct motif. BsmBI-linearized pTM11 was ligated to the BsmBI-restricted PCR fragment, and T7 transcripts were synthesized from the resulting template mixture. (C) MHV recombinants containing mutations in the 3′ UTR (solid rectangle) were generated by transfection of fMHV.v2-infected feline cells with synthetic donor RNA. The interspecies chimeric coronavirus fMHV.v2 (11) contains the ectodomain-encoding region of the feline infectious peritonitis virus (FIPV) S gene (gray rectangle), which renders it able to grow in feline cells but not in murine cells. Additionally, the order of the MHV structural protein genes downstream of S has been rearranged in fMHV.v2 to eliminate the possibility of unwanted secondary crossover events. Recombinants were selected as progeny that had regained the ability to grow in murine cells.
To generate a mixture of templates for the random mutagenesis of the oct motif, we constructed a plasmid, pTM11, that removed the 3′-most 86 nt of the 3′ UTR (including oct) and the poly(A) tail from pSG6 (Fig. 2B). A BsmBI site was also planted downstream of the 3′ UTR remnant in pTM11; when pTM11 was cleaved with BsmBI, which cuts at a distance from its recognition site, this created a sticky end at nt 87 to 90 of the 3′ UTR. PCR products containing the remainder of the 3′ UTR, including a randomized sequence at the position of oct and an 18-residue poly(A) tail, were generated using an upstream primer, 5′-GAACGGCGTCTCGCGCCCCCTGNNNNNNNNTCACATCAGGGTACTA-3′ (N = A, G, C, or T). When the PCR products were cleaved with BsmBI (site underlined), this created a complementary sticky end (in boldface) at nt 87 to 90 of the 3′ UTR. Ligation of the PCR products to pTM11 resulted in a collection of templates, with no vestige of the BsmBI sites or other exogenous sequence, that were used directly for in vitro transcription.
Oligonucleotides for PCR and DNA sequencing were obtained from Integrated DNA Technologies. For every constructed plasmid, the overall composition was monitored by restriction analysis, and the sequences of all ligation junctions and all regions created by PCR were verified by automated sequencing.
Targeted RNA recombination.
MHV mutants containing 3′ UTR mutations were obtained by targeted RNA recombination with host range-based selection, as described previously (Fig. 2C) (11, 12, 15, 19, 25, 35). Briefly, monolayers of feline FCWF cells were infected with fMHV.v2 and were then transfected by electroporation with capped donor RNAs that had been synthesized with a T7 mMessage mMachine transcription kit (Ambion), using PacI-truncated templates. The infected and transfected feline cells were plated into 10-cm2 wells, and progeny virus was harvested when syncytia or a cytopathic effect was apparent, typically at 18 to 24 h postinfection at 37°C. Recombinant candidates were identified and purified through two rounds of plaque titration on murine L2 cell monolayers at 37°C. A mutation was judged to be lethal if multiple targeted RNA recombination experiments yielded a clean background on mouse cells of no recombinants with fMHV.v2, in parallel with positive controls with wild-type (pSG6-derived) donor RNA that yielded a high frequency of recombinants.
For viable mutants, each purified recombinant candidate was used to infect a monolayer of 17Cl1 cells, and total cellular RNA was extracted at 18 to 24 h postinfection using Ultraspec reagent (Biotecx). RNA was reverse transcribed with a random hexanucleotide primer (Boehringer Mannheim) and avian myeloblastosis virus reverse transcriptase (Life Sciences); the 3′ UTR and partial N gene were then amplified by PCR under standard conditions using AmpliTaq polymerase (Roche). The resulting products were analyzed directly by agarose gel electrophoresis or were purified with Quantum-prep columns (Bio-Rad) prior to automated sequencing of the entire 3′ UTR. All viable isolated mutants were found to have the intended engineered mutation(s), and no extraneous mutations were found elsewhere in the 3′ UTR. For most mutants, two or more independent isolates were obtained; the Alb strain designation for one representative isolate of each viable mutant is noted in the figure that shows the 3′ UTR sequence of that mutant.
For comparisons of plaque sizes, titers of mutants and wild-type controls were determined in two groups. Plaques obtained on mouse L2 cell monolayers were stained with neutral red at 48 h postinfection at 37°C; plaques were photographed 18 h thereafter. Digital images of 20-cm2 dishes were printed at a magnification of ×3, and diameters of 25 to 30 well-separated plaques within the same area of a dish were measured, always in the same direction relative to the edge of the page. Throughout this work, the wild-type virus used for comparisons was Alb240, a well-characterized isogenic recombinant that was previously reconstructed from fMHV and pMH54 donor RNA (26).
For the measurement of growth kinetics, confluent monolayers of 17Cl1 cells (75 cm2) were inoculated at a multiplicity of 4.0 PFU per cell (wild type [Alb240], ΔHVR2 [Alb519], and Δoct [Alb445]) or at a multiplicity of 0.01 PFU per cell (wild type [Alb240] and ΔHVR2 [Alb519]) for 2 h at 37°C, with rocking every 15 min. Inocula were removed, monolayers were washed three times, and incubation was continued in fresh medium at 37°C. Sample aliquots of medium were then withdrawn at various times from 2 to 32 h postinfection, and infectious titers were subsequently determined.
Radiolabeling of viral RNA.
Intracellular viral RNA was metabolically labeled as described previously (11, 12, 16). Briefly, confluent 20-cm2 monolayers of 17Cl1 cells were infected with mutant or wild-type MHV at a multiplicity of 5 PFU per cell and were incubated at 37°C. Infected cells were starved from 2 h through 7 h postinfection in Eagle's minimal essential medium containing 5% dialyzed fetal bovine serum with 1/10 of the normal concentration of phosphate. Labeling was then carried out from 7 h through 9 h postinfection in 1 ml phosphate-free medium containing 100 μCi per ml [33P]orthophosphate (MP Biomedicals), 5% dialyzed fetal bovine serum, and 20 μg of actinomycin D (Sigma) per ml. Total cytoplasmic RNA was purified, and the amount of radioactivity incorporated into RNA was quantitated by spotting an aliquot onto DEAE paper filter (Whatman DE81). Filters were washed five times with 0.5 M Na2HPO4 and once each with water and ethanol, and then dried and counted in a scintillation counter. Samples containing equal amounts of radioactivity were analyzed by electrophoresis through 1% agarose containing formaldehyde and were visualized by fluorography.
Mouse infectivity studies.
Six-week-old, female BALB/c mice (Taconic Farms, Hudson, NY) were inoculated intranasally with a 20-μl inoculum (10 μl per nostril) under light anesthesia. For the morbidity and mortality study, there were eight mice per dose group (5 × 103, 5 × 104, or 5 × 105 PFU) and four mock-inoculated controls. Mock-inoculated mice were inoculated with supernatant from mock-infected cell cultures that were harvested at the same time as virus stocks. The highest dose (5 × 105 PFU) for both wild-type and ΔHVR2 viruses was in tissue culture supernatant diluted 1:1 with mock supernatant; lower doses of virus were diluted in tissue culture-grade phosphate-buffered saline containing 1% fetal bovine serum. Mice were weighed and observed for clinical signs daily for 29 days. Clinical disease was scored as follows: 0, normal; 0.5, mildly ruffled fur; 1.0, moderately ruffled fur; 2.0, “oily” fur; and 3.0, hunching, severe aggression, or neurological signs. A value of 0.5 was added to a score if there was 5% or greater weight loss observed on a given day, based on weight on the day of inoculation. Mice were housed four to a cage; mice that became severely aggressive were moved to separate cages. MHV seroconversion was monitored with a Murine ImmunoComb kit (Charles River Laboratories).
For the tissue tropism study, 20 mice per virus strain were inoculated with 5 × 104 PFU, and four mice were inoculated with diluent as mock controls. Four mice per virus strain were sacrificed on days 1, 3, 5, 7, and 9 postinoculation (p.i.). The four mock-inoculated mice were sacrificed on day 9 p.i. Nasal wash was obtained immediately postmortem by infusing 0.5 ml of phosphate-buffered saline-1% fetal bovine serum into the nasopharynx and collecting the wash from the nares. Blood was collected from the heart, and serum was separated by centrifugation. Brain (right sagittal half), lung, liver, and spleen were harvested and weighed. Buffer (M199 with Hanks' salts and l-glutamine, 0.05 M Tris, 1% bovine serum albumin, 0.35 g/liter sodium bicarbonate, 100 units/ml penicillin, 100 units/ml streptomycin, and 1 μg/ml amphotericin B [Fungizone], pH 7.4) was added to make a 10% homogenate for peripheral tissues or a 20% homogenate for brain. Tissues were homogenized by grinding with a zinc-plated 4.5-mm BB (Daisy Brand, Rogers, AR) in a Mixer Mill MM300 (QIAGEN, Valencia, CA) for 4 min at 24 cycles per second and were then frozen at −80°C until assayed. Homogenates were thawed and centrifuged at 3,800 × g and 4°C for 5 min prior to plaque assay. For each tissue type in which all virus titers were below the limit of detection, the potential presence of viral inhibitory activity in the homogenates was assessed. For this purpose samples of spleen, liver, lung, and serum from a mock-inoculated mouse were spiked with a known quantity of wild-type virus and assayed by plaque titration. In all cases no inhibitory activity was noted. All studies were approved by the Institutional Animal Care and Use Committee of the Wadsworth Center and followed criteria established by the National Institutes of Health.
RESULTS
Mutational truncations and substitutions in the HVR.
We constructed mutations in the 3′ UTR of the MHV genome by targeted RNA recombination (35). This method uses the inherent recombinational capacity of the viral polymerase to transfer mutations from transfected donor RNA to a recipient virus that can be selected against (Fig. 2C). In this study we used as the recipient virus the chimeric coronavirus fMHV.v2 (11), which grows only in feline cells, and we selected mutants on the basis of their having regained the ability to grow in murine cells.
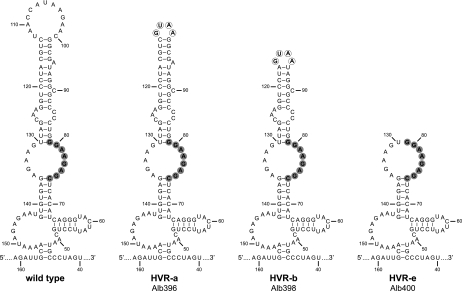
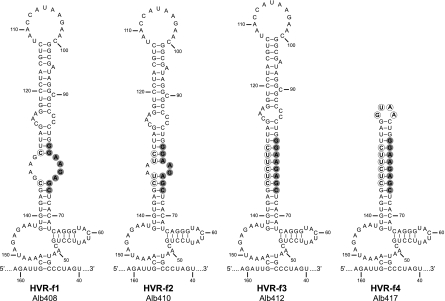
The framework for our analysis of the HVR was the complex RNA secondary structure that was previously described for the downstream 166 nt of the MHV genome (Fig. 1) (30). An RNA folding different from that shown in Fig. 1 has recently been proposed for the 3′-most 42 nt of the MHV genome (21), but it is not yet clear how this independently folded alternative structure might interact with the 166-nt structure. The work reported here does not address the furthest-downstream segment of the 3′ UTR. Initially, we created mutations in the terminal bulged stem-loop of the HVR, comprising nt 82 through 129, which prior work had suggested to be functionally important for DI RNA replication (30). In the first such mutant, the terminal loop was replaced with a tetraloop of the GNRA class, GUAA (Fig. 3, mutant HVR-a). Tetraloops are unusually stable secondary structural motifs that stabilize hairpins in a variety of RNAs, including rRNA (36). In a second mutant, HVR-b, the terminal loop and stem segment, as well as bulge base A95, were deleted and replaced with the same tetraloop. Finally, in a third mutant, HVR-e, the entire terminal bulged stem-loop was truncated, thereby placing the oct motif in a terminal loop. All three of these mutants exhibited wild-type-sized plaques and showed no obvious growth defect. The dispensability of such a large portion of the HVR led us to hypothesize that the principal, and perhaps only, role of the HVR is to display oct in a constrained single-stranded conformation, which is often a hallmark for protein recognition. This assumption led us to next construct mutations that would affect the accessibility of oct (Fig. 4). To progressively alter the arrangement of oct within the HVR, we changed the size and sequence of the opposite internal loop (nt 131 to 135), such that four (mutant HVR-f1), six (mutant HVR-f2), or all eight (mutant HVR-f3) nucleotides of the oct motif became part of the adjacent stem segments. An additional mutant, HVR-f4, was constructed with a truncated, tetraloop-capped version of the fully duplexed oct. Contrary to our expectations, none of these mutants was markedly defective; only mutant HVR-f3 formed plaques somewhat smaller than those of wild-type virus.
FIG. 3.
HVR truncation mutants. For each mutant, circled nucleotides are those that were changed from the wild-type sequence. The oct motif is highlighted in gray. Nucleotides are numbered according to the wild-type sequence, starting from the first base at the 3′ end of the genome, excluding poly(A).
FIG. 4.
HVR mutants that alter the accessibility of the oct motif. For each mutant, circled nucleotides are those that were changed from the wild-type sequence. The oct motif is highlighted in gray. Nucleotides are numbered according to the wild-type sequence, starting from the first base at the 3′ end of the genome, excluding poly(A).
Alteration of the sequence of oct.
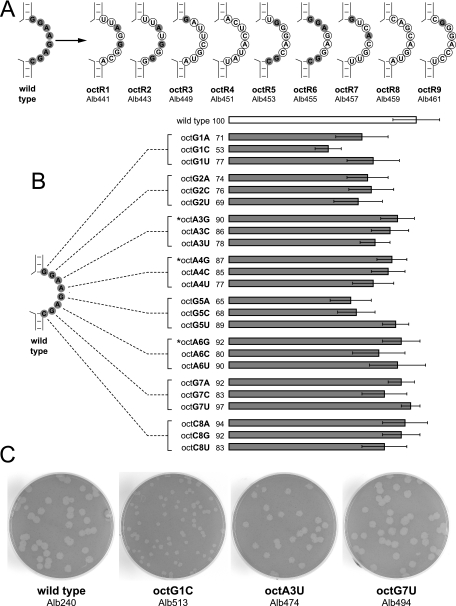
To further explore restrictions on oct, we generated mutants in which the oct motif was randomly mutagenized. To accomplish this, we ligated a truncated donor RNA transcription template to a PCR product in which all eight positions of oct had been randomized, as described in Materials and Methods (Fig. 2B). Transcripts obtained from the resulting collection of templates were used in targeted RNA recombination, and viable recombinants were isolated and sequenced throughout the 3′ UTR. Nine different random oct mutants were recovered in this manner, none of which had a significant phenotype relative to the wild type (Fig. 5A). Members of this set had as few as three or as many as all eight positions of the oct motif mutated from the canonical 5′GGAAGAGC3′. This clearly showed that at least some variations from the conserved oct sequence were tolerated for MHV replication in tissue culture, but we were unable to discern whether there was any pattern among the variant sequences that had been selected. To more systematically address this question, we sought to construct all 24 possible single-point mutants of oct, a set in which each nucleotide of the motif was changed to each of its three alternative residues. We were able to recover all of these single-point mutants, and collectively they were found to form plaques that spanned a range of sizes with respect to the wild type (Fig. 5B). The mutants could be grouped into three classes of plaque sizes (Fig. 5C): small, for which the octG1C mutant was the most extreme example; intermediate (70 to 85% of wild-type size), as exemplified by the octA3U mutant; and barely distinguishable from wild type, for which the octG7U mutant represented the upper end of the range. The distribution of plaque sizes (Fig. 5B) suggested trends in the relative importance of each of the eight positions of the oct motif, with G1, G2, and G5 being the most sensitive to mutation and A6, G7, and C8 being the least sensitive to mutation. Most notably, however, all 24 mutants were viable, and none was as dramatically impaired as certain other mutants that we have previously constructed with alterations either in the 3′ UTR (11) or in MHV structural genes (19, 25, 26). This implied that the oct motif, despite its strict conservation among all coronaviruses, has no sequence requirements that are essential for viral RNA synthesis.
FIG. 5.
oct motif point mutants. (A) Random, multiple-point mutants of oct. For each mutant, circled nucleotides are those that were changed from the wild-type sequence. Unchanged oct nucleotides are highlighted in gray. (B) Defined point mutants of oct. Each nucleotide in oct was changed to each of the three possible alternatives. Relative plaque sizes (diameters) of mutants are depicted as the percentage of the average diameter of wild-type plaques (± standard deviation). Mutants marked with an asterisk are those for which one naturally occurring variant coronavirus oct sequence has been reported. (C) Representative plaques for the three size classes of oct single-point mutants, compared to the wild type. Plaque titrations were performed on mouse L2 cells at 37°C. Monolayers were stained with neutral red at 48 h postinfection and were photographed 18 h later.
Deletions of oct and the HVR.
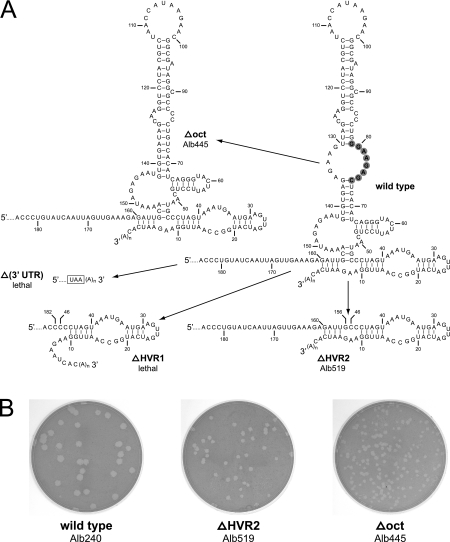
To more stringently test whether the oct motif or the HVR plays a critical role in viral replication, we made constructs deleting either oct alone or the whole HVR (including oct). In the first such mutant, Δoct, the oct motif (nt 74 through 81) plus the internal loop opposite oct (nt 130 through 136) were removed, creating a continuous helix from the two stem segments formerly on either side of oct (Fig. 6A). Remarkably, this mutant was viable, and it formed plaques that were, on average, 45% of the size of wild-type plaques (Fig. 6B). More surprising still was the viability of a second mutant, ΔHVR2, which contained a large deletion excising the entire HVR between nt 46 and 156. The ΔHVR2 mutant was actually found to be more fit than the Δoct mutant, forming plaques 60% of the size of wild-type plaques. It seemed somewhat incongruous that smaller mutations in the HVR, like those in the HVR-f3 mutant, the octG1C mutant, and the Δoct mutant, should be more inhibitory to the virus than the complete ablation of the HVR. However, this may be attributable to the creation of deleterious misfoldings or overly stable secondary structures in the 3′ UTRs of mutants that retained altered or residual versions of the HVR.
FIG. 6.
Mutants containing deletions encompassing the oct motif and the HVR. (A) At the right is the wild-type 3′ UTR, beginning at the nucleotide following the pseudoknot; the oct motif is highlighted in gray. Nucleotides in all structures are numbered according to the wild-type sequence, starting from the first base at the 3′ end of the genome, excluding poly(A). The Δoct mutant contains a deletion of the oct motif, nt 74 through 81, as well as the internal loop opposite oct, nt 130 through 136. The ΔHVR1 mutant contains a deletion of the region from nt 47 through 181, and the ΔHVR2 mutant contains a deletion of the region from nt 47 through 155. The Δ(3′ UTR) mutant contains a deletion of the entire 301-nt 3′ UTR, directly adjoining the N gene stop codon (boxed) to the poly(A) tail. (B) Plaques of the two viable deletion mutants, ΔHVR2 and Δoct, compared to those of an isogenic wild-type control. Plaque titrations were performed on mouse L2 cells at 37°C. Monolayers were stained with neutral red at 48 h postinfection and were photographed 18 h later.
By contrast with the ΔHVR2 mutant, we were unable to recover a recombinant with a more extensive mutation, ΔHVR1, in which the region between nt 47 and 181 was deleted (Fig. 6A). A total of 14 independent targeted RNA recombination experiments with donor RNA containing the ΔHVR1 mutation were conducted, in three separate sets. None of these trials yielded a mutant recombinant, whereas in the same trials wild-type donor RNA yielded robust numbers of control recombinants. Given the power of the host range-based selection with fMHV.v2 (11), this outcome established a strong criterion for the lethality of the ΔHVR1 mutation. Moreover, compared with the minimal effect of the ΔHVR2 mutation, this result suggested that there is an essential role for some portion of the region of nt 157 through 184, between the upstream pseudoknot and the HVR (Fig. 1). In addition, we attempted to delete the entire MHV 3′ UTR, prompted by results obtained with some picornaviruses (53). In this case, we conducted eight independent targeted RNA recombinations, in four separate trials, using donor RNA in which the N gene stop codon (UAA) was directly connected to the poly(A) tail (Fig. 6A). The negative outcomes of all of these experiments, as opposed to strong positive wild-type controls, indicated that complete removal of the 3′ UTR is lethal. This result was consistent with numerous previous analyses showing that the upstream bulged stem-loop and pseudoknot of the 3′ UTR are essential for viral replication (11, 15, 16, 55).
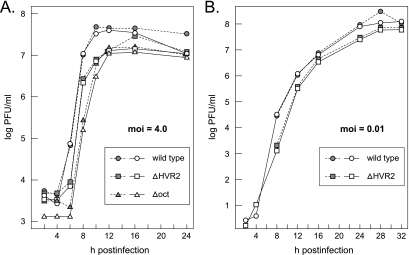
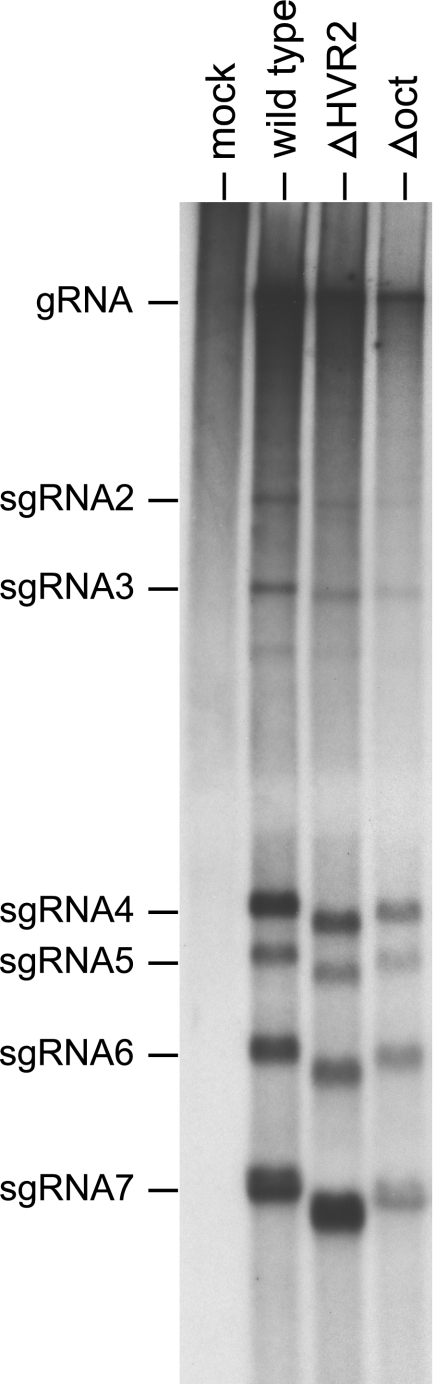
We more closely examined the phenotypes of the viable deletion mutants in single-step growth experiments (Fig. 7A). Overall, at a high multiplicity of infection (4.0 PFU per cell) both the ΔHVR2 mutant and the Δoct mutant exhibited growth kinetics comparable to that of the wild type. Single-step growth of the ΔHVR2 mutant was slightly delayed with respect to that of the wild type, and the Δoct mutant lagged behind the ΔHVR2 mutant as well as the wild type. Both deletion mutants grew to maximal titers that were two- to threefold lower than those of the wild type. To further compare the growth of the ΔHVR2 mutant and the wild type over multiple cycles of viral replication, infections were carried out at a low multiplicity (0.01 PFU per cell) (Fig. 7B). Under these conditions, the growth of the ΔHVR2 mutant lagged behind that of the wild type for the first 8 h of infection. However, by 12 h postinfection, and at all times thereafter, the mutant reached titers highly similar to those of the wild type. Metabolic labeling of RNA in infected cells revealed that the ΔHVR2 mutant and the Δoct mutant each produced the same set of sgRNA species, in the same relative proportions, as the wild type (Fig. 8). A noticeably faster mobility of the sgRNAs of the ΔHVR2 mutant relative to the wild type was observed, owing to the 109-nt deletion in the ΔHVR2 genome (and, consequently, in all of its sgRNAs). These results indicated that neither deletion mutant differed markedly from wild-type MHV in tissue culture. In particular, both mutants were fully competent in gRNA replication and in sgRNA transcription. This finding differs considerably from the severe inhibition of DI RNA replication and transcription that was previously reported to result from small deletions in oct and the HVR (17, 18).
FIG. 7.
Growth of the ΔHVR2 and Δoct mutants in tissue culture. (A) Single-step growth kinetics. Confluent monolayers of 17Cl1 cells were infected with wild-type, ΔHVR2, or Δoct viruses at a multiplicity of infection (moi) of 4.0 PFU per cell. At the indicated times postinfection, aliquots of medium were removed and infectious titers were determined on mouse L2 cells. Open and solid symbols represent results from two independent experiments. (B) Growth following a low multiplicity of infection. Confluent monolayers of 17Cl1 cells were infected with wild-type or ΔHVR2 viruses at a multiplicity of 0.01 PFU per cell. At the indicated times postinfection, aliquots of medium were removed and infectious titers were determined on mouse L2 cells. Open and solid symbols represent results from two independent experiments.
FIG. 8.
RNA synthesis by the ΔHVR2 and Δoct mutants. Infected or mock-infected 17Cl1 cells were metabolically labeled with [33P]orthophosphate in the presence of actinomycin D, and RNA was isolated and electrophoretically analyzed in 1% agarose containing formaldehyde as described in Materials and Methods.
In vivo attenuation of the ΔHVR2 mutant.
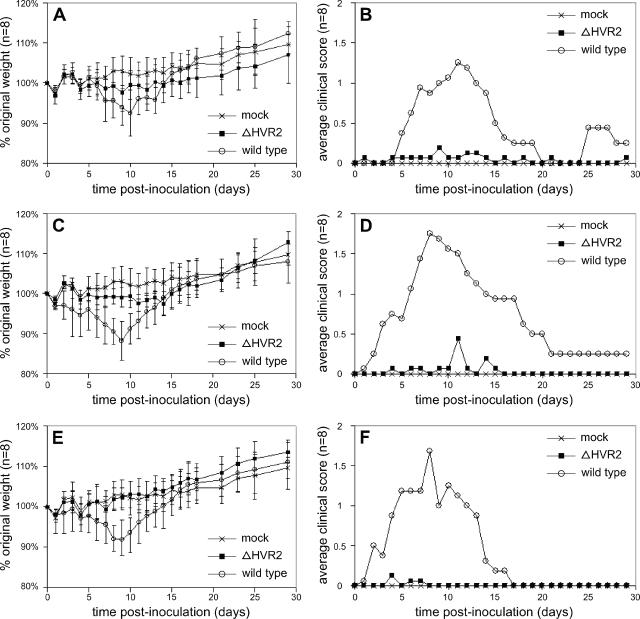
To examine the phenotype of the ΔHVR2 mutant in the infected host, we inoculated groups of 6-week-old female BALB/c mice intranasally with wild-type MHV or ΔHVR2 virus at 10-fold-increasing doses from 5 × 103 PFU to 5 × 105 PFU. Mice were weighed and scored for clinical signs daily for 29 days. A striking difference was observed between wild-type-virus-infected and mutant-infected animals. All mice infected with wild-type virus, at all doses, had substantial weight loss by the second week of infection (Fig. 9A, C, and E). By contrast, ΔHVR2-infected mice showed only minimal weight loss at all doses. Similarly, all mice infected with wild-type virus, at all doses, exhibited marked clinical signs of disease (Fig. 9B, D, and F). No clinical signs of disease were observed in ΔHVR2-infected mice at any dose (Table 1). Very similar results were obtained in a second, independent experiment (data not shown).
FIG. 9.
Reduced virulence of the ΔHVR2 mutant. Six-week-old, female BALB/c mice, in groups of eight, were inoculated intranasally with 5 × 103 PFU (A and B), 5 × 104 PFU (C and D), or 5 × 105 PFU (E and F) of wild-type or ΔHVR2 virus. A group of four mock-inoculated mice was inoculated with tissue culture supernatant. Mice were weighed and observed for clinical signs daily. (A, C, and E) Percentage of original weight (± standard deviation). (B, D, and F) Average clinical disease scores, as defined in Materials and Methods. The same mock-inoculated group is shown in all panels for comparison to each of the viral dose groups.
TABLE 1.
Morbidity of mice infected with wild-type and ΔHVR2 virusa
| Virus | Dose (PFU) | No. of mice:
|
Overall % morbidity | ||
|---|---|---|---|---|---|
| In group | With wt lossb | With clinical signsc | |||
| Mock | 4 | 0 | 0 | 0 | |
| Wild type (Alb240) | 5 × 103 | 8 | 7 | 8 | 100 |
| 5 × 104 | 8 | 8 | 8 | 100 | |
| 5 × 105 | 8 | 6 | 8 | 100 | |
| ΔHVR2 (Alb519) | 5 × 103 | 8 | 3 | 0 | 38 |
| 5 × 104 | 8 | 2 | 0 | 25 | |
| 5 × 105 | 8 | 3 | 0 | 38 | |
Six-week-old, female BALB/c mice were inoculated with virus or diluent. Mice were weighed and observed for clinical signs daily for 29 days. All mice survived. All mice inoculated with virus seroconverted; mock-inoculated mice remained seronegative.
Number of mice with ≥5% weight loss.
Number of mice with clinical signs, which included ruffled fur, hunching, aggression, “oily” fur, and circling (one mouse). Two mice in wild-type groups still had “oily” fur on day 29.
To study viral replication and tissue tropism, groups of mice were inoculated intranasally with wild-type MHV or ΔHVR2 virus at a dose of 5 × 104 PFU. At 1, 3, 5, 7, and 9 days p.i., four mice infected with each virus were sacrificed. Nasal wash and serum were obtained from each mouse, multiple tissues (brain, lung, spleen, and liver) were harvested, and infectious titers were determined for all samples. In the nasal washes (Fig. 10A) replication of wild-type virus was clearly detectable at 1 day p.i., but it was not clear whether the virus titers found for the mutant at 1 day p.i. were indicative of viral replication or represented the initial inoculum. However, virus titers in the upper respiratory tract were decreasing by 3 days p.i., and virus was completely cleared from all but one wild-type-inoculated mouse by 5 days p.i.
FIG. 10.
Reduced in vivo replication of the ΔHVR2 mutant. Six-week-old, female BALB/c mice, in two groups of 20, were inoculated intranasally with 5 × 104 PFU of wild-type or ΔHVR2 virus (the dose shown in Fig. 9C and D). Four mice in each group were sacrificed at 1, 3, 5, 7, and 9 days p.i., and tissues were harvested. Viral titers in nasal washes (A) and brains (B) were determined by plaque assay in L2 cells, as described in Materials and Methods. Titers for individual mice are represented by circles or squares. Geometric means of titers for each set of four mice are connected by solid lines or broken lines. The thicker, horizontal broken line indicates the limit of detection for each set of assays (10 PFU/ml for nasal washes and 50 PFU/g for brains).
In the brains of infected mice, replication was detectable for both viruses by 3 days p.i. (Fig. 10B), although wild-type titers were 800-fold higher than those of the ΔHVR2 mutant, and virus was detected in 100% and 50% of the wild-type- and mutant-inoculated mice, respectively. Viral replication in brains peaked at 5 days p.i., at which time the titers of wild-type virus were comparable to or greater than peak titers seen by other investigators following direct intracranial inoculation of mice with MHV-A59 (44, 51). Peak titers in brain for wild-type virus were, on average, 60-fold higher than those for the mutant. This difference correlated with the morbidity and aggression seen in wild-type-infected mice, but not in mutant-infected mice, at later times p.i. in this experiment and the one with the results shown in Fig. 9. By 7 days p.i., mutant titers were higher than the declining wild-type titers. Wild-type virus had largely been cleared from brains by 9 days p.i.; by contrast, mutant-infected animals still exhibited significant titers in brain at this time.
For both wild-type virus and the ΔHVR2 mutant, there was minimal or no spread to the lower respiratory tract or peripheral organs. The infectivity in all lung samples at every time point was undetectable, except for one wild type-infected mouse at day 7 p.i., which had a titer at the limit of detection (100 PFU/g). Likewise, infectious virus was undetectable in spleen samples, except for one wild-type-infected mouse at day 5 p.i., which had a titer at the limit of detection (100 PFU/g). Similarly, and in contrast to the case for intracranial inoculations (51), no viremia was observed, and there was no spread of infection to the liver. All titers in serum and liver were below the limits of detection (100 PFU/ml and 100 PFU/g, respectively). All mice that were sacrificed at 1, 3, and 5 days p.i. (and all mock-inoculated controls) were seronegative for MHV; all mice sacrificed at 7 and 9 days p.i. had seroconverted. Collectively, the results in Fig. 9 and 10 demonstrated that although the ΔHVR2 mutation had only marginal effects on MHV replication in tissue culture, it dramatically altered the replication and pathogenesis of the virus by the natural route of infection.
DISCUSSION
In this study, we have shown that the HVR within the MHV 3′ UTR is completely dispensable for viral replication in tissue culture. The MHV genome was found to tolerate a wide variety of substitutions and truncations in this region (Fig. 3 to 5). Most strikingly, we created a 109-nt deletion that removed the entire HVR, a span constituting more than one-third of the 3′ UTR (Fig. 6). The resulting ΔHVR2 mutant not only was viable but replicated almost as well as did wild-type virus, in either high-multiplicity, single-step infections or low-multiplicity, multiple-cycle infections in tissue culture (Fig. 7). These results are starkly different from those of analyses of other 3′ UTR structural elements. Many mutations that disrupted the upstream bulged stem-loop or pseudoknot were found to be lethal to the virus (11, 15, 16). Likewise, at least one alteration of the downstream minimal element for negative-strand RNA synthesis also could not be recovered in a viable virus (21). The extraneous nature of the HVR was particularly surprising, given that this region harbors the oct motif, 5′-GGAAGAGC-3′, which is a nearly invariant feature of coronavirus genomes. Remarkably, tissue culture replication of MHV was not drastically hampered by random mutagenesis of oct, by single point mutations at any of its eight positions, or by the complete excision of this motif in the Δoct mutant.
In distinct contrast to the minimally impaired phenotype of the ΔHVR2 mutant in vitro (Fig. 7 and 8), the in vivo effects caused by the removal of the HVR were readily discernible. The ΔHVR2 mutant was highly attenuated in mice inoculated by the intranasal route, which is thought to most closely resemble the natural mode of MHV infection. ΔHVR2-infected mice displayed no signs of clinical disease and underwent minimal weight loss compared to wild-type-infected mice (Fig. 9). ΔHVR2 virus infections, like wild-type infections, spread from the nasal epithelium to the brain. However, replication of the mutant virus in the brain was greatly reduced and persisted longer, compared to replication of the wild type (Fig. 10). This difference of in vivo replication between the mutant and the wild type was, we believe, far greater than that which could strictly be accounted for by cumulative effects of the small growth differences observed between the two viruses in tissue culture. Indeed, in multiple-cycle infections begun at a low multiplicity of infection, growth of the ΔHVR2 mutant did not fall progressively further behind that of the wild type as a function of time (Fig. 7B). We have recently found that an additional mutant, ΔHVR3, which has an even more extensive 3′ UTR deletion, has the same growth kinetics with respect to the wild type as does the ΔHVR2 mutant at both low and high multiplicities of infection (data not shown).
MHV causes disease that is strongly dependent on the genetic background, age, and immune status of the mouse host, as well as on the strain, dose, and mode of inoculation of the virus (7, 13, 43). From the standpoint of the virus, many aspects of pathogenesis map predominantly to the spike (S) protein (6). This has been elegantly demonstrated by the transfer of determinants for neurotropism, hepatotropism, or demyelination via the shuttling of S genes from one MHV strain to another (39, 41, 44). Since S protein governs receptor recognition and mediates cell-cell fusion, it is logical that it plays such a pivotal role in disease. Despite this, other viral factors clearly can influence pathogenesis; in some contexts, their effects are dominant over the tropism specified by S protein (5, 9, 20, 33, 40, 47). The potential role(s) of RNA elements in coronavirus pathogenesis, however, remain to be explored. Irrespective of the mechanism underlying the reduced virulence of the ΔHVR2 mutant, our finding has obvious implications for the design of live attenuated vaccines against coronaviruses of medical and veterinary importance.
Our results bear similarity to certain recent findings with picornaviruses, which have genomes and 3′ UTRs much smaller than those of coronaviruses. For both poliovirus and human rhinovirus, particular point mutations, small deletions, or insertions in the 3′ UTR appear to be harmful or lethal (45, 46, 48, 52). Yet, paradoxically, the entire 65-nt 3′ UTR of poliovirus or the entire 44-nt 3′ UTR of human rhinovirus can be deleted with only minimal loss of viability (3, 53). In tissue culture, the poliovirus Δ(3′ UTR) mutant, compared with the wild type, is much more impaired in cells of neuronal origin than in HeLa cells. In addition, the Δ(3′ UTR) mutant is at least 1,000-fold less neurovirulent than wild-type poliovirus in transgenic mice expressing the poliovirus receptor (4). It is currently not clear how close an analogy can be drawn between the MHV ΔHVR2 mutant and the poliovirus Δ(3′ UTR) mutant, but in both cases some elements in the 3′ UTR could conceivably play modulatory roles in RNA synthesis. Such roles, while not essential, may become rate-limiting under particular circumstances. For the poliovirus Δ(3′ UTR) mutant, there is a substantial selective reduction of positive-strand RNA synthesis, relative to that of the wild type, and the magnitude of this effect is cell type dependent (4). We have not yet examined individual stages of RNA synthesis in the ΔHVR2 mutant. It is possible that further work will reveal a selective effect on RNA synthesis that has a greater impact in vivo than in vitro. Alternatively, the HVR may serve to counteract some component of host defense. Such a host-related function would account for why the loss of the HVR is of little consequence in vitro but results in decreased replication in vivo. Our observations call for a more detailed examination of the influence of the HVR of the 3′ UTR on MHV pathogenesis.
It is reasonable to speculate that the role(s) of the HVR is mediated by host protein factors. Prior studies have investigated proteins binding to the 3′ UTR of MHV, using probes that included all or parts of the HVR. These studies have reached differing conclusions about how many, and which, proteins bind to this region (17, 18, 50, 59). The upstream end of the HVR (nt 129 through 154) binds a set of at least four host proteins that are possibly the same as a complex that binds to nt 1 through 42, at the downstream end of the genome (31, 59, 60). Constituents of the downstream complex have been identified as mitochondrial aconitase and the chaperones mtHSP70, HSP60, and HSP40 (37, 38). Overlapping the same portion of the HVR, a binding site for hnRNP A1 has been reported to lie between nt 90 and 170 (18). Additionally, a binding site for PTB has been localized to the negative strand of nt 53 through 149 (17). All of these host factors have been proposed to play critical roles in MHV RNA replication and transcription, based on the effects of binding-site mutations on DI RNA synthesis. However, there is a discrepancy between these latter conclusions and our results. Mutational disruption of the base pairing of helices A and B of the HVR (Fig. 1) was found to reduce, but not eliminate, DI RNA replication (30). Compensatory mutations that restored base pairing concomitantly returned DI RNA replication to wild-type levels. More significantly, small deletions, of nt 76 through 81 within oct and of nt 131 through 135 comprising the internal loop opposite oct, were seen to completely abolish DI RNA transcription (17); the second of these deletions also almost completely eliminated replication (18). The meaning of these DI RNA results needs to be reconsidered in light of our demonstration that similar, as well as much more extensive, HVR deletions yielded viruses that were fully competent in both gRNA replication and sgRNA transcription. Since DI RNAs replicate as a result of successful competition with helper viruses, DI RNAs are likely to be much more sensitive than genomic RNAs to small effects on replicative fitness brought about by alterations in the HVR. Nevertheless, it is clear from our results that none of the previously described host factor binding sites within the HVR plays an essential role in MHV RNA synthesis. Our findings therefore simplify the set of required cis-acting elements that must be considered to obtain an understanding of the basic mechanism of coronavirus RNA synthesis.
Acknowledgments
We thank Bilan Hsue and Todd Miller for construction of the plasmid precursor for the OctG1C mutant. We are grateful to the Molecular Genetics Core Facility of the Wadsworth Center for DNA sequencing.
This work was supported by Public Health Service grants AI 45695 and AI 060755 from the National Institutes of Health. The animal core of the Wadsworth Center, which was used for mouse infection experiments, is funded in part by National Institutes of Health grant U54AI7158 for the Northeast Biodefense Center.
Footnotes
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Boursnell, M. E. G., M. M. Binns, I. J. Foulds, and T. D. K. Brown. 1985. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J. Gen. Virol. 66:573-580. [DOI] [PubMed] [Google Scholar]
- 2.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, D. M., C. T. Cornell, G. P. Tran, J. H. Nguyen, and B. L. Semler. 2005. An authentic 3′ noncoding region is necessary for efficient poliovirus replication. J. Virol. 79:11962-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, M. M., K. C. MacNamara, L. San Mateo, H. Shen, and S. R. Weiss. 2004. Effects of an epitope-specific CD8+ T-cell response on murine coronavirus central nervous system disease: protection from virus replication and antigen spread and selection of epitope escape mutants. J. Virol. 78:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, A. R., R. L. Knobler, H. Powell, and M. J. Buchmeier. 1982. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology 119:358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compton, S. R., S. W. Barthold, and A. L. Smith. 1993. The cellular and molecular pathogenesis of coronaviruses. Lab. Anim. Sci. 43:15-28. [PubMed] [Google Scholar]
- 8.Dalton, K., R. Casais, K. Shaw, K. Stirrups, S. Evans, P. Britton, T. D. K. Brown, and D. Cavanagh. 2001. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 75:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A. M., H. Volders, C. A. Koetzner, P. S. Masters, and P. J. M. Rottier. 2002. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 76:12491-12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goebel, S. J., J. Taylor, and P. S. Masters. 2004. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 78:7846-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haring, J., and S. Perlman. 2001. Mouse hepatitis virus. Curr. Opin. Microbiol. 4:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton, R. M., and L. R. Pease. 1991. Recombination and mutagenesis of DNA sequences using PCR, p. 217-247. In M. J. McPherson (ed.), Directed mutagenesis, a practical approach. IRL Press, New York, NY.
- 15.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsue, B., and P. S. Masters. 1997. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 71:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, P., and M. M. C. Lai. 1999. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering conformation. J. Virol. 73:9110-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, P., and M. M. C. Lai. 2001. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′ untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst, K. R., L. Kuo, C. A. Koetzner, R. Ye, B. Hsue, and P. S. Masters. 2005. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 79:13285-13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iacono, K. T., L. Kazi, and S. R. Weiss. 2006. Both spike and background genes contribute to murine coronavirus neurovirulence. J. Virol. 80:6834-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, R. F., M. Feng, P. Liu, J. J. Millership, B. Yount, R. S. Baric, and J. L. Leibowitz. 2005. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570-14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonassen, C. M., T. Kofstad, I. L. Larsen, A. Lovland, K. Handeland, A. Follestad, and A. Lillehaug. 2005. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos). J. Gen. Virol. 86:1597-1607. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y.-N., Y. S. Jeong, and S. Makino. 1993. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology 197:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapps, W., B. G. Hogue, and D. A. Brian. 1987. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology 157:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, Y.-J., and M. M. C. Lai. 1993. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J. Virol. 67:6110-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, Y.-J., C.-L. Liao, and M. M. C. Lai. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Q., R. F. Johnson, and J. L. Leibowitz. 2001. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 75:12105-12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Q., W. Yu, and J. L. Leibowitz. 1997. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology 232:74-85. [DOI] [PubMed] [Google Scholar]
- 32.Luytjes, W., H. Gerritsma, and W. J. M. Spaan. 1996. Replication of synthetic interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology 216:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacNamara, K. C., M. M. Chua, J. J. Phillips, and S. R. Weiss. 2005. Contributions of the viral genetic background and a single amino acid substitution in an immunodominant CD8+ T-cell epitope to murine coronavirus neurovirulence. J. Virol. 79:9108-9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masters, P. S., and P. J. M. Rottier. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 287:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, P. B. 1999. Structural motifs in RNA. Annu. Rev. Biochem. 68:287-300. [DOI] [PubMed] [Google Scholar]
- 37.Nanda, S. K., and J. L. Leibowitz. 2001. Mitochondrial aconitase binds to the 3′ untranslated region of the mouse hepatitis virus genome. J. Virol. 75:3352-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanda, S. K., R. F. Johnson, Q. Liu, and J. L. Leibowitz. 2004. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′ untranslated region of the murine hepatitis virus genome. Arch. Virol. 149:93-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navas, S., S.-H. Seo, M. M. Chua, J. Das Sarma, E. Lavi. S. T. Hingley, and S. R. Weiss. 2001. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 75:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navas, S., and S. R. Weiss. 2003. Murine coronavirus-induced hepatitis: JHM genetic background eliminates A59 spike-determined hepatotropism. J. Virol. 77:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ontiveros, E., T. S. Kim, T. M. Gallagher, and S. Perlman. 2003. Enhanced virulence mediated by the murine coronavirus, mouse hepatitis virus strain JHM, is associated with a glycine at residue 310 of the spike glycoprotein. J. Virol. 77:10260-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, D., C. A. Koetzner, T. McMahon, Y. Zhu, and P. S. Masters. 1995. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J. Virol. 69:5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlman, S. 1998. Pathogenesis of coronavirus-induced infections. Adv. Exp. Med. Biol. 440:503-513. [PubMed] [Google Scholar]
- 44.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierangeli, A., M. Bucci, P. Pagnotti, A. M. Degener, and R. Perez Bercoff. 1995. Mutational analysis of the 3′-terminal extra-cistronic region of poliovirus RNA: secondary structure is not the only requirement for minus strand RNA replication. FEBS Lett. 374:327-332. [DOI] [PubMed] [Google Scholar]
- 46.Pilipenko, E. V., K. V. Poperechny, S. V. Maslova, W. J. Melchers, H. J. Slot, and V. I. Agol. 1996. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (′kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 47.Rempel, J. D., S. J. Murray, J. Meisner, and M. J. Buchmeier. 2004. Mouse hepatitis virus neurovirulence: evidence of a linkage between S glycoprotein expression and immunopathology. Virology 318:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarnow, P., H. D. Bernstein, and D. Baltimore. 1986. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc. Natl. Acad. Sci. USA 83:571-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schreiber, S. S., T. Kamahora, and M. M. C. Lai. 1989. Sequence analysis of the nucleocapsid protein gene of human coronavirus 229E. Virology 169:142-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd, S., and B. L. Semler. 1996. Structure-infectivity analysis of the human rhinovirus genomic RNA 3′ non-coding region. Nucleic Acids Res. 24:2133-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, A. K., L Wang, L. W. Sneed, and E. W. Collisson. 1993. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 28:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams, G. D., R. Y. Chang, and D. A. Brian. 1999. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 73:8349-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wise, A. G., M. Kiupel, and R. K. Maes. 2006. Molecular characterization of a novel coronavirus associated with epizootic catarrhal enteritis (ECE) in ferrets. Virology 349:164-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo, P. C. Y., S. K. P. Lau, C.-m. Chu, K.-h. Chan, H.-w. Tsoi, Y. Huang, B. H. L. Wong, R. W. S. Poon, J. J. Cai, W.-k. Luk, L. L. M. Poon, S. S. Y. Wong, Y. Guan, J. S. M. Peiris, and K.-Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, H.-Y., J. S. Guy, D. Yoo, R. Vlasak, E. Urbach, and D. A. Brian. 2003. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology 315:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, W., and J. L. Leibowitz. 1995. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J. Virol. 69:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, W., and J. L. Leibowitz. 1995. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology 214:128-138. [DOI] [PMC free article] [PubMed] [Google Scholar]