Summary
We screened for mutants of Arabidopsis thaliana that displayed enhanced disease resistance to the powdery mildew pathogen Erysiphe cichoracearum and identified the edr3 mutant, which formed large gray lesions upon infection with E. cichoracearum and supported very little sporulation. The edr3-mediated disease resistance and cell death phenotypes were dependent on salicylic acid signaling, but independent of ethylene and jasmonic acid signaling. In addition, edr3 plants displayed enhanced susceptibility to the necrotrophic fungal pathogen Botrytis cinerea, but showed normal responses to virulent and avirulent strains of Pseudomonas syringae pv. tomato. The EDR3 gene was isolated by positional cloning and found to encode Arabidopsis dynamin-related protein 1E (DRP1E). The edr3 mutation caused an amino acid substitution in the GTPase domain of DRP1E (proline 77 to leucine) that is predicted to block GTP hydrolysis, but not GTP binding. A T-DNA insertion allele in DRP1E did not cause powdery mildew-induced lesions, suggesting that this phenotype is caused by DRP1E being locked in the GTP-bound state, rather than by a loss of DRP1E activity. Analysis of DRP1E–green fluorescent protein fusion proteins revealed that DRP1E is at least partially localized to mitochondria. These observations suggest a mechanistic link between salicylic acid signaling, mitochondria and programmed cell death in plants.
Keywords: disease resistance, powdery mildew, salicylic acid, programmed cell death, senescence, mitochondria
Introduction
Powdery mildews are obligate biotrophic pathogens that infect numerous plant species, including many economically important crops such as barley (Hordeum vulgare), wheat (Triticum aestivum) and tomato (Solanum lycopersicon) (Schulze-Lefert and Vogel, 2000). In Arabidopsis, accession Col-0 is highly susceptible to the powdery mildew strain Erysiphe cichoracearum UCSC (Adam and Somerville, 1996). A number of Arabidopsis mutants displaying enhanced disease resistance to powdery mildew have been characterized (Frye and Innes, 1998; Tang et al., 2005a; Vogel and Somerville, 2000; Vogel et al., 2002). These mutants can be grouped into two broad classes based on the presence or absence of mildew-induced lesions (Frye et al., 2001; Tang et al., 2005a; Vogel and Somerville, 2000; Vogel et al., 2002, 2004). The edr1 and edr2 mutants typify the former class (Frye and Innes, 1998; Tang et al., 2005a). In these mutants, fungal growth is inhibited at a very late stage of the infection process and resistance correlates with a more rapid activation of host defenses relative to wild-type plants, including programmed cell death (PCD) (Frye and Innes, 1998; Tang et al., 2005a). Another feature of these mutants is an enhanced senescence response upon exposure to exogenous ethylene, a natural plant hormone that regulates various developmental programs in plants as well as several defense responses (Chen et al., 2005). Loss-of-function mutations in EDR1 and EDR2 confer the above phenotypes, suggesting that EDR1 and EDR2 are negative regulators of plant defense and cell death (Frye et al., 2001; Tang et al., 2005a).
Resistance mediated by the edr1 and edr2 mutations requires the plant hormone salicylic acid (SA), as mutations that reduce levels of SA, or block its perception, restore susceptibility to powdery mildew (Frye et al., 2001; Tang et al., 2005a,b). In contrast, mutations that block the perception of the plant hormones ethylene and jasmonic acid (JA) do not affect resistance mediated by edr1 or edr2 (Frye et al., 2001; Tang et al., 2005a,b). These observations are consistent with the general finding that SA-induced defenses are most effective against biotrophic pathogens, while defenses induced by JA and ethylene are most effective against necrotrophic pathogens (Thomma et al., 1998, 1999). Indeed, several papers have reported antagonistic interactions between these signaling pathways (Cui et al., 2005; Kunkel and Brooks, 2002; Thaler et al., 2002).
EDR1 encodes a protein kinase with high similarity to CTR1, a negative regulator of the ethylene signaling pathway (Chen et al., 2005; Frye et al., 2001). The C-terminal third of EDR1 alone displays kinase activity in vitro and overexpression of a kinase-deficient form of full-length EDR1 induces a dominant negative phenotype that mimics the edr1 mutant (Tang and Innes, 2002; Tang et al., 2005b). These observations indicate that the kinase domain of EDR1 is central to its role in regulating PCD. EDR2 encodes a novel protein consisting of a putative pleckstrin homology (PH) domain and a steroidogenic acute regulatory protein-related lipid-transfer (START) domain, suggesting that EDR2 regulates defense responses through lipid signaling (Tang et al., 2005a).
Programmed cell death in plants is thought to play an important role in defense against biotrophic pathogens by depriving such pathogens of a nutrient source (Goodman and Novacky, 1994). Compared with our knowledge of PCD in animal cells, however, the regulation of PCD in plant cells is poorly understood. In animal systems, the most intensively studied form of PCD is referred to as apoptosis. The key regulator of apoptotic cell death is the mitochondrion, which collects information from various signal transduction cascades, integrates this with information on cellular metabolites and then triggers PCD by release of cytochrome C into the cytosol (Ferri and Kroemer, 2001). It appears that mitochondria may also function as a trigger of PCD in plants, as a change in mitochondrial membrane potential has been shown to be an early marker of PCD triggered by several different stimuli (Yao et al., 2004). Interestingly, the EDR2 protein contains an N-terminal mitochondrial targeting sequence, suggesting a link between edr2-mediated cell death and mitochondrial function (Tang et al., 2005a).
In an effort to further investigate the regulation of PCD and other defense responses in plants, we conducted a mutant screen for additional edr1-like mutants that displayed mildew-induced lesions and enhanced resistance. Here we describe the identification and characterization of the edr3 mutant and the isolation of the EDR3 gene, which was found to encode Arabidopsis Dynamin-related Protein 1E.
Results
The edr3 mutant displays enhanced disease resistance to E. cichoracearum
We screened ethyl methanesulfonate (EMS) mutagenized Col-0 plants (M2 generation) with the UCSC1 strain of E. cichoracearum for enhanced disease resistance. From a screen of 12 000 M2 plants derived from 3000 M1 parents, we identified three edr1-like mutants (Tang et al., 2005a) that we have designated edr2, edr3 and edr4. The edr2 mutant has already been described (Tang et al., 2005a) and the edr4 mutant remains to be fully characterized. Here we describe the edr3 mutant and EDR3 gene.
In contrast to wild-type Col-0 plants, edr3 mutant plants displayed very little powder and formed large lesions 8 days after infection with E. cichoracearum (Figure 1a). To ascertain at what stage fungal development was being arrested, we stained infected leaves with trypan blue, which stains fungal structures and dead plant cells. No obvious difference between wild-type and edr3 leaves was observed during the first 3 days after inoculation, and no epidermal cell death was observed in either wild-type or edr3 mutant leaves. The spores of E. cichoracearum germinated approximately 24 h after inoculation and developed extensive branched hyphae on both Col-0 leaves and edr3 leaves by day 3 (data not shown). However, by day 5 and day 7, fewer conidiophores formed on edr3 leaves than on Col-0 leaves (Figure 1). Similar to the previously identified edr1 and edr2 mutants, the edr3 mutant displayed large patches of dead mesophyll cells by 5 days after infection (Figure 1c), with cell death becoming extensive by 7 days after infection (Figure 1e). Such mesophyll cell death was not observed on Col-0 leaves (Figure 1b,d), and was not observed on edr3 mutants prior to powdery mildew infection (data not shown). These observations indicate that growth of E. cichoracearum was being arrested at a late stage of the infection process, possibly due to nutrient deprivation resulting from massive mesophyll cell death.
Figure 1.
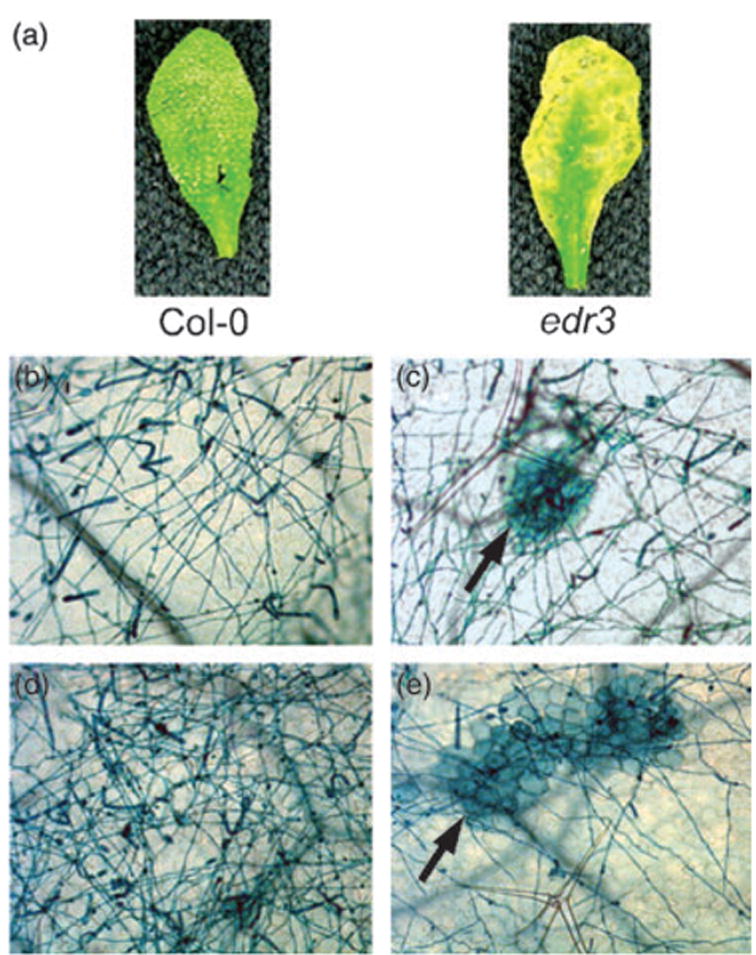
Enhanced resistance of the edr3 mutant to E. cichoracearum
(a) Leaves were removed from wild-type Col-0 and edr3 mutant plants and photographed 8 days after inoculation.
(b–e) Trypan blue-stained leaves from wild-type and edr3 plants 5 days (b, c) and 7 days (d, e) after infection with E. cichoracearum. The arrows in panels (c) and (e) indicate patches of dead mesophyll cells.
The edr3 mutant displays normal responses to virulent and avirulent strains of P. syringae pv. tomato strain DC3000
To further characterize the edr3 mutant, we investigated whether edr3 mutant plants displayed altered responses to virulent and avirulent strains of P. syringae pv. tomato. Both Col-0 and edr3 were susceptible to the virulent strain DC3000, but resistant to the avirulent strain DC3000 (avrRpt2). The disease phenotypes displayed by edr3 and wild-type Col-0 plants were indistinguishable and there were no significant differences in bacterial growth (data not shown). These data suggest that EDR3 does not function in regulating defense responses against virulent or avirulent P. syringae strains.
The edr3 mutant displays enhanced susceptibility to Botrytis cinerea
We also inoculated the edr3 mutant with a necrotrophic fungal pathogen, Botrytis cinerea, the causal agent of gray mold. Botrytis cinerea causes symptoms of soft rot on more than 200 different plant species, including Arabidopsis (Ferrari et al., 2003). Wild-type Col-0 plants are susceptible to this pathogen and mutations in the ethylene (ein2) and JA (jar1) pathways enhance susceptibility to B. cinerea, while the SA pathway mutations sid2 and pad4 have no effect on resistance to B. cinerea (Ferrari et al., 2003). The edr3 mutant displayed enhanced susceptibility to this pathogen, producing significantly larger necrotic lesions than wild-type Col-0 plants (Figure 2a,b).
Figure 2.
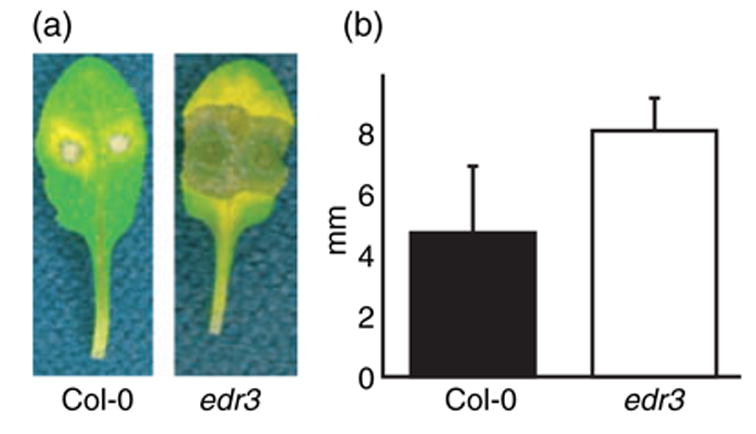
The edr3 mutant displays enhanced susceptibility to B. cinerea
(a) Leaves from wild-type Col-0 and edr3 mutant plants were detached, placed in Petri dishes and inoculated with B. cinerea. Leaves were photographed 3 days after inoculation.
(b) Lesion sizes induced by B. cinerea. Bars represent the mean and standard deviation of values obtained from eight leaves 3 days after infection. The lesion sizes in Col-0 and edr3 are significantly different according to Student’s t-test (P < 0.05). This experiment was repeated twice with similar results.
The edr3 mutant displays a normal ethylene-induced senescence response
In addition to resistance to powdery mildew, the previously identified edr1 and edr2 mutants display an enhanced ethylene-induced senescence phenotype (Frye et al., 2001; Tang et al., 2005a). Unlike edr1 and edr2 mutant plants, edr3 plants displayed no visible difference from wild-type Col-0 plants when treated with 100 μl l−1 ethylene (data not shown). This observation suggests that edr3 may function in a pathway distinct from edr1 and edr2.
Enhanced disease resistance mediated by the edr3 mutation requires SA signaling, but not JA and ethylene signaling
To assess the roles of SA in edr3-mediated disease resistance, we crossed edr3 to the SA-deficient mutant sid2-2 (Nawrath and Metraux, 1999). The edr3-mediated powdery mildew resistance and cell death were suppressed by the sid2-2 mutation (Figure 3), indicating that edr3-mediated disease resistance is dependent on SA. To further assess the requirement for SA-induced defenses, we created edr3/npr1 and edr3/pad4 double mutants. Mutations in NPR1 block most SA-induced defense responses (Cao et al., 1994; Delaney et al., 1995), while pad4 mutations reduce accumulation of SA induced by some, but not all, pathogen strains (Zhou et al., 1998). Consistent with the results of the edr3/ sid2 analysis, both the npr1 (allele nim1-1) and the pad4-1 mutations suppressed the edr3-mediated resistance to powdery mildew and cell death (Figure 3).
Figure 3.

Suppression of the edr3 resistance phenotype by the sid2, npr1 and pad4 mutations.
The indicated double mutants were inoculated with E. cichoracearum and single representative leaves were removed and photographed 8 days after inoculation.
To investigate whether ethylene and JA play a role in edr3-mediated disease resistance, we crossed the edr3 mutant to the ethylene-insensitive mutant ein2-1 (Guzman and Ecker, 1990) and to the jasmonate-insensitive mutant coi1-1 (Xie et al., 1998). In contrast to mutations that suppress the SA defense pathway, the ein2 and coi1 mutations did not suppress edr3-mediated resistance (data not shown).
Taken together, the above data indicate that edr3-mediated powdery mildew resistance depends on SA-induced defense responses and is independent of defenses induced by ethylene and JA. The edr1- and edr2-mediated resistance phenotypes have also been shown to be dependent on SA-induced defenses and independent of ethylene and JA signaling by similar double mutant analyses (Frye et al., 2001; Tang et al., 2005a,b).
The edr3 mutant does not constitutively express the SA-inducible defense gene PR-1
Because edr3-mediated resistance depends on SA signaling, we assessed whether the SA-inducible defense gene PR-1 was constitutively expressed. Constitutive expression of PR-1 would indicate that the edr3 mutation may simply disrupt cellular physiology, leading to an elevation of SA levels that would provide a trivial explanation for the enhanced resistance to powdery mildew. However, using RNA gel-blot analysis, we observed no enhanced expression of PR-1 in edr3 plants, either before or after inoculation with powdery mildew (Figure 4). This result indicates that the edr3-mediated resistance is not caused by constitutive expression of SA-inducible defense genes. This conclusion is consistent with the lack of enhanced resistance to P. syringae observed in edr3 plants.
Figure 4.
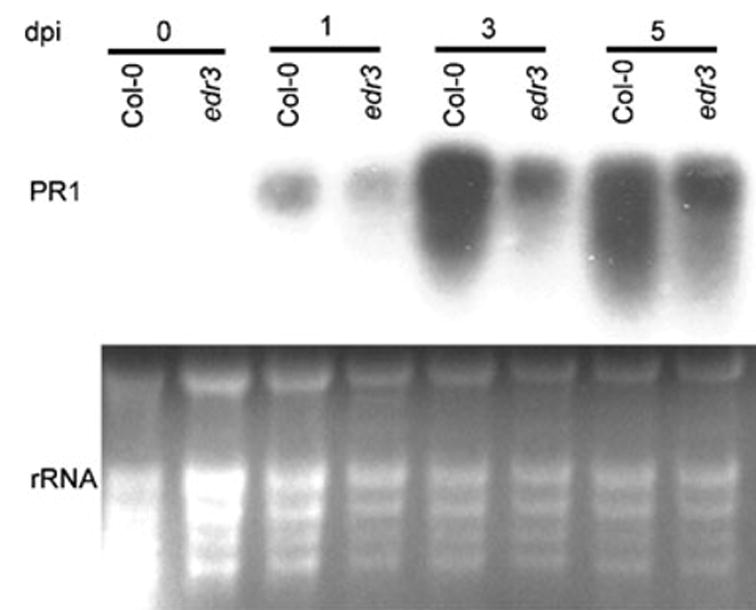
PR-1 expression is not enhanced in the edr3 mutant.
Plants were inoculated with E. cichoracearum and leaves were collected at the indicated time points for RNA isolation, which was then used in an RNA gel blot. The blot was hybridized with a PR-1 (At2g14610) cDNA probe. The relative amounts of RNA loaded in each lane are shown by the ethidium bromide-stained gel, which reveals the rRNA bands. dpi, days post-infection.
edr1 is epistatic to edr3
Although the edr1 and edr3 mutations both confer a lesion phenotype induced by powdery mildew that is dependent on SA signaling, the lesions on these two mutants have a qualitatively distinct appearance. In the edr1 mutant, the lesions are generally yellow-brown in color and appear dry, while in the edr3 mutant, the lesions are grayer and water-soaked in appearance, suggesting that these lesions may be induced by different pathways. To assess the relationship between lesions induced by edr1 and edr3 we crossed edr1 with edr3 and characterized the phenotype of the edr1/edr3 double mutant. The double mutant displayed the edr1-like lesion phenotype upon infection with powdery mildew, indicating that edr1 is epistatic to edr3, at least at the level of mildew-induced lesions (data not shown).
Identification of the EDR3 gene
To map the EDR3 gene, the edr3 mutant (Columbia genotype, Col-0) was crossed to Arabidopsis ecotype Landsberg erecta (Ler) and scored for the mildew-induced lesion phenotype in the F2 generation. Approximately 25% of the F2 plants displayed the lesion phenotype, indicating that edr3 is recessive. Initially, the EDR3 gene was mapped to a region between the microsatellite markers CIW4 and nga6 on chromosome 3 in a population of 48 resistant F2 plants. To further localize the EDR3 gene, we developed new markers at intervals between these two markers using Monsanto Col-0 and Ler polymorphism data (http://www.arabidopsis.org/Cereon/index.jsp). Five hundred and six resistant F2 plants representing 1012 meioses were scored, which allowed us to localize the edr3 mutation to a region spanned by bacterial artificial chromosome (BAC) clone F27H5 (GenBank accession no. AL163852). This analysis defined a 40-kb interval containing EDR3 (Figure 5a).
Figure 5.
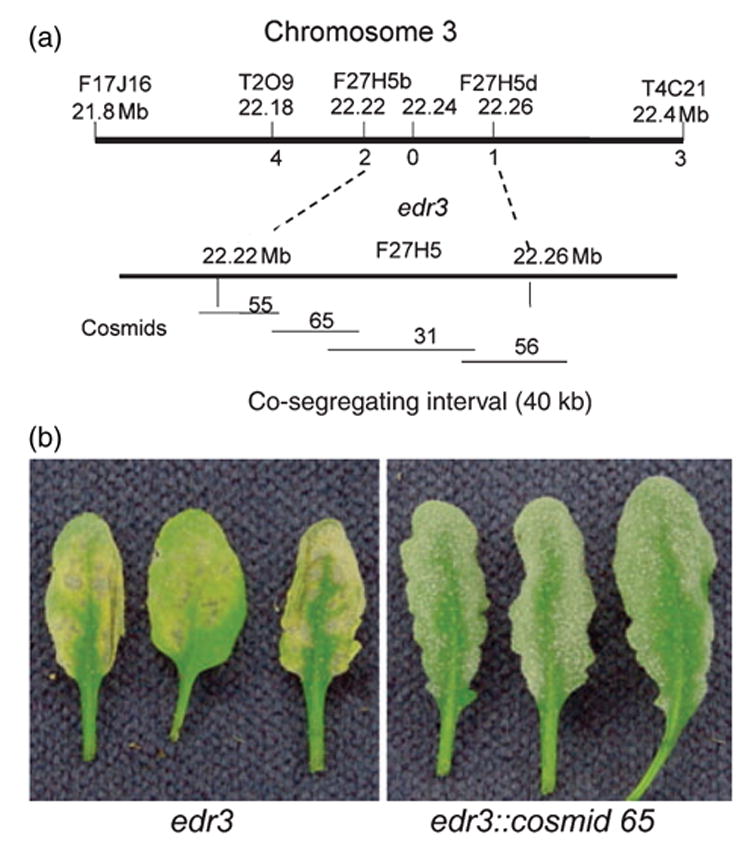
Positional cloning of the EDR3 gene.
(a) Genetic and physical mapping of the EDR3 region. The top bar represents the combined genetic and physical map data with marker names and physical positions indicated above the bar. Numbers immediately below the bar indicate the number of recombination events between the indicated markers and the edr3 mutation out of 1012 meioses scored.
(b) Complementation of the edr3 mutation by Agrobacterium-mediated transformation.
To identify the EDR3 gene, we constructed a cosmid library using BAC F27H5 DNA. Four cosmid subclones of BAC F27H5, which overlapped and covered the entire EDR3 interval, were identified and used to rescue the edr3 phenotype. Among these four subclones, only cosmid clone 65 complemented the edr3 mutation, with nine out of ten T1 plants displaying the wild-type susceptible phenotype (Figure 5b). Self-progeny from these lines were obtained and assayed for resistance to powdery mildew. The susceptible phenotype co-segregated with the T-DNA.
Sequencing of the vector-insert junction fragments of cosmid 65 revealed that it contained four full-length genes: At3g60190 (encodes Arabidopsis dynamin-related protein 1E, DRP1E), At3g60200 (encodes an unknown protein), At3g60210 (encodes a putative protein chaperonin 10) and At3g60220 (encodes a putative ring finger transcription factor). We sequenced all four of these genes from the edr3 mutant and found a single mutation, a C → T transition, in AT3g60190, which caused an amino acid change (P77L) in the predicted open reading frame. No mutations were found in the other three genes.
To confirm that At3g60190 is EDR3, we transformed two additional cosmid clones, A4 (7.1–24.5 kb of BAC F27H5) and A64 (23.9–42.1 kb of BAC F27H5) into the edr3 mutant. Clone A4 contained the complete At3g60190 gene, but lacked full-length copies of the other genes on clone 65, while A64 contained a truncated At3g60190 gene along with all three other full-length genes on clone 65. Clone A4 complemented the edr3 mutant phenotype, while clone A64 did not (data not shown), confirming that At3g60190 is EDR3.
EDR3 encodes a member of the dynamin-related protein family
The EDR3 gene consists of 15 exons and 14 introns and encodes dynamin-related protein 1E (DRP1E, also known as ADL4 and ADL1E; Hong et al., 2003a), which is 624 amino acid residues long, has a predicted mass of 69.8 kDa and a pI of 7.5. Dynamin-related protein 1E belongs to a plant-specific subclass of dynamin-related proteins (DRP1), consisting of five members in Arabidopsis (A, B, C, D, E) (Hong et al., 2003a). This class is characterized by having an N-terminal GTPase domain, a central ‘dynamin 2‘ domain and a C-terminal GTPase effector domain (GED), a typical structure for plant dynamin-related proteins. However, this class lacks a PH domain and a proline-rich domain, which are found in classical animal dynamin-like proteins. Based on work on animal dynamins, the plant DRP1 proteins should be able to form polymeric structures that wrap around membranes to facilitate membrane tubulation and pinching off of vesicles (Praefcke and McMahon, 2004), processes that are essential to vesicle trafficking and membrane compartmentalization.
The edr3 mutation causes a P77L substitution in the G2 motif of the GTPase domain of DRP1E. Mutations in this domain of mammalian dynamin 1 block GTP hydrolysis without significantly affecting GTP binding (Marks et al., 2001). Such mutations also block endocytosis and lead to the formation of long tubular invaginations from the plasma membrane in COS-7 fibroblasts (Marks et al., 2001), indicating that GTP hydrolysis by dynamin is required for pinching off of vesicles during endocytosis.
Because the edr3 mutation is likely to alter DRP1E function, rather than eliminate it, we obtained a T-DNA insertion in DRP1E to assess whether loss of DRP1E function causes an edr3-like phenotype. The SALK T-DNA insertion line 060080 contains an insertion at amino acid position 366 of DRP1E, which is 258 amino acids from the C-terminus and is located within the central dynamin 2 domain. This allele is thus unlikely to produce a functional protein. We confirmed the location of the T-DNA insertion by PCR-based amplification of the T-DNA left border and direct sequencing of the PCR product. Plants homozygous for the T-DNA insertion displayed no noticeable morphological or growth defects (data not shown). They also showed normal susceptibility to infection by E. cichoracearum (data not shown). This observation indicates that the edr3 mutant phenotype is probably a gain-of-function phenotype, despite the fact that it is recessive and can be complemented by a single copy of a wild-type transgene.
Localization of DRP1E
To gain more insight into the function of DRP1E we investigated the subcellular localization of the DRP1E protein using a green fluorescent protein (GFP) fusion to the N-terminus of DRP1E. Expression of this fusion protein in transgenic Arabidopsis Col-0 plants under control of the constitutive cauliflower mosaic virus 35S promoter revealed that GFP:DRP1E localizes to punctate spots in Arabidopsis root cells (Figure 6a), indicating that DRP1E associates with an organelle. To assess whether these spots were mitochondria, we stained transgenic Arabidopsis roots with the mitochondrial-specific fluorescent dye MitoTracker Red CMXRos (Figure 6b). The MitoTracker fluorescence and GFP fluorescence co-localized (Figure 6c), indicating that DRP1E associates with mitochondria, at least part of the time.
Figure 6.
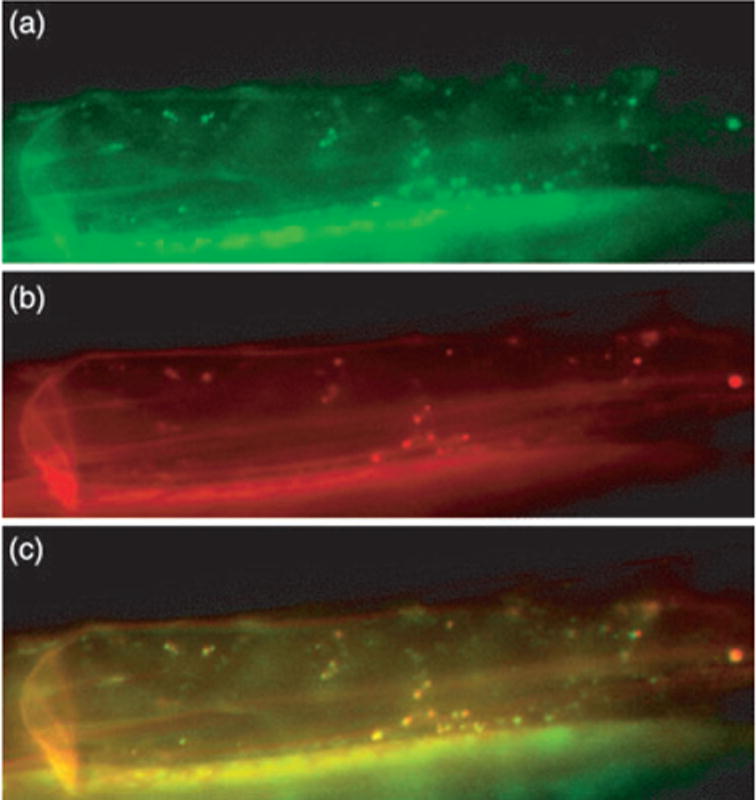
GFP:DRP1E localizes to mitochondria.
(a–c) Roots of transgenic Arabidopsis expressing a 35S-GFP:DRP1E transgene: (a) GFP fluorescence, (b)MitoTracker fluorescence, (c) merge of images in (a) and (b).
Discussion
We have shown that a missense mutation in the GTP hydrolysis site of the dynamin-related protein DRP1E causes an enhanced cell death response upon infection with powdery mildew. Dynamins are a well-studied family of proteins that mediate membrane fission and/or fusion, and are found in all eukaryotes (Praefcke and McMahon, 2004). They are involved in many different cellular processes including division of organelles, budding and transport of vesicles, cytokinesis and pathogen defense.
Three papers have been published on the function and localization of Arabidopsis DRP1E (Hong et al., 2003b; Jin et al., 2003; Kang et al., 2003a), but these papers reached different conclusions. Jin et al. 2003 reported that DRP1E is localized to punctate spots and that loss-of-function mutations cause abnormal elongation of mitochondria. Based on these observations, they concluded that DRP1E was required for mitochondrial fission, similar to the human dynamin-related protein Drp1. Although Kang et al. 2003a also reported that DRP1E localized to punctate spots, they could detect no morphological abnormalities in the mitochondria of drp1e mutants. We also examined mitochondrial morphology in both the edr3 and the DRP1E T-DNA insertion line using transmission electron microscopy and observed no clear differences from wild-type plants (data not shown). Thus, our data and those of Kang et al. 2003a do not support a role for DRP1E in mitochondrial fission. However, we found that at least a portion of DRP1E is physically associated with mitochondria, consistent with the proposal of Jin et al. 2003 that DRP1E functions in mitochondria. Based on this observation, we hypothesize that DRP1E may be involved in delivering vesicles to mitochondria, and that the edr3 mutant allele may affect the structure and function of the mitochondrial membrane such that it more readily triggers PCD.
Kang et al. 2003a also showed that a double mutant containing mutations in DRP1A and DRP1E is embryo lethal, and displays defects in assembly of the cell plate, formation of the cell wall and recycling of the plasma membrane. Furthermore, immunolocalization of DRP1E showed it to be most abundant at growing cell plates in dividing cells. From this they concluded that DRP1A and DRP1E function redundantly in expansion of polar cells and biogenesis of the cell plate, probably via their role in formation of vesicles and fusion of vesicles with target membranes. Supporting this conclusion, Hong et al. 2003b reported that a DRP1E:GFP fusion protein expressed in dividing BY2 tobacco cells localized to the newly forming cell plate at a late stage of telophase, and was concentrated at the growing edges of the plate. In addition, the GFP fluorescence at the cell plate produced a tubular pattern, suggesting that it was associated with a vesicular network. Consistent with our data, Hong et al. also observed numerous punctate spots throughout the cell cytoplasm.
Hong et al. 2003b also showed that DRP1E can interact with the soybean (Glycine max) phragmoplastin protein using a yeast two-hybrid assay. Phragmoplastin belongs to the same subfamily of plant dynamin-like proteins as the Arabidopsis DRP1 proteins (Hong et al., 2003a); thus, this observation suggests that DRP1E can probably form heterooligomers with other DRP1 family members.
There are five DRP1 family members in Arabidopsis (Hong et al., 2003a). Of these, DRP1D and DRP1E are the most similar, with DRP1A and DRP1B being the most distant from DRP1E (Hong et al., 2003a). However, DRP1A and DRP1E share the most similar expression patterns based on promoter:GUS fusion analyses (Kang et al., 2003a,b). Independent T-DNA insertion mutations in all five genes have been identified, and of these, drp1A insertions have the strongest phenotype, with insertions in DRP1B, DRP1D and DRP1E having no observable phenotype (Kang et al., 2001, 2003a,Kang et al., b). Homozygous drp1a seeds are shriveled, and mutant seedlings arrest shortly after germination with severely stunted roots (Kang et al., 2001). These mutant seedlings can be rescued in tissue culture, however, by addition of sucrose, and rescued plants display normal vegetative development and flower normally, but produce very few viable seed (Kang et al., 2001). Because DRP1A and DRP1E have similar amino acid sequences and the drp1A/ drp1E double mutant has a much more severe phenotype than either single mutant, it appears that these two proteins have partially redundant functions. This may account for the lack of a phenotype in the drp1E T-DNA insertion line.
Because a loss-of-function mutation in DRP1E does not cause an edr3-like phenotype, we conclude that the P77L amino acid substitution caused by the edr3 mutation alters, rather than eliminates, the function of DRP1E. Proline 77 is a highly conserved residue in the G2 region of the GTPase domain of DRP1E. This region of DRP1E (SGIVTRRPLV) is identical to the G2 region of human dynamin 1 (accession no. NP_004399), which functions in clathrin-mediated endocytosis and other vesicular trafficking processes (Praefcke and McMahon, 2004). The central threonine of this sequence functions as the catalytic residue for GTP hydrolysis (Praefcke and McMahon, 2004). In animal dynamins, GTP hydrolysis is stimulated by self-oligomerization in the presence of membranes. Dynamin oligomerization leads to formation of membrane tubules, with the dynamin forming a coiled tube around the membrane. A ‘poppase’ model has been proposed to explain how dynamins function in the pinching off of membrane vesicles. Under this model, GTP hydrolysis causes a change in the helical pitch of the dynamin tube, effectively elongating the tube and popping off the vesicle (Praefcke and McMahon, 2004). Mutations in the catalytic threonine of the G2 region of dynamin cause a marked reduction in GTP hydrolysis, which leads to a block in vesicle scission and a build up of membrane tubules (Marks et al., 2001). In contrast, a complete loss of dynamin function blocks endocytosis and vesicle formation without build up of tubules. These observations may explain the difference in phenotypes between the edr3 and T-DNA insertion alleles of DRP1E.
Similar recessive gain-of-function phenotypes caused by mutations in an ATP-binding site have also been described for the cytosolic heat shock protein HSP90.2 in Arabidopsis (Hubert et al., 2003). Three different amino acid substitutions in the ATP binding site of HSP90.2 cause a destabilization of the RPM1 protein, and hence loss of RPM1-mediated disease resistance, while a null mutation in HSP90.2 has no effect on RPM1-mediated resistance (Hubert et al., 2003). Similar to DRP1E, HSP90.2 belongs to a small family of highly similar proteins that can dimerize. Thus, it appears that mutations in the ATP-binding site can somehow prohibit functional compensation by other family members, whereas a complete null mutation is compensated.
Although dynamin-related proteins in plants have been intensively studied (Hong et al., 2003a,b; Jin et al., 2001, 2003; Kang et al., 1998, 2001, 2003a, Kang et al., b), there have been no reports that they play a role in defense responses and PCD. In animal cells, however, dynamin-related proteins are key regulators of PCD. For instance, in HeLa cells, moderate overexpression of dynamin 2 induces expression of the transcription factor p53, which triggers apoptosis (a form of PCD) (Fish et al., 2000). This observation suggests that dynamin 2 can function as a signal transducer involved in transcriptional regulation of apoptosis (Fish et al., 2000). A second member of the dynamin family, dynamin-related protein 1 (DRP1), has also been shown to regulate apoptosis in human cells. DRP1 moves from the cytosol to mitochondria during apoptosis and regulates mitochondrial fission and swelling (Frank et al., 2001). Inhibition of DRP1 by overexpression of a dominant negative mutant blocks the release of cytochrome C from mitochondria (a hallmark of apoptosis in mammalian cells) and blocks cell death (Frank et al., 2001), implicating DRP1 in regulation of the permeability of the mitochondrial membrane. More recently, DRP1 was shown to be both necessary and sufficient for the induction of mitochondrial fragmentation and programmed cell death during development of Caenorhabditis elegans, indicating an important role for DRP1 in the cell death pathway in C. elegans (Jagasia et al., 2005).
Although the mechanism of edr3-mediated resistance and cell death needs to be further studied, our finding that a mutation in DRP1E leads to a pathogen-inducible cell death phenotype assigns a new function for a dynamin-related protein in plants. Furthermore, our data suggest a mechanistic link between SA signaling, mitochondrial function and programmed cell death.
Experimental procedures
Plant growth conditions and mutant screening
Arabidopsis thaliana plants were grown in growth rooms under a 9-h light/15-h dark cycle at 23°C as described previously (Frye and Innes, 1998).
Ethyl methanesulfonate-mutagenized Col-0 plants (M2 generation) were inoculated with E. cichoracearum and scored for disease responses 8 days after inoculation. Plants displaying no visible powder or necrotic lesions were selected and allowed to set seeds. Approximately 12 000 M2 plants derived from 3000 M1 plants were screened (Tang et al., 2005a).
Pathogen infections
Erysiphe cichoracearum strain UCSC1 was maintained on hyper-susceptible A. thaliana pad4-2 mutant plants, with passage of asexual spores (conidia) onto fresh plants every 8–10 days. Plants were inoculated between 4 and 6 weeks of age. To inoculate plants, spores were transferred directly from diseased plants to healthy plants by gently brushing the leaves together. Disease phenotypes were scored 8 days after inoculation.
Fungal structures and dead plant cells were stained using alcoholic trypan blue (Koch and Slusarenko, 1990). Samples were observed as described previously (Frye and Innes, 1998).
Inoculation of A. thaliana plants with P. syringae pv. tomato strain DC3000 and measurement of bacterial growth within leaves was performed as described previously (Frye and Innes, 1998).
Inoculation of A. thaliana plants with B. cinerea was performed as described by Ferrari et al. 2003. Lesion size induced by botrytis was determined by measuring the major axis of the necrotic area 3 days after infection.
Ethylene-induced senescence assay
Six-week-old plants were placed in a sealed chamber containing 100 μl l−1 ethylene for 3 days and then visually inspected for chlorotic (yellow) leaves.
RNA gel-blot analysis
Total RNA was isolated from Arabidopsis leaf tissue using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). A total of 5 μg of RNA was separated on a denaturing formaldehyde–agarose gel and transferred to Hybond N nylon membranes (Amersham Pharmacia Biotech, Buckinghamshire, UK). RNA gel-blots were hybridized with a [32P]-labeled PR-1 (At2g14610) cDNA probe and washed at 65°C using Church buffer (Ashfield et al., 1998).
Genetic and physical mapping of EDR3
Genetic mapping was accomplished using an F2 population derived from a cross between the edr3 mutant (Columbia genotype, Col-0) and Landsberg erecta (Ler). F2 seeds were planted and scored for disease resistance to E. cichoracearum as described above. Genomic DNA was isolated from 48 resistant F2 plants and scored with published microsatellite markers. This initial mapping localized the edr3 mutation between molecular markers CIW4 and nga6 on chromosome 3. We then developed our own molecular markers at intervals between these two markers using Monsanto Col-0 and Ler polymorphism data (http://www.arabidopsis.org/Cereon/index.jsp; primer sequences available upon request). Five hundred and six resistant F2 plants representing 1012 meioses were scored. Ultimately, the edr3 mutation was localized to BAC clone F27H5. This analysis defined a 40 kb region that co-segregated with the edr3 mutation.
Cosmid library construction and assembly of cosmid contigs
Bacterial artificial chromosome clone F27H5 (GenBank accession no. AL163852) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). The binary vector pCLD04541 (Bent et al., 1994) and F27H5 BAC DNA were isolated using a HiSpeed kit (Qiagen) following the manufacturer’s instructions. F27H5 BAC DNA was partially digested using the restriction enzyme Sau3AI and ligated to BamHI-digested pCLD04541. The ligation mix was packaged using Gigapack III Plus packaging extract (Stratagene, La Jolla, CA, USA) and transfected into Escherichia coli strain DH5α. Positive clones were selected on Luria–Bertani (LB) medium with 10 μg ml−1 tetracycline (Sigma, St Louis, MO, USA).
Overlapping cosmid clones were identified by PCR-based library screening with specific primer pairs to internal sequences of BAC F27H5. Four cosmid clones that covered the defined edr3 co-segregating region of BAC F27H5 were selected.
Plant transformation
The selected cosmid clones were purified using a plasmid miniprep kit (Qiagen) and transformed into Agrobacterium tumefaciens strain GV3101 by electroporation with selection on LB plates containing 50 μg ml−1 kanamycin sulfate (Sigma). Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected by growing on 0.5× Murashige and Skoog salts plus 0.8% agar and 50 μg ml−1 kanamycin. Transformants were transplanted to soil 7 days after germination and were inoculated with E. cichoracearum when 5 weeks old. T1 plants displaying wild-type phenotypes were allowed to self and the T2 generation was tested for segregation of susceptibility to powdery mildew. Co-segregation of the T-DNA insert with powdery mildew disease was examined by PCR using primers specific to the NPTII gene on the T-DNA.
Sequencing of candidate genes
The cosmid that complemented the edr3 mutant phenotype was analyzed by sequencing of the junctions between the insert and the vector using primers T3 and T7. This cosmid contained the sequence from 17.9 to 33.0 kb of BAC F13C5. Genes contained within this stretch of DNA were amplified from the edr3 mutant by PCR and directly sequenced. All sequencing reactions were performed using BigDye Terminator Kits (Applied Biosystems, Foster City, CA, USA) and separated on an ABI 3730 automated DNA sequencer (Applied Biosystems).
To confirm the annotation of EDR3, we amplified EDR3 from a Col-0 cDNA library by PCR using primers covering the beginning and end of the predicted EDR3 open reading frame. The PCR products were sequenced directly and DNA sequences were assembled using the SEQUENCHER program (Gene Codes, Ann Arbor, MI, USA).
Construction of double mutants
Double mutants were created by standard genetic crosses and verified by PCR and sequencing as described in Tang et al. 2005a. The edr3, pad4-1, sid2-2 and ein2-1 mutations were in the Col-0 background, while npr1 (nim1-1) was in the Wassilewskija (Ws) background and coi1-1 was in the Col-6 background.
Localization of EDR3
A C-terminal GFP fusion to a full-length EDR3 cDNA was constructed. EDR3 cDNA was amplified by PCR using primers that incorporated restriction sites EcoRI and BamHI (5′-AACCGAATTCATGACGACTATGGAGAGTTTGATTGG- 3′ and 5′-AGCTGGATCCTCATCTTACCCAAGCAACAGCATCAA- 3′). The PCR product was then digested and inserted into EcoRI- and BamHI-digested pEGAD vector, which contains a 35S Cauliflower Mosaic Virus promoter (Cutler et al., 2000). The construct was verified by sequencing and transformed into Agrobacterium strain GV3101 by electroporation. For stable transformation, transgenic plants (in the Col-0 background) were selected by spraying with 0.01% glufosinate- ammonium (Basta) (Farnam, Phoenix, AZ, USA).
For co-localization of DRP1E and mitochondria, seeds of Arabidopsis transgenic plants expressing 35S-GFP:DRP1E were sown on 0.8% agar plates and treated at 4°C for 2 days, then grown at 23°C under continuous light for 8 days. The roots were cut into small pieces, vacuum infiltrated for 5 min and incubated at 23°C for 1 h in 4 μM MitoTracker Red CMXRos (Molecular Probes, Eugene, OR, USA). The roots were then rinsed twice with water. Samples were observed and photographed using a Nikon e800 microscope (Melville, NY, USA). Excitation and emission wavelengths were 460–500 nm and 505 nm, respectively for visualization of GFP and 530–550 nm and 590–650 nm for visualization of MitoTracker Red.
Identification of a DRP1E T-DNA insertion mutant
A T-DNA insertion line was obtained from the Arabidopsis Biological Resource Center at Ohio State University (SALK060080). Homozygous individuals were identified using PCR with a T-DNA specific primer and primers specific to DRP1E that flanked the T-DNA insertion. The location of the insertion site was confirmed by direct sequencing of the PCR product.
Acknowledgments
We thank J. G. Turner for providing coi1-1 seeds, F. M. Ausubel for providing sid2-2 seeds, J. Parker for providing pad4-1 seeds, T. Delaney for providing nim1-1 seeds and J. Dewdney for the B. cinerea strain. We also thank the Arabidopsis Biological Resource Center at Ohio State University for providing the F27H5 BAC clone, seeds of T-DNA insertion line SALK060080 and ein2-1 seeds. This work was supported by the National Institutes of Health, grant number R01 GM063761 to RWI.
References
- Adam L, Somerville SC. Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 1996;9:341–356. doi: 10.1046/j.1365-313x.1996.09030341.x. [DOI] [PubMed] [Google Scholar]
- Ashfield T, Danzer JR, Held D, Clayton K, Keim P, Saghai Maroof MA, Webb PM, Innes RW. Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet. 1998;96:1013–1021. [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon S, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583– 1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Ann Bot. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA. 2005;102:1791– 1796. doi: 10.1073/pnas.0409450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602– 6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003;35:193–205. doi: 10.1046/j.1365-313x.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- Ferri KF, Kroemer G. Mitochondria – the suicide organelles. Bioessays. 2001;23:111–115. doi: 10.1002/1521-1878(200102)23:2<111::AID-BIES1016>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Fish KN, Schmid SL, Damke H. Evidence that dynamin- 2 functions as a signal-transducing GTPase. J Cell Biol. 2000;150:145–154. doi: 10.1083/jcb.150.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frye CA, Innes RW. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell. 1998;10:947–956. doi: 10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA. 2001;98:373–378. doi: 10.1073/pnas.98.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RN, Novacky AJ. The Hypersensitive Reaction in Plants to Pathogens. St Paul, MN: American Phytopathological Society; 1994. [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Bednarek SY, Blumwald E, et al. A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol Biol. 2003a;53:261–265. doi: 10.1023/b:plan.0000007000.29697.81. [DOI] [PubMed] [Google Scholar]
- Hong Z, Geisler-Lee CJ, Zhang Z, Verma DP. Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol Biol. 2003b;53:297–312. doi: 10.1023/b:plan.0000006936.50532.3a. [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Bae H, Kim SJ, Jin YH, Goh CH, Kim DH, Lee YJ, Tse YC, Jiang L, Hwang I. The Arabidopsis dynamin-like proteins ADL1C and ADL1E play a critical role in mitochondrial morphogenesis. Plant Cell. 2003;15:2357–2369. doi: 10.1105/tpc.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Jin JB, Piao HL, Pih KT, Jang HJ, Lim JH, Hwang I. Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol Biol. 1998;38:437–447. doi: 10.1023/a:1006099718761. [DOI] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell. 2003a;15:899– 913. doi: 10.1105/tpc.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Rancour DM, Bednarek SY. The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J. 2003b;35:1–15. doi: 10.1046/j.1365-313x.2003.01775.x. [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Vogel J. Closing the ranks to attack by powdery mildew. Trends Plant Sci. 2000;5:343–348. doi: 10.1016/s1360-1385(00)01683-6. [DOI] [PubMed] [Google Scholar]
- Tang D, Innes RW. Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J. 2002;32:975– 983. doi: 10.1046/j.1365-313x.2002.01482.x. [DOI] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 2005a;44:245–257. doi: 10.1111/j.1365-313X.2005.02523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Christiansen KM, Innes RW. Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol. 2005b;138:1018–1026. doi: 10.1104/pp.105.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Fidantsef AL, Bostock RM. Antagonism between jasmonate- and salicylate-mediated induced plant resistance: effects of concentration and timing of elicitation on defense-related proteins, herbivore, and pathogen performance in tomato. J Chem Ecol. 2002;28:1131–1159. doi: 10.1023/a:1016225515936. [DOI] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999;121:1093–1102. doi: 10.1104/pp.121.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell. 2002;14:2095–2106. doi: 10.1105/tpc.003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004;40:968–978. doi: 10.1111/j.1365-313X.2004.02264.x. [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yao N, Eisfelder BJ, Marvin J, Greenberg JT. The mitochondrion – an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J. 2004;40:596–610. doi: 10.1111/j.1365-313X.2004.02239.x. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]


