Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8) is associated with Kaposi's sarcoma (KS) as well as primary effusion lymphomas (PEL). The expression of viral proteins capable of inactivating the p53 tumor suppressor protein has been implicated in KSHV oncogenesis. However, DNA-damaging drugs such as doxorubicin are clinically efficacious against PEL and KS, suggesting that p53 signaling remains intact despite the presence of KSHV. To investigate the functionality of p53 in PEL, we examined the response of a large number of PEL cell lines to doxorubicin. Two out of seven (29%) PEL cell lines harbored a mutant p53 allele (BCBL-1 and BCP-1) which led to doxorubicin resistance. In contrast, all other PEL containing wild-type p53 showed DNA damage-induced cell cycle arrest, p53 phosphorylation, and p53 target gene activation. These data imply that p53-mediated DNA damage signaling was intact. Supporting this finding, chemical inhibition of p53 signaling in PEL led to doxorubicin resistance, and chemical activation of p53 by the Hdm2 antagonist Nutlin-3 led to unimpaired induction of p53 target genes as well as growth inhibition and apoptosis.
Primary effusion lymphoma (PEL) or body cavity-based lymphoma is a rapidly progressive disease often arising in the context of human immunodeficiency virus (HIV)/AIDS, but it is also seen in HIV-negative transplant recipients and in the elderly. PEL exists as a fluid tumor of B-cell origin within the serous cavities of the body. Current multidrug chemotherapies against PEL, such as CHOP (cyclophosphamide, doxorubicin [Dox], vincristine, and prednisone) target proliferating cells and typically include a DNA-damaging agent such as doxorubicin. These therapies are initially successful (64). However, the immunocompromised status of PEL patients makes them highly susceptible to the myelosuppressive side effects of high-dose chemotherapy regimens. As a result, the median survival time for those diagnosed with PEL is typically less than 3 months (1, 12), and there exists an urgent need for novel therapies against PEL.
All PEL tumor cells are infected with Kaposi's sarcoma-associated herpesvirus (KSHV) (13, 49). KSHV is a member of the gamma-herpesviruses and was first isolated in the AIDS-associated malignancy Kaposi's sarcoma (KS) (15). KSHV remains largely latent in PEL and expresses only a limited set of genes, including Kaposin/K12, v-cyclin/orf72, the latency-associated nuclear antigen (LANA)/ORF73, v-FLIP/ORF71, and the viral micro-RNAs (19, 26) (8, 23, 35, 51, 53, 58, 59). PEL are dependent upon KSHV for survival, as loss of the KSHV genome results in cell death (30). These results suggest that one or more KSHV oncogenes are essential for PEL survival. Following the precedence of the small DNA tumor viruses such as simian virus 40 and human papilloma virus, KSHV too was hypothesized to contain viral genes that curb p53 function and thereby mediate viral oncogenesis.
p53 has been deemed the most frequently mutated gene in human cancer, with the majority of alterations localizing to the protein's DNA binding domain (reviewed in reference 32). In response to DNA-damaging chemotherapeutics, p53 is activated and functions as a sequence-specific transcription factor for a well-defined set of p53-responsive promoters. This in turn leads to a variety of cellular outcomes, including cell cycle arrest and apoptosis (reviewed in reference 69). p53 stability is tightly regulated by the ubiquitin ligase activity of the human Mdm2 homologue, Hdm2 (reviewed in reference 34). Overexpression of Hdm2 is frequently noted in human cancers and provides a mechanism to inactivate p53 epigenetically (45). The Mdm2-p53 binding site is well defined (44), and recently novel compounds, such as Nutlin-3, that inhibit the Hdm2-p53 interaction have been proposed as therapeutics for tumors maintaining wild-type p53 status (33, 68). Nutlin-3, unlike doxorubicin, does not induce cellular DNA damage, thus allowing for the direct assessment of p53 function independent of any other effects that may be induced by DNA-damaging drugs. Neither the efficacy of Nutlin-3 for PEL treatment nor the functional status of p53 in the context of KSHV-positive tumor cells has been previously studied.
To examine p53 functionally in KSHV-associated cancers, we employed the largest set of patient-derived PEL lines published to date. As previously reported for clinical PEL and KS biopsies (14, 38, 46), we found that p53 mutations were infrequent in PEL even after continued passage in culture (two of seven lines). The wild-type p53 genotype translated into a wild-type p53 phenotype, as PEL cells responded to physiologically relevant levels of doxorubicin. PEL treatment with doxorubicin led to p53 phosphorylation, activation of downstream target genes, and subsequent cell cycle arrest, consistent with an intact p53 signaling pathway. Moreover, PEL lines responded to Nutlin-3 in a p53- and Hdm2-dependent manner, evidencing a functional p53-Hdm2 pathway.
MATERIALS AND METHODS
Cell culture and materials.
PEL cells and cell culture conditions were previously described (Table 1) (2, 5, 7, 10, 13, 29, 42). Doxorubicin and Nutlin-3 (racemic) were obtained from Sigma and diluted in dimethyl sulfoxide (DMSO). Pifithrin-α was obtained from Alexis Biochemicals and stored in DMSO.
TABLE 1.
Analysis of PEL lines
p53 sequencing.
cDNA was prepared from all PEL cell lines using the RNeasy mini kit (QIAGEN), 100 U of Superscript II reverse transcriptase (Life Technologies), and random hexanucleotide primers (Amersham Pharmacia Biotech). The resulting cDNA was used to PCR amplify two regions of the p53 DNA binding domain spanning amino acids (aa) 126 to 224 and aa 225 to 331. The primers were as follows: aa 126 to 224 forward, 5′-TCTCCTTCCTCTTCCTACAG-3′; reverse, 5′-ACCCCAGTTGCAAACCAGAC-3′; aa 225 to 331 forward, 5′-GGGCCTGTGTTATCTCCTAG-3′; reverse, 5′-GAGGTCCCAAGACTTAGTAC-3′. PCR was performed using the GeneAmp High Fidelity PCR system (Applied Biosystems) according to the manufacturer's instructions. The resulting PCR products were cloned into pBluescript II KS(+) (Stratagene) and sequenced.
MTT/cell viability assays.
On day 0, cells were seeded at 3 × 105 cells/1.5 ml complete media in 12-well dishes, and drugs and/or was vehicle added. Each day thereafter, 50 μl of cells was removed and incubated in a 96-well plate with 0.1 mg/ml MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide; Sigma) for 4 h at 37°C and 5% CO2. The reaction was stopped, and formazan crystals were dissolved in a 0.1 N HCl-isopropanol solution. MTT metabolism (at an optical density of 570 nm [OD570]) was read and normalized to cell density (OD690). All assays were performed in triplicate. Graphs display the average growth relative to the untreated sample at day 1. Error bars represent the standard deviation. Alternatively, cell viability was determined from the 50-μl samples via trypan blue exclusion. The 50% inhibitory concentration (IC50) was calculated using a sigmoidal dose-response fit of the MTT assay results on day 4 and GraphPad Prism 4 software (GraphPad Software).
Western blotting.
Cells were seeded in two flasks each containing 40 ml complete media at a concentration of 2 × 105 cells/ml. The following morning, 0.025 μg/ml doxorubicin was added to both flasks, and 20 ml of one culture was immediately harvested (t = 0 h). Eight, 24, and 48 h postdrug addition, 20 ml of culture was removed and centrifuged, and the cell pellet stored at −80°C. The resulting pellets were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 50 nM Tris [pH 8.0]) containing 1 nM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail (Sigma), 10 nM sodium fluoride, 1 mM sodium vanadate, and 60 mM β-glycerophosphate. Lysates were quantified using the BCA Protein Assay kit (Pierce Inc.), and proteins were separated on SDS-12% polyacrylamide gels. Gels were transferred to Hybond-P (Amersham Biosciences) and probed for the indicated protein products. Primary antisera utilized were the following: p53 phosophoserine 15 (1:1,000; Cell Signaling), total p53 (1:1,000; Santa Cruz Biotechnologies), p21 (1:500; Santa Cruz), CDK6 (1:1,000; Santa Cruz), and Hdm2 (1:500; Ab-1; Calbiochem). Following incubation with horseradish peroxidase-conjugated secondary antibodies (goat-α-mouse immunoglobulin G; Santa Cruz; and goat-α-rabbit immunoglobulin G; Vector Laboratories Inc.) and ECL substrate (Pierce Inc.), antibody-antigen complexes were visualized via autoradiography. Quantification of protein expression was conducted using ImageQuant software. Hdm2 levels were normalized to CDK6 (loading control). The relative level of p21 was determined using ImageJ software (Wayne Rashband, National Institutes of Health). Specifically, p21 band density was first normalized to CDK6 (loading control) and then the quantity was determined relative to 0 h for each cell line.
Flow cytometry.
Cells were seeded and treated as described for the Western blotting experiments. At 0, 8, 24, and 48 h postdrug addition, cells were harvested by centrifugation, fixed, and stained as we previously described (20, 52). Cells were analyzed on a Becton-Dickinson FACscan flow cytometer. At least 10,000 cells were analyzed per sample and graphed using FlowJo software (Tree Star Inc.).
Array design and implementation.
Quantitative real-time PCR (QPCR) primers were designed using Primer 3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi [57]) according to our standard parameters (47). Where possible, primer sets were chosen to span an intron so as to avoid background signal from any residual genomic DNA (Table 2). Real-time QPCR analysis was conducted as previously published (21, 22). The fold induction was determined for each time point using the following calculation: fold change = 1.8(dCt1 − dCt2), where dCt1 and dCt2 are the cycle thresholds normalized to that of GAPDH at times 1 and 2, respectively). Data were clustered using ArrayMiner to produce a heat map diagram.
TABLE 2.
Primers for real-time QPCR p53 target gene analysis
| Target | Sense primer (5′ to 3′) | Antisense primer (5′ to 3′) | Size (bp) |
|---|---|---|---|
| p53 | AGGCCTTGGAACTCAAGGAT | TGAGTCAGGCCCTTCTGTCT | 140 |
| p21 | GGAAGACCATGTGGACCTGT | GGCGTTTGGAGTGGTAGAAA | 146 |
| PUMA | GACGACCTCAACGCACAGTA | CTGGGTAAGGGCAGGAGTC | 113 |
| Bcl-2 | ATGTGTGTGGAGAGCGTCAA | ACAGTTCCACAAAGGCATCC | 136 |
| Bax | GGGGACGAACTGGACAGTAA | CAGTTGAAGTTGCCGTCAGA | 122 |
| Bad | CGGAGGATGAGTGACGAGT | CCACCAGGACTGGAAGACTC | 123 |
| APO-1 | ATAAGCCCTGTCCTCCAGGT | TGGAAGAAAAATGGGCTTTG | 120 |
| KILLER | GATGGTCAAGGTCGGTGATT | CCCCACTGTGCTTTGTACCT | 79 |
| cdc25c | TCCTTAAAGGCGGCTACAGA | GGGCAGTAGCTCTGTGGTTC | 70 |
| EZH2 | TTCATGCAACACCCAACACT | CTCCCTCCAAATGCTGGTAA | 96 |
| p53R2 | GCCAGGACTCACTTTTTCCA | TCAGGCAAGCAAAGTCACAG | 71 |
| PIG3 | ACGCTGAAATTCACCAAAGG | AACCCATCGACCATCAAGAG | 107 |
| Gadd45 | GGAGGAAGTGCTCAGCAAAG | ATCTCTGTCGTCGTCCTCGT | 135 |
| 14-3-3α | ACCCAATTCGTCTTGGTCTG | GCTTCATCAAATGCCGTTTT | 105 |
| Glyceraldehyde-3-phosphate dehydrogenase | CCAGGTGGTCTCCTCTGACTTCTC | ATACCAGGAAATGAGCTTGACA | 107 |
Annexin V flow cytometry.
BCBL-1, BCP-1, and BC-3 cells were seeded in duplicate at a density of 2 × 105 cells/ml in a total of 40 ml. The cultures were immediately treated with either 5 μM Nutlin-3 (racemic) or vehicle (DMSO) alone. Forty-eight hours later, the cells were counted, spun down, and washed three times in ice-cold phosphate-buffered saline. Each pellet was resuspended in Annexin V binding buffer (0.01 M HEPES, pH 7.4, 14 mM NaCl, 2.5 mM CaCl2) to reach a final concentration of 1 × 107 cells/ml. Five 100-μl aliquots were generated from each condition, and four were incubated with 5 μl of fluorescein isothiocyanate-conjugated Annexin V antibody (Caltag Laboratories, Burlingame, CA) for 15 min at room temperature. The remaining sample was used as a control. Flow cytometry was performed on ∼10,000 cells per sample using a FacScan instrument and analyzed using FloJo software.
RESULTS
p53 (aa 126 to 331) is wild type in PEL based on sequence analysis.
As p53 is rarely mutated in KS, we hypothesized that other KSHV-associated tumor types would retain wild-type p53 status. To test this hypothesis, we examined the genotypes of seven culture-adapted PEL cell lines. This extensive panel of cultures is representative of all cells widely utilized in experimental research today (Table 1). All seven lines were KSHV positive. Two cell lines (BC-1 and JSC-1) were coinfected with Epstein-Barr virus (EBV) (10, 13). To determine the mutation status of p53, we employed high-fidelity PCR on PEL-derived cDNA with primers that cover the previously identified hot spot region (aa 126 to 331). The corresponding PCR products were cloned into pBluescript, and at least eight clones were sequenced per cell line. BCBL-1 cells were determined to be heterozygous for the M246I mutation as previously reported (38). BCP-1 cells contained a homozygous insertion at codon 262. All other cell lines contained wild-type p53 sequence. These data demonstrate that p53 mutations were present in ≤30% (two of seven) of PEL, all of which grow permanently in culture and form tumors in nude mice (Table 1 reference 65).
PEL growth is effectively limited by doxorubicin.
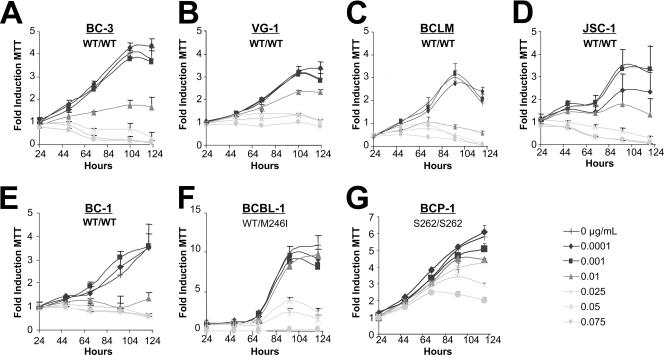
The current standard of care for PEL treatment includes chemotherapeutics such as anthracyclines (e.g., doxorubicin, daunorubicin, and mitoxantrone). These agents function through DNA intercalation and oxidative radical formation to induce the p53-dependent DNA damage pathway (reviewed in reference 28). Since p53 was wild type by sequence in PEL, we assessed the ability of doxorubicin (Dox) to limit cellular viability. Each cell line was seeded at equal density in 12-well dishes, and Dox and/or vehicle (water) was added to the culture at the indicated dosages. At designated times thereafter, a sample was removed from each culture and the cellular viability was assessed using an MTT assay. All treatments were performed in triplicate wells, and results of the MTT assays directly paralleled those obtained by trypan blue exclusion counts (data not shown). The dosages of doxorubicin utilized (0.001 to 0.075 μg/ml) were clinically relevant, as patient serum concentrations of the drug have been reported to be as high as 11 μg/ml following administration at 60 to 70 mg/m2 (6). As seen in Fig. 1, PEL lines harboring the wild-type p53 allele (BC-3, VG-1, BCLM, BC-1, and JSC-1) were susceptible to Dox (complete response at 0.01 to 0.025 μg/ml). This phenotype was independent of EBV status, as both the JSC-1 and BC-1 cell lines also responded to Dox treatment. A higher tolerance for Dox was observed in an allele-specific, dose-dependent manner in both BCP-1 and BCBL-1 cells. Significantly, BCP-1 proliferation continued in the presence of 0.075 μg/ml Dox. Utilizing data from day 4, wherein the optimal growth differential was observed for all lines, the IC50 was calculated for each cell line. Doxorubicin susceptibility directly correlated with p53 status (Table 1). These results demonstrate that PEL lines containing the wild-type p53 genotype also exhibit a wild-type p53 phenotype as measured by the cell response to the DNA-damaging agent Dox.
FIG. 1.
PEL lines are sensitive to doxorubicin. (A to G) PEL-derived cell lines were seeded in triplicate at 2 × 105 cells/ml. The indicated dosage of doxorubicin (in micrograms/milliliter) and/or vehicle (double-distilled H2O) was added to each sample. Twenty-four hours postseeding and treatment, a sample was removed and cell survival was scored by MTT assay. Similar MTT measurements were performed every 24 h as indicated. Raw MTT numbers were normalized to the average reading of the untreated sample at day 1. Data shown represent three independent points, with error bars indicating the standard deviations. WT, wild type.
p53 mediates DNA damage-induced growth arrest in PEL.
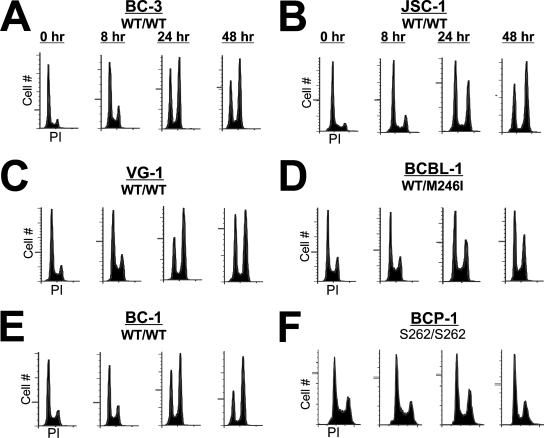
To assess whether Dox treatment resulted in growth arrest, cell cycle distribution was determined by propidium iodide (PI) incorporation. PEL lines were seeded at equal density and treated with an inhibitory concentration of Dox (0.025 μg/ml), and samples were analyzed at 0, 8, 24, or 48 h following treatment. As seen in Fig. 2, all PEL lines treated with Dox demonstrated marked arrest at the G1/S and G2/M checkpoints, indicative of a p53-mediated DNA damage response. By 48 h, the G2/M population of cells harboring wild-type p53 had increased dramatically (between 14 and 38%). In contrast, accumulation of BCP-1 and BCBL-1 cells, which contain a mutant p53 allele, in G2/M was significantly reduced (less than 10% increase). Analysis of the sub-G1 population by this method revealed little apoptotic induction in any of the cell lines within the time period examined. This observation may be explained by the relatively slow growth rate of the PEL lines in culture and/or a molecular mechanism that delays apoptosis. Together, these data support the growth analyses performed in Fig. 1 and demonstrate that DNA damage in p53 wild-type PEL lines resulted in cell cycle arrest.
FIG. 2.
Doxorubicin induces cell cycle arrest in PEL lines. (A to F) PEL lines were seeded at 2 × 105 cells per ml and treated the following day with 0.025 μg/ml doxorubicin. Cells were harvested 0, 8, 24, and 48 h posttreatment. The cells were fixed, and DNA content was analyzed by propidium iodide (PI) flow cytometry. Shown are histograms representing the cell cycle profile of ≥10,000 cells. WT, wild type.
The p53 signaling cascade is fully functional in response to doxorubicin.
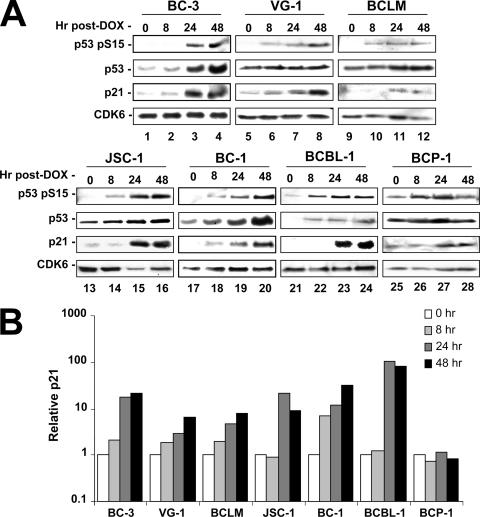
To further evaluate p53 function in PEL, we examined the ability of upstream DNA damage sensors such as ATM to activate and initiate p53 signaling in Dox-treated cells. Specifically, PEL lines were seeded at equal density, treated with an inhibitory concentration of Dox (0.025 μg/ml), and sampled at 0, 8, 24, or 48 h following treatment. Cell pellets were lysed, and equal protein concentrations were run on SDS-polyacrylamide gels. The resulting gels were transferred to nitrocellulose and probed as indicated. p53 phosphorylation on serine 15 is an indicator of upstream DNA damage signaling (3, 9) and was readily induced as early as 8 h post-Dox treatment (Fig. 3). Interestingly, BCP-1 cells showed constitutive, elevated levels of phosphoserine 15. However, p21 was not induced, suggesting that the S262 insertion leads to a defect in downstream signaling. All cell lines containing wild-type p53 displayed marked induction of the p53 target gene, p21 (cip1/waf-1) (24). These data are quantified in Fig. 3B. CDK6 served as a loading control and remained constant across all samples and all time points. All cells containing at least one wild-type copy of p53 responded to drug treatment, whereas the BCP-1 cell line (homozygous mutant) showed no elevation of p53 signaling as measured by p21 induction following Dox treatment. These data demonstrate that both upstream and downstream p53-dependent signaling was functional in p53 wild-type PEL irrespective of the presence of KSHV.
FIG. 3.
Doxorubicin treatment of PEL lines activates the p53-mediated DNA damage response. (A) PEL lines were seeded at 2 × 105 cells per ml in a total of 2 volumes of 40 ml each. The following morning, the cells were treated with 0.025 μg/ml doxorubicin and an initial aliquot of 20 ml was immediately obtained (t = 0 h). At the indicated time points following drug addition, cells were collected and pellets were frozen. Lysates from the time course were quantified, and equal amounts of protein were loaded for SDS-polyacrylamide gel electrophoresis analysis. Gels were transferred and blotted for the indicated protein products. Here, CDK6 is utilized as a loading control, p21 as a marker for p53 transcriptional activity, and phosphoserine 15 as an indicator of upstream DNA damage signaling. (B) Using densitometry, p21 expression was normalized to CDK6 (loading control), and the relative induction over levels observed at 0 h was determined for each time point.
Functional p53 is required to mediate DNA damage-induced inhibition of PEL growth.
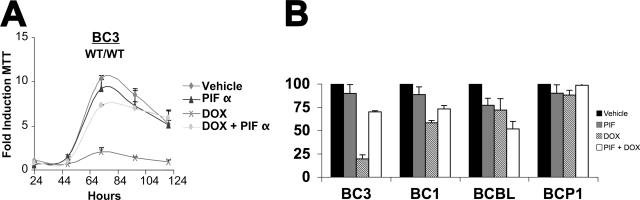
To test the hypothesis that activation of p53 is necessary to mediate cell cycle arrest in response to DNA-damaging agents, we conducted Dox treatment in the presence of the p53 inhibitor pifithrin α (p-fifty three inhibitor α [PIFα]) (43). BC-3 (wild type/wild type) cells were pretreated overnight with PIFα or vehicle. Dox (0.025 μg/ml) and/or vehicle was then added, and cell viability was assessed by MTT assays. Figure 4A shows our results graphed as the average fold growth increase relative to the untreated sample on day 1. PIFα-treated, Dox-exposed BC-3 cells demonstrated resistance to doxorubicin and proliferated similarly to the untreated controls. By contrast vehicle-treated, Dox-exposed BC-3 cells ceased to proliferate. PIFα or vehicle alone had no effect. These data imply that p53 is required to inhibit cellular proliferation following Dox treatment of PEL. Figure 4B shows the relative growth retardation as a percent of vehicle control (black bars) for a set of four PEL cell lines at 72 h posttreatment. PIFα alone (gray bar) had no or minor toxicity. Dox (striped bars) reduced cell growth significantly in the wild-type BC-3 and BC-1 cells, less in the heterozygous p53 BCBL-1 cells, and not at all in the homozygous p53 BCP-1 cells. PIFα (open bars) counteracted the effect of Dox in the wild-type p53 BC-3 and BC-1 cells but not the p53 mutant cells. In the p53 heterozygous BCBL-1, PIFα seemed to augment Dox, while in the p53 homozygous mutant p53 BCBP-1 cells PIFα had no effect.
FIG. 4.
PEL response to doxorubicin is p53 dependent. (A) BC-3 cells were seeded in triplicate at 2 × 105 cells per ml in a 12-well dish with the indicated amount of pifithrin-alpha (PIF) or vehicle (DMSO). Following 24 h of pretreatment, 0.025 μg/ml doxorubicin (DOX) or vehicle (double-distilled H20) was added to the media. MTT assays were then performed every 24 h post-DOX addition as described in the legend to Fig. 1. Cell growth was normalized to the MTT reading of vehicle-treated cells on day 1. Points represent the average reading of three independent samples with error bars indicating the standard deviations. (B) Relative growth (percent vehicle control) at 72 h of culture for the indicated PEL cell lines either mock treated (black bars), PIF treated (gray bars), DOX treated (striped bars), or PIF and DOX treated (open bars). WT, wild type.
Activation of p53 is sufficient to inhibit PEL growth independent of DNA damage.
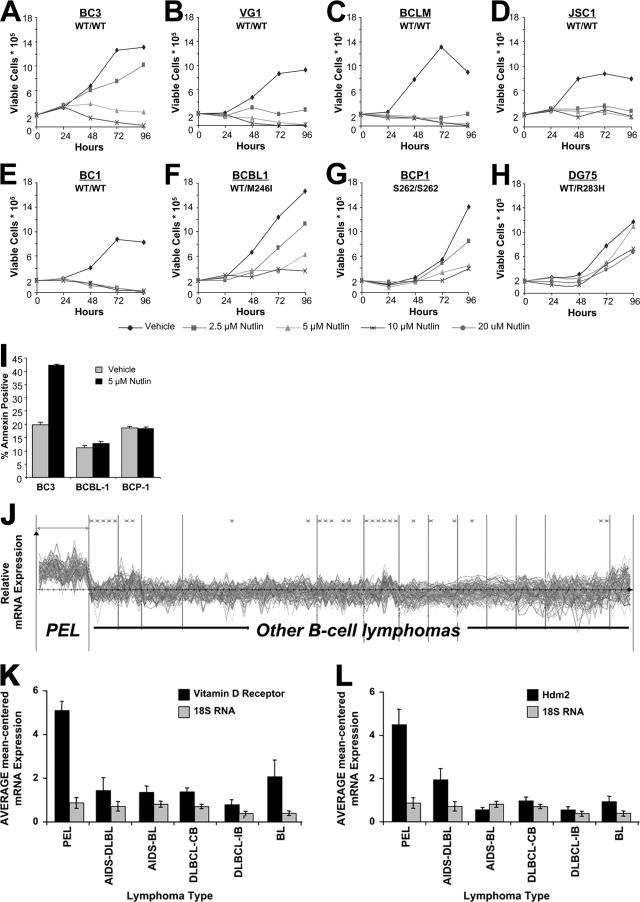
To test the hypothesis that activation of p53 is sufficient to mediate the cell cycle arrest in PEL, we used a recently discovered chemical activator of p53, Nutlin-3. Nutlins are selective compounds which target the Hdm2-p53 interaction to prevent p53 degradation (67, 68). To test the efficacy of Nutlin-3 in PEL, we conducted cell viability assays. Specifically, we seeded our panel of cells at equal density and added racemic Nutlin-3. Next, cell viability was determined through trypan blue exclusion counts. Nutlin-3 significantly reduced cell proliferation in the majority of PEL lines harboring wild-type p53 (VG-1, BCLM, JSC-1, and BC-1) at low dosage (2.5 μM) (Fig. 5). BC-3 cells displayed resistance to low-dose Nutlin-3 but ceased proliferation readily at a 5 μM dose. Growth inhibition by Nutlin-3 was compromised at low doses in PEL cells harboring a mutant p53 allele (BCP-1 and BCBL-1). As expected, DG75 B-cell lymphoma cells which harbored mutant p53 (17), but neither KSHV nor EBV, were completely resistant to Nutlin-3. The p53 dependence of Nutlin-3 was even more striking when we used Annexin V positivity as a measure for cell death (Fig. 5I). Thus, Nutlin-3 response follows p53 status in a gene-dose-dependent manner, i.e., the susceptibility to Nutlin increased by the following relationship: homozygous mutant p53 (BCP-1 and S262/S262) < heterozygous mutant p53 (BCBL-1 and M246I) < homozygous wild-type p53 PEL. This validates Nutlin-3 as a p53-dependent treatment alternative for PEL. Moreover, use of Nutlin-3 in these studies demonstrates that p53 activation, independent of DNA damage, is sufficient to cause cell cycle arrest in PEL.
FIG. 5.
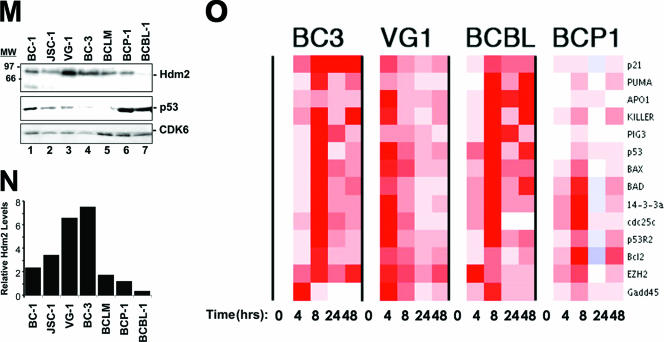
Nutlin-3 efficacy in PEL is Hdm2 and p53 dependent. (A to H) PEL lines were seeded at 2 × 105 cells/ml in a total of 5 ml. The indicated dosage of Nutlin-3 (racemic) or vehicle (DMSO) was then added to the cells. Twenty-four, 48, 72, and 96 h post-Nutlin-3 addition, cell viability was determined using trypan blue exclusion. DG75 cells do not contain KSHV or EBV. (I) BCBL-1, BCP-1, and BC-3 cells were seeded in media containing 5 μM Nutlin or vehicle (DMSO) alone. Forty-eight hours later, samples were analyzed in quadruplicate for Annexin V positivity. Bar graphs represent the average Annexin V-positive population in each sample. Error bars show the standard deviation. WT, wild type. (J) Transcription profile analysis of Hdm2 transcript levels in PEL. Using Affymetrix data from 101 B-cell tumors and controls previously recorded by Klein et al. (40), we identified mRNAs that classify PEL away from all other tumors. These markers include the previously identified vitamin D receptor (K) and Hdm2 (L) and are shown using a bar graph representation of the average of median-centered mRNA levels for each tumor type. Each group represents ≥6 examples. (BL, Burkitt's lymphoma; DLBL, diffuse large B-cell lymphoma; CB, centroblastic; IB, immunoblastic). (M) PEL lines grown in complete media were harvested, and lysates were quantified. Equal protein quantities were separated by SDS-polyacrylamide gel electrophoresis, transferred, and immunoblotted for Hdm2, p53, and CDK6 (loading control) expression (top panel). Band intensities were quantified using ImageQuant software and normalized to CDK6. (N) The relative expression of Hdm2. (O) PEL lines were seeded, treated with 5 μM Nutlin-3, and harvested at increasing time points as described in the legend to Fig. 3. Following RNA isolation, cDNA was generated using random hexamer-primed reverse transcription. cDNA was subjected to quantitative real-time PCR for the indicated targets. Data were collected, and the relative change in gene expression was determined by calculating the difference between dCt1 and dCt2. Data were analyzed using ArrayMiner. Red represents gene induction relative to the median of all genes and data points in the array.
Nutlin efficacy directly correlates with Hdm2 status (50). To examine whether increased Hdm2 expression levels can explain the robust response of PEL to Nutlin, we first examined Hdm2 mRNA levels in PEL using data from Klein et al. (40), which represents the transcriptional profile of over 60 B-cell tumors. Hdm2 mRNA levels served as a class prediction marker for PEL, clearly separating PEL samples from all others in the study (Fig. 5J, top panel). In fact, the predictive value of Hdm2 mRNA for the classification of PEL was as high as that of a previously validated marker, the vitamin D receptor (36). To verify Hdm2 protein expression we performed Western blotting analysis. Hdm2 levels were high and relatively consistent across all PEL lines (Fig. 5K), with the exception of BCBL-1 cells, which showed lower Hdm2 levels (Fig. 5L, lane 7). In sum, intact p53 signaling as well as elevated levels of Hdm2 contribute to the marked sensitivity of PEL to Nutlin-3.
Previous reports demonstrate that the induction of p53 target genes in response to Nutlin-3 treatment is tumor type dependent (16, 41, 60, 67). To test the hypothesis that the p53 transcriptional response is consistent across multiple PEL tumors, we developed a quantitative real-time QPCR array for 14 experimentally validated p53 target genes (Table 2). PEL were treated with 5 μM Nutlin-3 (an inhibitory concentration for all cell lines), and cells were harvested 0, 8, 24, and 48 h thereafter. RNA was isolated, reverse transcribed, and subjected to real-time QPCR. All PEL lines harboring the wild-type p53 allele showed induction of p53 target genes in response to Nutlin-3, which peaked at 8 h (Fig. 5M). By contrast, target gene induction was limited in the p53 mutant BCP-1 cells. Neither nontemplate control nor reverse transcriptase-omitting reactions generated PCR products as determined by gel electrophoresis and melting curve analysis (data not shown). These findings suggest that the p53 status of the BCP-1 line (S262/S262) limits p53 transcriptional capabilities. The observation that BCBL-1 cells robustly induce all p53 target genes is consistent with the previously reported phenotype of the M246I mutation (56) and indicates that BCBL-1 cells may have developed apoptotic resistance through additional mutations. Together, these findings show that p53 signaling is fully functional in PEL, engaging multiple pathways to mediate the therapeutic effects of Nutlin-3 and DNA-damaging agents.
DISCUSSION
DNA-damaging chemotherapeutic regimens are clinically efficacious for the KSHV-associated malignancies KS and PEL. Typically, the tumor response to chemotherapy is dependent upon functional p53. However, an in-depth analysis of p53 function in PEL has been missing, because multiple proteins of the PEL-associated human tumor virus KSHV were shown to inhibit p53 in ectopic expression experiments (27, 31, 48, 55, 61, 62), thus asserting the dogma that all DNA tumor viruses inactivate p53. If this were true, the presence of KSHV would severely limit the treatment options for PEL and other KSHV-associated cancers. PEL should not be responsive to CHOP, which is in contrast to the clinical experience (64). To test the hypothesis that p53 is fully functional in PEL despite the presence of viral oncogenes, we assessed p53 status and function in response to the prototypical DNA-damaging agent doxorubicin. Analyzing the most extensive panel of PEL lines published to date, we find that, similar to KS (38), p53 mutations are rarely detected in PEL (Table 1). DNA damage-induced p53 activation is intact in PEL harboring wild-type p53, as evidenced by target gene induction and cell cycle arrest following treatment with clinically relevant doses of doxorubicin (Fig. 1, 2, and 3). These data imply that in PEL the p53 status dictates the response to chemotherapeutics regardless of KSHV viral latent proteins.
We corroborated our initial observations by modifying p53 signaling using the well-characterized activator Nutlin-3 (Fig. 5) or the p53 inhibitor pifithrin-alpha (Fig. 4). The phenotypic response to each of these drugs, as well as the p53-dependent transcriptional response, correlated tightly with p53 status, adducing that DNA damage-induced p53 signaling is functional. How, then, can we reconcile these data with reports demonstrating disruption of p53 signaling by lytic and latent KSHV viral proteins? KSHV lytic proteins have been shown to inhibit p53 function through coactivator sequestration (Rta/ORF50) (18, 31), repression of ATM-mediated DNA damage signaling (v-IRF) (62), or other unknown mechanisms (K-bZIP) (48). Neither Rta/ORF50, K-bZIP, nor vIRF-1 is expressed during latency in PEL (26). These transcripts are only detectable upon viral reactivation. Herpesvirus reactivation leads to replication of the viral genomic DNA, which is resolved through recombination into unit-length pieces that are packed into the virion. This process generates many DNA double-strand ends, and it is therefore highly plausible that KSHV evolved to inhibit the p53/ATM response during the lytic phase of the viral life cycle.
The KSHV latency-associated nuclear antigen (LANA) also binds and inhibits p53 function in reporter assays (27, 71). Conversely, p53 can inhibit the LANA promoter (37). Unlike the lytic proteins, LANA is constitutively expressed in all PEL cells (19, 23, 66). LANA, like p53, is a relatively “sticky” protein, binding to many partners in vitro. Due to the multitude of LANA binding partners (≥10) and functions (63), it has been considerably more difficult to establish the specificity and functionality of the p53-LANA complex. LANA binds to the histone core components H2A, H2B, and H1, thereby tethering the viral episome to host chromatin (4). This interaction results in the characteristic nuclear speckled pattern observed using LANA-specific monoclonal antibodies (39, 54). It is conceivable that some p53 would be sequestered into these complexes, which also contain Ku70, Ku80, and PARP-1. The results presented here show that the nature of the LANA-p53 complex is such that enough functional p53 is available to mediate PEL cell cycle inhibition in response to DNA damage or to Nutlin-3. In fact, upon Nutlin-3 treatment p53 and LANA occupy distinct, nonoverlapping nuclear compartments (Fig. S1 in the supplemental material).
The rarity of p53 mutations observed herein expands upon an earlier report (46), which similarly did not detect p53 mutations in primary PEL biopsies. The p53 mutations in two of the cell lines examined (Table 1) may reflect the origins of the patient samples. BCP-1 cells (p53, S262/S262) were derived from an HIV-negative, 94-year-old man with previous history of both KS and colon cancer (5). This individual was treated for 3 years with prolonged chemotherapy before succumbing to the disease, whereupon the BCP-1 cell line was isolated. Likewise, BCBL-1 (wild type/M246I) cells were derived from an HIV-positive patient who underwent prior chemotherapeutic treatment with doxorubicin (42). Unlike other primary effusions, which typically fail to grow after a period of 14 days upon explantation in culture, BCBL-1 cells grew rapidly with no discernible lag phase (B. Herndier and D. Ganem, personal communication). BCLM and JSC-1 were isolated prior to therapeutic administration and maintained wild-type p53 function. Thus, long-term PEL culture is compatible with p53 activity, and only prolonged treatment with DNA-damaging agents in vivo appears to select for p53 mutations.
The M246I and S262 mutations identified in these PEL lines were previously reported in other cancers. The M246I mutation (BCBL-1) was initially identified in the H23 lung cancer cell line and was reported to maintain DNA binding to consensus p53 binding elements (56). However, transactivation of nonconsensus elements by p53 M246I was limited, resulting in decreased apoptotic induction. While the Mdm2 promoter (consensus element) was effectively induced by M246I, PIG3 (nonconsensus) transactivation was significantly reduced. This is consistent with the phenotype of BCBL-1 cells, which demonstrated intense induction of p53 target genes following Nutlin-3 treatment yet failed to undergo significant apoptosis (Fig. 5). Far less is known about the p53 S262 insertion mutant. Previously identified in pancreatic cancer (11), a characterization of the mutant has yet to be conducted. Our findings suggest that S262 has little transactivation potential, as few genes were induced in the Nutlin-3 array studies. Additionally, BCP-1 cells (S262/S262) demonstrated a low background level of all p53 transcripts assayed, further implying that this signaling pathway is disrupted (data not shown). Understanding how individual p53 mutations contribute to therapeutic response will be essential to providing individualized PEL therapy.
Structural studies of the p53-Mdm2 interaction have revealed evidence that three key residues (Phe, Trp, and Lev) of p53 bind to a deep cavity on the Mdm2 surface (44). Nutlins represent the most potent and selective inhibitors of this interaction described to date. Nutlins are cis-imidizoline analogs, capable of penetrating cellular membranes, and thus can be administered orally (68). Nutlin-3 was well tolerated in murine xenograph studies (up to 200 mg/g of body weight twice daily), wherein the growth of p53-positive tumors was inhibited (67). Recent studies implicate Nutlin-3 as a novel therapeutic for B-cell chronic lymphocytic leukemia (B-CLL) and acute myeloid leukemia (AML) (16, 41, 60). Interestingly, these previous studies utilized the purified, active Nutlin-3a enantiomer. Herein, we studied the effects of the racemic mixture (containing a 1:1 ratio of active [3a] versus inactive [3b] enantiomer). By comparison, PELs were more sensitive to Nutlin-3 treatment than B-CLL and AML (Fig. 5; 2.5 to 5.0 μM racemic versus 4.7 and 5 μM Nutlin-3a, respectively), perhaps reflecting their phenotypically elevated Hdm2 expression. PELs harboring the wild-type p53 allele exhibited significant apoptosis following Nutlin-3 treatment; however, PEL lines with homozygous, mutant p53 failed to proliferate only at increased doses of drug (Fig. 5). BC-3 cells deviated somewhat from the phenotype of other p53 wild-type cell lines, and further studies are under way to clarify this phenotype. One possible explanation is that BC-3 cells express a high level of HdmX (unpublished observation). Increased HdmX levels would explain their partial resistance to Nutlin-3, since Nutlin-3 fails to block the p53-HdmX interaction (50). Alternatively, Nutlin-3 may affect other targets than p53 (25, 70). A key finding of this study is that Hdm2 is overexpressed in PEL. Moreover, Hdm2 mRNA levels can be utilized to classify PEL away from diffuse large B-cell lymphoma and other B-cell-lymphoproliferative diseases (Fig. 5). The underlying mechanism for this phenotype is currently under investigation. Regardless, these data explain the dramatic susceptibility of this tumor type to Nutlin-3.
The use of a novel targeted p53 real-time QPCR array allowed us to molecularly characterize PEL response to Nutlin-3. Activation of the p53 transcriptome following Nutlin-3 addition differs between PEL and other hematological malignancies. In B-CLL, Nutlin-3a activates Mdm2, p21, and PUMA expression but not BAX (16). In AML, Nutlin-3a activates Mdm2, p21, and NOXA but not PUMA or BAX (41). Here we find that in PEL, Nutlin-3 rapidly (<8 h) activates multiple p53 targets, including p21, NOXA, PUMA, and BAX (Fig. 5). Independent of the specific transcriptional targets activated, the cellular outcome, namely growth inhibition and apoptosis, is the same for PEL, B-CLL, and AML. PEL is a rapidly progressing disease, wherein the immunocompromised status of the patient often influences the physician's ability to administer therapy. Our findings suggest that determination of p53 and Hdm2 status is critical to the assessment of potential therapeutic regimens. KSHV oncogenes did not influence the ability of PEL to respond to DNA-damaging agents. However, they may modulate p53 function at steady-state growth or during KSHV lytic replication. Nutlins and other p53-activating compounds may prove highly efficacious for the treatment of PEL patients.
Supplementary Material
Acknowledgments
We thank D. Ganem, R. Ambinder, D. Stadden, E. Cesarman, and W. Harrington for providing the cell lines utilized in these studies. We thank B. Damania and K. Knudsen for critical reading of the manuscript as well as members of the Dittmer laboratory, L. Mayo, and A. Baldwin for their helpful discussions. We especially thank Thomas W. Marshall and James E. Bear for providing the equipment and expertise to generate the confocal images shown in Fig. S1 in the supplemental material.
This work was supported by NIH grants CA109232 and CA700580 to D.P.D., the UNC Lineberger Comprehensive Cancer Center, and the Leukemia and Lymphoma Society of America. C.E.P. is supported by NIH training grant CA009156.
Footnotes
Published ahead of print on 22 November 2006.
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Ansari, M. Q., D. B. Dawson, R. Nador, C. Rutherford, N. R. Schneider, M. J. Latimer, L. Picker, D. M. Knowles, and R. W. McKenna. 1996. Primary body cavity-based AIDS-related lymphomas. Am. J. Clin. Pathol. 105:221-229. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 3.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 4.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856-861. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff, C., S. J. Gao, L. E. Healy, S. Matthews, A. J. Thomas, L. Coignet, R. A. Warnke, J. A. Strauchen, E. Matutes, O. W. Kamel, P. S. Moore, R. A. Weiss, and Y. Chang. 1998. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood 91:1671-1679. [PubMed] [Google Scholar]
- 6.Bramwell, V. H., D. Morris, D. S. Ernst, I. Hings, M. Blackstein, P. M. Venner, E. I. Ette, M. W. Harding, A. Waxman, and G. D. Demetri. 2002. Safety and efficacy of the multidrug-resistance inhibitor biricodar (VX-710) with concurrent doxorubicin in patients with anthracycline-resistant advanced soft tissue sarcoma. Clin. Cancer Res. 8:383-393. [PubMed] [Google Scholar]
- 7.Brander, C., P. O'Connor, T. Suscovich, N. G. Jones, Y. Lee, D. Kedes, D. Ganem, J. Martin, D. Osmond, S. Southwood, A. Sette, B. D. Walker, and D. T. Scadden. 2001. Definition of an optimal cytotoxic T lymphocyte epitope in the latently expressed Kaposi's sarcoma-associated herpesvirus kaposin protein. J. Infect. Dis. 184:119-126. [DOI] [PubMed] [Google Scholar]
- 8.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casey, G., Y. Yamanaka, H. Friess, M. S. Kobrin, M. E. Lopez, M. Buchler, H. G. Beger, and M. Korc. 1993. p53 mutations are common in pancreatic cancer and are absent in chronic pancreatitis. Cancer Lett. 69:151-160. [DOI] [PubMed] [Google Scholar]
- 12.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 13.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 14.Chadburn, A., E. Hyjek, S. Mathew, E. Cesarman, J. Said, and D. M. Knowles. 2004. KSHV-positive solid lymphomas represent an extra-cavitary variant of primary effusion lymphoma. Am. J. Surg. Pathol. 28:1401-1416. [DOI] [PubMed] [Google Scholar]
- 15.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 16.Coll-Mulet, L., D. Iglesias-Serret, A. F. Santidrian, A. M. Cosialls, M. de Frias, E. Castano, C. Campas, M. Barragan, A. Fernandez de Sevilla, A. Domingo, L. T. Vassilev, G. Pons, and J. Gil. 2006. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood 107:4109-4114. [DOI] [PubMed] [Google Scholar]
- 17.Crook, T., G. A. Parker, M. Rozycka, S. Crossland, and M. J. Allday. 1998. A transforming p53 mutant, which binds DNA, transactivates and induces apoptosis reveals a nuclear:cytoplasmic shuttling defect. Oncogene 16:1429-1441. [DOI] [PubMed] [Google Scholar]
- 18.Damania, B., J. H. Jeong, B. S. Bowser, S. M. DeWire, M. R. Staudt, and D. P. Dittmer. 2004. Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J. Virol. 78:5491-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dittmer, D. P. 2003. Transcription profile of Kaposi's sarcoma-associated herpesvirus in primary Kaposi's sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 63:2010-2015. [PubMed] [Google Scholar]
- 22.Dittmer, D. P., C. M. Gonzalez, W. Vahrson, S. M. DeWire, R. Hines-Boykin, and B. Damania. 2005. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J. Virol. 79:8637-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 25.Elison, J. R., D. Cobrinik, N. Claros, D. H. Abramson, and T. C. Lee. 2006. Small molecule inhibition of HDM2 leads to p53-mediated cell death in retinoblastoma cells. Arch. Ophthalmol. 124:1269-1275. [DOI] [PubMed] [Google Scholar]
- 26.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76:6213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 28.Fridman, J. S., and S. W. Lowe. 2003. Control of apoptosis by p53. Oncogene 22:9030-9040. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh, S. K., C. Wood, L. H. Boise, A. M. Mian, V. V. Deyev, G. Feuer, N. L. Toomey, N. C. Shank, L. Cabral, G. N. Barber, and W. J. Harrington, Jr. 2003. Potentiation of TRAIL-induced apoptosis in primary effusion lymphoma through azidothymidine-mediated inhibition of NF-kappa B. Blood 101:2321-2327. [DOI] [PubMed] [Google Scholar]
- 30.Godfrey, A., J. Anderson, A. Papanastasiou, Y. Takeuchi, and C. Boshoff. 2005. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood 105:2510-2518. [DOI] [PubMed] [Google Scholar]
- 31.Gwack, Y., S. Hwang, H. Byun, C. Lim, J. W. Kim, E. J. Choi, and J. Choe. 2001. Kaposi's sarcoma-associated herpesvirus open reading frame 50 represses p53-induced transcriptional activity and apoptosis. J. Virol. 75:6245-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris, S. L., and A. J. Levine. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899-2908. [DOI] [PubMed] [Google Scholar]
- 33.Issaeva, N., A. Friedler, P. Bozko, K. G. Wiman, A. R. Fersht, and G. Selivanova. 2003. Rescue of mutants of the tumor suppressor p53 in cancer cells by a designed peptide. Proc. Natl. Acad. Sci. USA 100:13303-13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwakuma, T., and G. Lozano. 2003. MDM2, an introduction. Mol. Cancer Res. 1:993-1000. [PubMed] [Google Scholar]
- 35.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenner, R. G., K. Maillard, N. Cattini, R. A. Weiss, C. Boshoff, R. Wooster, and P. Kellam. 2003. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA 100:10399-10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katano, H., Y. Sato, and T. Sata. 2001. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92:3076-3084. [DOI] [PubMed] [Google Scholar]
- 39.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein, U., A. Gloghini, G. Gaidano, A. Chadburn, E. Cesarman, R. Dalla-Favera, and A. Carbone. 2003. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood 101:4115-4121. [DOI] [PubMed] [Google Scholar]
- 41.Kojima, K., M. Konopleva, T. McQueen, S. O'Brien, W. Plunkett, and M. Andreeff. 2006. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Mdm2 and Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood 108:993-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komanduri, K. V., J. A. Luce, M. S. McGrath, B. G. Herndier, and V. L. Ng. 1996. The natural history and molecular heterogeneity of HIV-associated primary malignant lymphomatous effusions. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 13:215-226. [DOI] [PubMed] [Google Scholar]
- 43.Komarov, P. G., E. A. Komarova, R. V. Kondratov, K. Christov-Tselkov, J. S. Coon, M. V. Chernov, and A. V. Gudkov. 1999. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285:1733-1737. [DOI] [PubMed] [Google Scholar]
- 44.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 45.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 46.Nador, R. G., E. Cesarman, A. Chadburn, D. B. Dawson, M. Q. Ansari, J. Sald, and D. M. Knowles. 1996. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpes virus. Blood 88:645-656. [PubMed] [Google Scholar]
- 47.Papin, J., W. Vahrson, R. Hines-Boykin, and D. P. Dittmer. 2004. Real-time quantitative PCR analysis of viral transcription. Methods Mol. Biol. 292:449-480. [DOI] [PubMed] [Google Scholar]
- 48.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patton, J. T., L. D. Mayo, A. D. Singhi, A. V. Gudkov, G. R. Stark, and M. W. Jackson. 2006. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 66:3169-3176. [DOI] [PubMed] [Google Scholar]
- 51.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petre-Draviam, C. E., E. B. Williams, C. J. Burd, A. Gladden, H. Moghadam, J. Meller, J. A. Diehl, and K. E. Knudsen. 2005. A central domain of cyclin D1 mediates nuclear receptor corepressor activity. Oncogene 24:431-444. [DOI] [PubMed] [Google Scholar]
- 53.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 54.Rainbow, L., G. Platt, G. Simpson, R. Sarid, S. Gao, H. Stoiber, C. Herrington, P. Moore, and T. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth, J., P. Koch, A. Contente, and M. Dobbelstein. 2000. Tumor-derived mutations within the DNA-binding domain of p53 that phenotypically resemble the deletion of the proline-rich domain. Oncogene 19:1834-1842. [DOI] [PubMed] [Google Scholar]
- 57.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 58.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Secchiero, P., E. Barbarotto, M. Tiribelli, C. Zerbinati, M. G. di Iasio, A. Gonelli, F. Cavazzini, D. Campioni, R. Fanin, A. Cuneo, and G. Zauli. 2006. Functional integrity of the p53-mediated apoptotic pathway induced by the non-genotoxic agent nutlin-3a in B-cell chronic lymphocytic leukemia (B-CLL). Blood 107:4122-4129. [DOI] [PubMed] [Google Scholar]
- 61.Seo, T., J. Park, D. Lee, S. G. Hwang, and J. Choe. 2001. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus binds to p53 and represses p53-dependent transcription and apoptosis. J. Virol. 75:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin, Y. C., H. Nakamura, X. Liang, P. Feng, H. Chang, T. F. Kowalik, and J. U. Jung. 2006. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J. Virol. 80:2257-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Si, H., S. C. Verma, and E. S. Robertson. 2006. Proteomic analysis of the Kaposi's sarcoma-associated herpesvirus terminal repeat element binding proteins. J. Virol. 80:9017-9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonelli, C., M. Spina, R. Cinelli, R. Talamini, R. Tedeschi, A. Gloghini, E. Vaccher, A. Carbone, and U. Tirelli. 2003. Clinical features and outcome of primary effusion lymphoma in HIV-infected patients: a single-institution study. J. Clin. Oncol. 21:3948-3954. [DOI] [PubMed] [Google Scholar]
- 65.Staudt, M. R., Y. Kanan, J. H. Jeong, J. F. Papin, R. Hines-Boykin, and D. P. Dittmer. 2004. The tumor microenvironment controls primary effusion lymphoma growth in vivo. Cancer Res. 64:4790-4799. [DOI] [PubMed] [Google Scholar]
- 66.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 67.Tovar, C., J. Rosinski, Z. Filipovic, B. Higgins, K. Kolinsky, H. Hilton, X. Zhao, B. T. Vu, W. Qing, K. Packman, O. Myklebost, D. C. Heimbrook, and L. T. Vassilev. 2006. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. USA 103:1888-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vassilev, L. T., B. T. Vu, B. Graves, D. Carvajal, F. Podlaski, Z. Filipovic, N. Kong, U. Kammlott, C. Lukacs, C. Klein, N. Fotouhi, and E. A. Liu. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844-848. [DOI] [PubMed] [Google Scholar]
- 69.Vogelstein, B., and K. W. Kinzler. 2001. Achilles' heel of cancer? Nature 412:865-866. [DOI] [PubMed] [Google Scholar]
- 70.Wade, M., E. T. Wong, M. Tang, L. T. Vassilev, and G. M. Wahl. 2006. Hdmx modulates the outcome of p53 activation in human tumor cells. J. Biol. Chem. 281:33036-33044. [DOI] [PubMed] [Google Scholar]
- 71.Wong, L. Y., G. A. Matchett, and A. C. Wilson. 2004. Transcriptional activation by the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J. Virol. 78:10074-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.