Abstract
Coarse-grained molecular dynamics simulations are used to explore the interaction with a phospholipid bilayer of the voltage sensor (VS) domain and the S4 helix from the archaebacterial voltage-gated potassium (Kv) channel KvAP. Multiple 2-μs self-assembly simulations reveal that the isolated S4 helix may adopt either interfacial or transmembrane (TM) locations with approximately equal probability. In the TM state, the insertion of the voltage-sensing region of S4 is facilitated via local bilayer deformation that, combined with side chain “snorkeling,” enables its Arg side chains to interact with lipid headgroups and water. Multiple 0.2-μs self-assembly simulations of the VS domain are also performed, along with simulations of MscL and KcsA, to permit comparison with more “canonical” integral membrane protein structures. All three stably adopt a TM orientation within a bilayer. For MscL and KcsA, there is no significant bilayer deformation. In contrast, for the VS, there is considerable local deformation, which is again primarily due to the lipid-exposed S4. It is shown that for both the VS and isolated S4 helix, the positively charged side chains of S4 are accommodated within the membrane through a combination of stabilizing interactions with lipid glycerol and headgroup regions, water, and anionic side chains. Our results support the possibility that bilayer deformation around key gating charge residues in Kv channels may result in “focusing” of the electrostatic field, and indicate that, when considering competing models of voltage-sensing, it is essential to consider the dynamics and structure of not only the protein but also of the local lipid environment.
Keywords: coarse grained, membrane protein, molecular dynamics, potassium channel, S4 helix
Voltage-gated potassium (Kv) channels play a key role in excitable cells (1). Recent structural (i.e., x-ray) and biophysical (2–4) studies have focused on the voltage sensor (VS) domain. Structural data suggest that the VS forms a relatively independent domain, and a high-resolution structure of an isolated VS domain from the archaebacterial channel KvAP has been determined (5, 6). The VS appears capable of adopting different orientations relative to the pore domain of Kv channels (7). The structure of the VS in a mammalian channel, Kv1.2, is similar to that of the isolated KvAP VS, and packs rather loosely against the pore domain (8, 9). In support of the VS domain as an independent unit, homologous domains have been found in two non-Kv proteins: a voltage-sensitive phosphatase (10) and a voltage-sensitive proton (Hv) channel (11, 12). It is thought that the VS, or elements of its structure, move in response to change in voltage across a cell membrane. However, the exact nature and extent of such VS motion remains unresolved (6, 9, 13–17). In this context, it is crucial to understand how a VS domain interacts with the surrounding lipid bilayer.
Central to the function of the VS is the S4 helix. The VS is composed of four transmembrane (TM) helices, S1–S4. S4 is unusual for a TM helix in that it contains several basic amino acid side chains that act as the primary voltage-sensing elements. The pattern of basic side chains within S4 is conserved in Kv channels, and also in voltage-activated calcium and sodium channels, in the voltage-sensitive phosphatase, and in the Hv channel. This observation raises the question of how the S4 helix is accommodated within a lipid bilayer. Are the charges of S4 completely shielded from interactions with the lipid by other regions of the VS protein, or does local reorganization of the lipid bilayer accommodate the voltage-sensing S4 helix?
A related question is that of how the S4-containing VS domain is inserted into a bilayer. Given the two-state model of membrane protein folding (18) and its recent modifications (19, 20), one may ask whether the S4 region of the Kv sequence may form a stable isolated helix within a bilayer. An S4 helix derived from KvAP may be biosynthetically inserted into a membrane (21), and S4 helix peptides also insert into a bilayer in vitro.†
Molecular dynamics (MD) simulations provide a computational approach to exploring the interactions of membrane proteins with their lipid bilayer environment (23–25). Atomistic MD simulations have been used to explore the interactions of a TM S4 helix with a lipid bilayer (26), of a Kv1.2 channel with a lipid bilayer (27), and of the KvAP VS with a detergent micelle (28). However, such simulations do not readily access long (≈1 μs) time scales, which may be necessary to allow local lipid rearrangement around S4 and the VS. Coarse-grained (CG) MD simulations (29–33) enable longer time scales to be addressed and have proved useful in enabling us to model the dynamics of lipid bilayers (29, 30, 34) and the interactions between lipid bilayers and membrane proteins (35–38). Here we use CG-MD self-assembly simulations to explore the interaction of the S4 helix from the KvAP VS, and of the intact KvAP VS domain, with a phospholipid bilayer. We demonstrate that both may be accommodated in a membrane via local bilayer deformation, which allows favorable interactions between the charged side chains of S4 and the lipid headgroup region and associated water molecules. The implications of this with respect to channel gating are explored.
Results
CG Self Assembly Simulations.
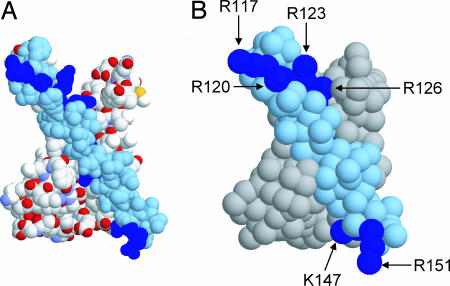
In the CG model, instead of representing each atom in a protein or lipid molecule (29), particles corresponding to approximately four atoms are used, and are parameterized to capture the hydrophobicity/hydrophilicity, charge, and H-bonding properties of their constituent atoms. This model was derived from the work on CG lipid systems by Marrink et al. (29). We have recently adapted and tested this for application to a number of proteins and peptides (37, 38). As in ref. 29, water particles correspond to four water molecules. The process of coarse graining is illustrated for the KvAP VS domain protein (Fig. 1). A similar process may be applied to the isolated S4 helix of the KvAP VS. The overall shape of the protein is preserved in the CG model. In particular, the relative surface exposure of the basic side chains (Fig. 1B) is close to that in the starting atomistic structure (Fig. 1A) for both the isolated S4 helix and for the helix in the VS.
Fig. 1.
The VS domain (PDB ID 1ORS) from KvAP. (A) Atomistic structure of the VS, showing non-H atoms only in space-filling format with the S4 helix in (pale) blue and the Arg side chains of S4 in deep blue. (B) CG model of the VS, shown in space-filling format with the S4 helix in pale blue and the Arg residues of S4 in deep blue.
S4 Helix Simulations.
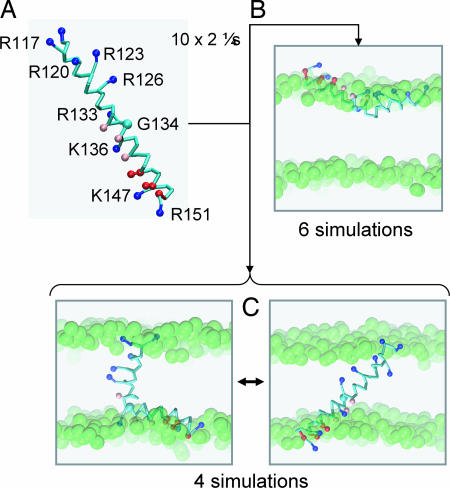
The isolated S4 helix (Fig. 2A) is divided into two regions by a central glycine (G134). The N-terminal half contains the main voltage-sensing arginine side chains (R117, R120, R123, R126, and R133). The C-terminal half contains a number of cationic (Arg, Lys) and anionic (Asp) side chains. Thus S4 is relatively polar for a TM helix. It should be noted that S4 adopts different conformations in different crystal structures. Thus, in the first KvAP structure [Protein Data Bank (PDB) ID 1ORQ], the S4 helix is interrupted by a short loop around G134, before forming another helical region contiguous with the S5 helix (5, 6). In comparison, in a later KvAP structure (PDB ID 2A0L), the S4 helix is longer (extending to residue 140), and is ended by a long, unstructured loop (the “S4–S5 linker”) (7). However, S4 from the isolated VS domain (PDB ID 1ORS) is composed of a single, continuous helix (from residues 115–148), as shown in Fig. 2A, and this continuous helix was the starting point for our CG simulations. Given that S4 (albeit a shorter version) can insert biosynthetically into a membrane (21), and the subsequent debate concerning the significance of this result (39), we explored how S4 inserts into a lipid bilayer in CG simulations.
Fig. 2.
Interactions of the S4 helix with a PC bilayer. (A) CG model of the S4 helix (residues 115–153) from KvAP showing the location of the side chain particles for basic (blue), acidic (red), and polar (pink) residues. This model, in a box of 256 DPPC molecules and 3,150 water particles, was the starting point for five CG-MD simulations, each of duration 2 μs. (B) Snapshots from the end of the S4 CG-MD simulations showing the S4 helix located at the lipid/water interface. (C) Snapshots from the end of the S4 CG-MD simulations showing the S4 helix inserted in the bilayer and switching between an extended helix and kinked helix conformation. The glycerol backbone particles are shown as green spheres.
Because we anticipated that S4 might adopt either a TM or an interfacial location, we ran an ensemble of 10 CG-MD simulations (each of 2-μs duration) of the self-assembly of S4 with a dipalmitoyl-phosphatidylcholine (DPPC) bilayer, to obtain a representative sample of the behavior of the helix in a bilayer environment. The initial configuration of the simulation system was a preformed S4 helix (Fig. 2A) surrounded by 256 randomly positioned CG lipid molecules and ≈3,000 water particles. The bilayer formed after ≈0.1 μs, and the S4 helix was indeed able to adopt two alternative orientations relative to a lipid bilayer. In six of the 10 simulations, the S4 helix adopted an interfacial location, whereas in the other four simulations a TM orientation was adopted across the bilayer (Fig. 2B). In any given simulation, we did not observe transitions between the interfacial and TM orientation once either had been reached, suggesting that the energy barrier for interconversion between these may be too high to be crossed on a microsecond time scale. Thus, in vivo insertion of S4 into a preformed bilayer must require longer time scales, and presumably, additional translocon-related machinery (40).
Further examination of those simulations in which the S4 helix adopted a TM orientation reveals a dynamic equilibrium between two conformations. In one conformation, the TM helix is tilted relative to the bilayer (by ≈45°), whereas, in the other, there is a kink in the middle of S4 (the kink angle varies between ≈0° and ≈60°) such that its N-terminal segment (containing the four key Arg residues) is TM and the C-terminal segment adopts a more interfacial location. Switching between kinked and unkinked conformations occurred on a ≈100-ns time scale. The kink in S4 is close to the G134 residue, i.e., the middle of the helix. Interestingly, this G134 is the location of a kink in the original full-length structure of KvAP (PDB ID 1ORQ) (5, 6), but the kink at this glycine is not observed in the isolated VS (PDB ID 1ORS) from which the CG model was derived (or in the later KvAP structure, i.e., 2A0L). Thus, in our simulations the TM configuration of the isolated S4 helix may be capturing an inherent flexibility which is responsive to the local environment, and which may therefore have a functional role in voltage-sensing. The kinking and tilting of the S4 helix implies that there is a degree of “tension” in trying to accommodate the long (35 residues) and multiply charged S4 helix in a lipid bilayer. When S4 is kinked, the voltage-sensing Arg-containing N-terminal region of S4 is more often inserted, as this is hydrophobic relative to the C-terminal half of S4. Even when S4 is tilted but unkinked, the C-terminal region is pushed up toward the interface. Visualization of the simulations suggests that the Arg-containing N-terminal half of S4 is accommodated in the bilayer via local bilayer deformation which, combined with side chain “snorkeling” (41, 42), enables the Arg side chains to interact with lipid headgroup particles (see below).
It is significant that S4 adopts a TM orientation in ≈50% of the simulations. This compares well with the biosynthetic insertion data (21), which suggested ΔG ≈ 0 for interconversion between an inserted (TM) and noninserted location of S4. More recently, oriented CD studies of an S4 peptide suggested it can insert in either an interfacial or a TM orientation (22). These comparisons lend confidence to the accuracy of the CG-MD method. Our observation of a combination of S4 helix kink and tilt when inserted into the bilayer suggests stabilization of S4 is complex. Experimental studies of insertion (21, 22) used a shorter S4 sequence than that in our simulations, complicating more detailed comparisons. NMR studies on a related S4 peptide (derived from the Shaker Kv channel sequence; ref. 43) suggested a predominantly interfacial location.
Although our results are consistent with recent biosynthetic insertion data, one might question whether the TM vs. interfacial orientations obtained here could be “metastable” artifacts of the self-assembly procedure. This is somewhat difficult to address because, even though the simulation times are extensive (a total of 20 μs of data were obtained), transitions between TM and interfacial orientations are not observed. However, the simulation method has been validated via application of the CG self-assembly protocol to a variety of other peptides for which experimental data on helix orientations in a membrane were available, resulting in correct predictions for transmembrane, interfacial, and mixed interfacial/transmembrane peptides (38). Nevertheless, it might be argued that comparison of these with such an unusually complex peptide as the S4 helix remains problematic. Therefore, we carried out a series of in silico mutant S4 simulations. Specifically, we mutated the side chain of Leu-128, located in the center of the S4 helix to an Arg side chain. An ensemble of 10 2-μs self-assembly simulations for the S4-L128R mutant revealed that, in all cases, this helix adopted an interfacial orientation after bilayer formation. Thus, as expected, an increase in helix polarity results in a decrease in the percentage of membrane insertion. This finding confirms the sensitivity of the CG self-assembly process, and suggests that a correlation between the results of such simulations and biosynthetic insertion equilibria might not be unreasonable. Interestingly, in some cases, an apparently metastable location of the helix in a TM orientation was evident for up to a few hundred nanoseconds before it adopted an interfacial location and the bilayer was formed. To further investigate this finding, we extracted snapshots from the simulations of the wild-type S4 helix in which a TM orientation was observed. After this, we converted these S4 helices into the more polar L128R mutant, and carried out three 2-μs simulations starting the mutant in a TM state. In each case, we observed rapid peptide deformation, resembling the normal S4 helix while in its Gly-134-kinked state as described above, suggesting a high degree of “tension” in the TM orientation for the L128R S4 mutant. Over the subsequent 10–50 ns, the additional Arg side chain at position 128 “dragged” the N-terminal half of the S4 helix toward the bilayer interface, before causing the peptide to completely exit the hydrophobic region of the membrane and adopt an interfacial location by within ≈60 ns. Therefore, at least for the more polar S4-L128R mutant helix, transitions between metastable states are observable on the time scales of our CG simulations, suggesting that the distribution of TM vs. non-TM orientations observed for the wild-type S4 helix reflects a genuine equilibrium.
VS Simulations.
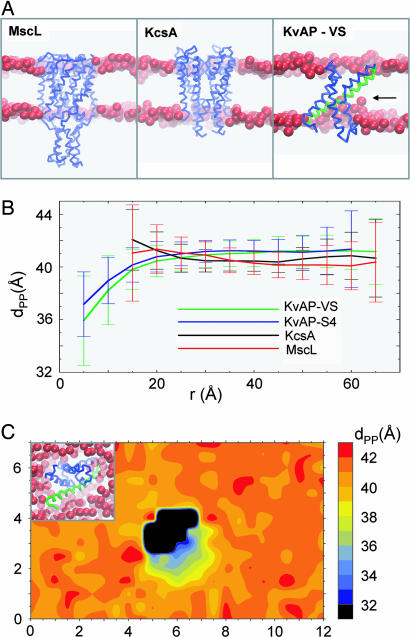
We extended the S4 studies to a more “physiological” simulation of an intact VS domain in a bilayer. Three 0.2-μs self-assembly simulation were run for the isolated KvAP VS domain in a 256-lipid DPPC bilayer. Experience with ≈40 different membrane proteins suggests this protocol will allow self-assembly/relaxation of a phospholipid bilayer around a membrane protein. In particular, it should be noted that successful bilayer insertion has been achieved using this protocol for more complex membrane proteins, including multidomain proteins e.g., the sugar-transporter LacY (38) and the ABC transporter BtuCD, and membrane proteins with large extracellular domains, e.g., Ca2+-ATPase, and the nicotinic acetylcholine receptor (nAChR) (see http://sbcb.bioch.ox.ac.uk/cgdb; P.J.B. K. Scott, K. Balali-Mood, and M.S.P.S., unpublished data). Moreover, CG self-assembly has been carried out for a number of monotopic membrane proteins, locating resulting these proteins at the bilayer/water interface (K. Balali-Mood, P.J.B., and M.S.P.S., unpublished data) in good agreement with experimental data (44). Furthermore, the CG method has been shown to correctly locate voltage sensor toxins (e.g., SGTx) at the membrane/water interface, in agreement both with atomistic simulations and experimental data (45). Therefore, this protocol provides a relatively robust method for simulating lipid self-assembly around the transmembrane regions of membrane proteins. To compare with the VS domain, simulations were also run for the TM domains of MscL and KcsA as example of more “canonical” integral membrane protein/ion channel structures. All three membrane proteins (MscL, KcsA, and KvAP-VS) assembled into a bilayer in their anticipated TM orientations (see Fig. 3A). MscL and KcsA stably adopted a TM orientation in the lipid bilayer, with no suggestion of any significant bilayer deformation. The final Cα root mean square deviations of the proteins from their initial structures were 3.4 ± 0.2 Å and 1.9 ± 0.1 Å for MscL and KcsA, respectively. The corresponding value for the three KvAP-VS simulations was 2.3 ± 0.3 Å. These values are comparable to those seen in atomistic MD simulations of membrane proteins (23). This finding demonstrates that the harmonic restraining potentials used in the CG model were sufficient to maintain the protein tertiary structure, reproducing the behavior of corresponding atomistic simulations (as seen in previous comparisons, e.g., LacY in ref. 38).
Fig. 3.
CG-MD simulations of membrane protein insertion into a lipid (DPPC) bilayer. (A) Three membrane proteins inserted in a bilayer via CG-MD self-assembly simulations: MscL, KcsA, and the KvAP VS domain. In each case, the protein is shown as a Cα trace, and the glycerol backbone particles of the bilayer are represented as red spheres. For KvAP, the S4 helix is highlighted in green, whereas significant bilayer deformation is indicated by a horizontal arrow. (B) Local deformation of the lipid bilayer measured as the average distance (±SD) between upper and lower P particles (dPP) vs. the distance of the particles in the xy plane from the centre of mass of the corresponding protein (r). The curve for S4 corresponds to analysis of a simulation in which S4 adopted a TM orientation (similar results were obtained from the other such simulations). (C) Local deformation of the lipid bilayer around the KvAP VS measured as the average distance between upper and lower P particles (dPP) as a function of position in the xy plane. The central black object corresponds to the area occupied by the protein.
By visualization, it is evident that there is considerable local deformation of the bilayer by the VS, in marked contrast with the other two membrane proteins/ion channels. To quantify such deformation we estimated the mean distance across the bilayer between opposing headgroup P particles (corresponding to the phosphate groups of the lipid molecules) to give dPP as a function of radial distance of the lipids from the centre of mass of the membrane protein (Fig. 3B). For MscL and KcsA, there is a suggestion of a very small degree of protein/bilayer mismatch immediately adjacent to the protein but there is no significant deformation of the bilayer. In contrast, for the VS there is a considerable (≈5 Å) local deformation of the bilayer. Close to the center of the VS protein, the Ps are “pulled” into the protein near the S4 helix (aided by snorkeling of the Arg and Lys side chains). An almost identical degree of mismatch is also evident for the glycerol backbone particles. This effect is gradually diminished toward the ends of the lipid molecules, so that the deformation is ≈3–4 Å for the second dipalmitoyl tail particles, and ≈0–1 Å for the fourth (i.e., terminal) tail particles. Significantly, a very similar pattern of local deformation is seen for the intact VS and for the isolated S4 helix, with a bilayer deformation of ≈4 Å for the headgroup P particles. Deformation of the bilayer by the VS can also be visualized as an average dPP value as a function of position in the membrane (xy) plane (Fig. 3C). This finding reveals that the deformation is most pronounced around the (lipid exposed) S4 helix of VS, with little/no deformation on the opposite, more hydrophobic face of the protein.
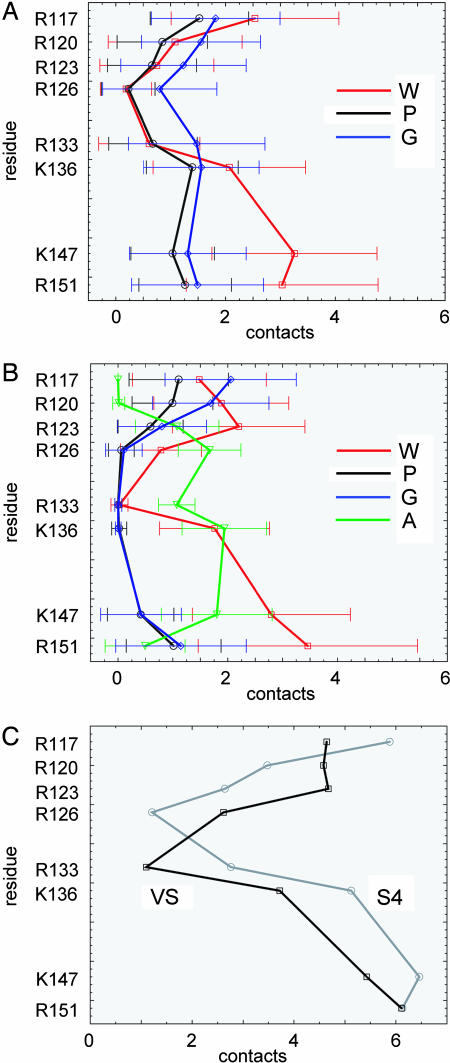
To examine how the S4 helix is stabilized by lipid and water contacts, interparticle contacts of <6 Å to P (i.e., phosphate), G (i.e., glycerol), and W (i.e., water) particles from S4 Arg and Lys side chain particles were analyzed. (This analysis was carried out for the equilibrated period of each simulation only. Thus, for the isolated S4 helix simulations in which the peptide was in a TM orientation, and for all VS domain simulations, the first 100–200 ns were discarded to allow for bilayer formation and equilibration of protein–lipid interactions.) For the isolated S4 helix (Fig. 4A) it is evident that there are significant water contacts to the C-terminal region of S4. For the N-terminal Arg-rich region, all three classes of contact (W, P, and G) are present, even extending to the R126 and R133 side chains that are “buried” in the middle of the lipid bilayer. For the VS (Fig. 4B), the N-terminal 4 Arg residues also form all three (W, P, and G) classes of contact. For the central (R133 and K136) and C-terminal segments, in addition to water contacts, there are significant contacts to anionic side chains of the remainder of the VS protein.
Fig. 4.
Analysis of S4/lipid and S4/water contacts in S4, averaged over all bilayer-inserted S4 and VS simulations. (A) Contacts (interparticle distance <6 Å) between the basic side chains of the isolated S4 helix and the water (W, red), PO4 (P, black), and glycerol (G, blue) particles. For each side chain, the mean (±SD) number of contacts over the second half of the simulation is shown. (B) Contacts between the basic side chains of the S4 helix within the KvAP VS domain and: water (W, red), PO4 (P, black), glycerol (G, blue), and anionic side chain (A, green) particles. For each side chain, the mean (±SD) number of contacts over the second half of the simulation is shown. (C) Total contacts (with all particles) of the basic side chains of S4, and of S4 within the KvAP domain (black).
Details of interaction between the protein and the other system components differed between simulations of the isolated S4 helix and of the VS domain. However, if one analyzes the total interactions (Fig. 4C) with protein, the pattern becomes more similar. Thus, in both systems, the four key Arg residues form at least one stabilizing interaction each, and all but one (R126 in S4, and R133 in the VS) of the eight Arg and Lys residues form at least two interactions. Nevertheless, there remain some differences. In particular, there are slightly more interactions between the N-terminal half of the helix in the VS simulation than the isolated S4 simulation, due primarily to greater solvation by water. On the other hand, there are slightly more interactions with the C-terminal half in the S4 simulation than in the VS simulation; the lack of protein–protein contacts for the isolated S4 leads to greater helix flexibility, enabling the C-terminal half of S4 to interact more strongly with the lipid headgroups as a result of the helix kinking/tilting described above.
It is also of interest to examine the time scale of interactions, by analyzing how long each phosphate, glycerol, water, and anionic side chain particle was bound to a particular S4 side chain before disassociation over the course of all simulations. From this analysis, it was evident that for both the S4 and VS simulations, no tightly bound water particles were present. In comparison, for both the glycerol and phosphate side chain particles, both short (<5 ns) and long (>5 ns) time scale interactions were present. For both the VS and isolated S4 helix simulations, up to ≈15% of each simulation included long time scale interactions between Arg/Lys side chains and phosphate particles, increasing up to ≈20% for interactions with glycerol backbone particles. These longer time scale interactions presumably facilitate the local bilayer deformation described above.
For the VS, although the strongest interactions with lipid headgroups occur for the most N-terminal Arg side chains, both short and long time scale interactions are gradually abolished toward the center of the helix. Presumably to counteract this, long time scale interactions with anionic side chains are evident for all but the first two N-terminal Arg residues. This finding is in agreement with spin-label side-chain accessibility and mobility measurements on KvAP, which suggested that the S4 helix resides on the outer perimeter of the protein against the membrane, and that the two most N-terminal Arg side chains are exposed on the membrane surface, whereas the following two side chains are buried within the protein (46). In our studies, the protein–protein contacts are made with several specific Asp and Glu side chain particles, and last for over ≈90% of each simulation. In particular, Arg-133, which makes zero contacts with water, glycerol, or phosphate groups, lies in the center of the S4 helix, and makes a permanent contact with Asp-62 on the S2 helix. Interestingly, this salt bridge is also maintained throughout long time scale atomistic simulations of the VS domain in a detergent micelle, and is similarly the primary constriction to water penetration (28).
Discussion
These results provide further evidence that the S4 helix, and in particular the S4 helix within the VS domain structure, is able to (meta)stably span a phospholipid bilayer. This spanning is achieved by local deformation of the lipid bilayer (and consequent penetration of water) combined with conformational changes of the protein in the form of tilting/kinking of the backbone and Arg/Lys snorkeling. Bilayer deformation was suggested by earlier atomistic simulation studies of S4 (26) and of the intact Kv1.2 channel (27), both of which relied on brief (≈10 ns) simulations in which the helix or channel was preinserted in a bilayer. In the current simulations, we used a coarse-grained approach to simulate self-assembly on a microsecond time scale, thus enhancing sampling of lipid molecule configurations around the S4 helix. Local bilayer deformation will enable “focusing” of the electrostatic field around key gating charge residues of S4 (26, 27). These results are relevant to our understanding of mechanisms of Kv channel gating. In particular, they indicate that when considering competing models (4, 16, 47, 48) of voltage-sensing, it is essential to allow for the molecular scale deformability of the lipid bilayer, and not to treat it as a hydrophobic slab of fixed and uniform thickness surrounding the channel protein. Indeed, Freites et al. (26) used a simulation of a preinserted S4 helix to propose that the membrane serves as a “structural extension” of the protein. The self-assembly simulations presented here of both S4 and the VS domain confirm this hypothesis.
Additionally, we have demonstrated that the VS domain can exist stably as an independent domain in a lipid bilayer. This finding is important in explaining how a homologous VS domain may be exploited in a voltage-sensitive enzyme (e.g., the voltage-sensitive phosphatase from Ciona intestinalis; ref. 10) or as a “combined” sensor and pore domain in voltage-gated proton channels (11, 12).
One should reflect briefly on methodological limitations. The CG approach lacks the detail of atomistic simulations. However, it has been tested extensively against a number of α-helical membrane peptides and proteins with respect to surface vs. TM orientations, and shown to yield good agreement with experimental data (38). It has also been shown to agree with extended (50–100 ns) atomistic MD simulations of OmpA, Glycophorin A (37), and LacY (38) in terms of protein–lipid interactions. Therefore, we are confident of its ability to reproduce the qualitative nature of membrane protein/lipid interactions in a number of systems.
We are especially conscious that the high polarity (for a transmembrane segment) of the S4 helix would be expected to make the thermodynamics of membrane partitioning quite dependent on the CG interaction potentials. However, we have additionally demonstrated the sensitivity of the CG self-assembly protocol to the peptide sequence by carrying out simulations of a more polar S4 “mutant” helix, for which TM insertion is eliminated. Moreover, in artificially constructed systems containing “metastable” mutant S4 helices in the TM orientation, rapid transitions to the energetically more favorable interfacial location are observed. Furthermore, preliminary studies in which the Arg potential is made slightly more polar (i.e., hydrophilic) suggest that the self-assembly simulations of the VS and of isolated S4 are robust to changes in the CG potential. Thus, there is a slightly lower rate of isolated S4 helix insertion and an increase in protein/bilayer mismatch for both S4 and the VS of ≈1–2 Å. Although this may seem surprising given that the experimental ΔG for S4 insertion is ≈0 (21), one should consider that such an increase in polarity is distributed over the entire helix. This result may be contrasted with the situation described by our “mutant S4” simulations, in which an additional Arg residue introduced into the center of the helix resulted in bilayer insertion being completely abolished. Therefore, the tendency for insertion depends not only on the overall peptide polarity, but also on the position of the associated polar residues. This finding is reminiscent of results obtained in studies of recognition of a range of synthetic TM helix sequences by the translocon (49), and further supports our previous results that the background hydrophobicity of the S4 helix is sufficient to offset the bilayer strain necessary for inserting multiple charged side chains into the low dielectric membrane environment. It should also be noted that the elastic properties of the bilayer model have been shown to be of the same order as those calculated from both atomistic simulations and from experimental data (29), providing us with further confidence in the observed bilayer deformation around the S4 helix and KvAP under a range of conditions. It will be of interest to develop quantitative approaches for comparison and refinement of the CG potential function against recent atomistic free-energy profiles of amino acid side chains across lipid bilayers (50, ‡).
In summary, our demonstration of bilayer deformation on a microsecond time scale helps to explain some experimental observations apparently incompatible with biophysical principles: a highly charged helix embeds itself within a low dielectric, but malleable, membrane environment, which consequently may provide the exquisite sensitivity to membrane potential necessary for channel gating.
Methods
Simulations were performed by using GROMACS (www.gromacs.org) (51, 52). CG simulations were performed as described in ref. 37, with CG parameters for lipid molecules (dipalmitoyl-phosphatidylcholine, DPPC), Na+ and Cl− ions, and water molecules as in ref. 29, and for amino acids as in ref. 37. A CG peptide or protein model was generated from the corresponding atomistic structure and was composed of a chain of backbone particles with attached side chain particles. For details of protein bond and angle potentials, see supporting information (SI) Text and SI Figs. 5 and 6. Lennard–Jones interactions were shifted to zero between 9 and 12 Å, and electrostatics were shifted to zero between 0 and 12 Å, with a relative dielectric constant of 20. The nonbonded neighbor list was updated every 10 steps. All simulations were performed at constant temperature, pressure, and number of particles. The temperature of the protein, lipid, and solvent were each coupled separately using the Berendsen algorithm (53) at 323 K, with a coupling constant τT = 40 ps. The system pressure was anisotropically coupled using the Berendsen algorithm at 1 bar with a coupling constant τP = 40 ps and a compressibility of 1 × 10−5 bar−1. The time step for integration was 40 fs.
Protein coordinates were extracted from the following PDB entries and converted to equivalent CG models: 1ORS (KvAP); the S4 helix from 1ORS; 1MSL (MscL); and 1K4C (KcsA). For the S4 peptide simulations, the distance between backbone particles was restrained to mimic secondary structure H-bonds in the atomistic structure (37), using a harmonic distance restraint with an equilibrium length of 6 Å and a force constant of 10 kJ mol−1Å−2. For the three integral proteins (i.e., the KvAP VS domain, MscL, and KcsA), the tertiary structure of the protein was maintained by using an elastic network model (54, 55). Harmonic restraints were applied between all backbone particles within 7 Å of one another, each with a force constant of 10 kJ mol−1Å−2 and an equilibrium bond length equal to that in the starting structure.
The S4 peptide was placed in a box of dimension (100 Å)3, whereas the VS domain was placed in a box of dimension (110 Å)3. The MscL and KcsA simulation boxes were each 100 × 100 × 120 Å3. Each CG model was energy minimized using <100 steps of the steepest decent method, to relax any steric conflicts within the protein. Subsequently, each system was combined with randomly positioned CG DPPC lipid molecules (256 lipids for S4 peptide and VS domain; 249 lipids for MscL; 253 lipids for KcsA). Each system was solvated with CG water particles, and sodium or chloride counterions were added to preserve overall electrical neutrality. Each system was then energy minimized again, for a further <100 steps, to relax any steric conflicts between protein, lipid, and solvent. Production simulations were then performed on Linux workstations. Analyses were performed by using GROMACS tools and locally written code. Visualization used VMD (56) and RasMol (22).
Supplementary Material
Acknowledgments
We thank Zara Sands for discussions concerning this work. This work was supported by the Biotechnology and Biological Sciences Research Council, the Membrane Protein Structure Initiative consortium (www.mpsi.ac.uk), and the Wellcome Trust.
Abbreviations
- TM
transmembrane
- MD
molecular dynamics
- CG
coarse grained
- VS
voltage sensor
- PDB
Protein Data Bank.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Fernández-Vidal, M., Castro-Roman, F., White, S. H. (2006) Biophys J 90:241A (abstr.).
This article contains supporting information online at www.pnas.org/cgi/content/full/0606822104/DC1.
Dorairaj, S., Allen, T. W. (2006) Biophys J90:213A (abstr).
References
- 1.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 2.Bezanilla F. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 3.Yellen G. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- 4.Swartz KJ. Nat Rev Neurosci. 2004;5:905–916. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, Mackinnon R. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Ruta V, Chen J, Lee AG, Mackinnon R. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 7.Lee SY, Lee A, Chen J, MacKinnon R. Proc Natl Acad Sci USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long SB, Campbell EB, MacKinnon R. Science. 2005;309:897–902. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 9.Long SB, Campbell EB, MacKinnon R. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 10.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 11.Ramsey IS, Moran MM, Chong JHA, Clapham DE. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki M, Takagi M, Okamura Y. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 13.Starace DM, Bezanilla F. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 14.Chanda B, Asamoah OK, Blunck R, Roux B, Bezanilla F. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 15.Posson DJ, Ge P., Miller C, Bezanilla F, Selvin PR. Nature. 2005;436:848–851. doi: 10.1038/nature03819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruta V, Chen J, MacKinnon R. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 17.Phillips LR, Milescu M, Li-Smerin Y, Midell JA, Kim JI, Swartz KJ. Nature. 2005;436:857–860. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- 18.Popot JL, Engelman DM. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 19.Engelman DM, Chen Y, Chin C, Curran R, Dixon AM, Dupuy A, Lee A, Lehnert U, Mathews E, Reshetnyak Y, Senes A, Popot JL. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 20.Bowie JU. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 21.Hessa T, White SH, von Heijne G. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 22.Sayle RA, Milner-White EJ. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 23.Ash WL, Zlomislic MR, Oloo EO, Tieleman DP. Biochim Biophys Acta. 2004;1666:158–189. doi: 10.1016/j.bbamem.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Deol SS, Bond PJ, Domene C, Sansom MSP. Biophys J. 2004;87:3737–3749. doi: 10.1529/biophysj.104.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deol SS, Domene C, Bond PJ, Sansom MSP. Biophys J. 2006;90:822–830. doi: 10.1529/biophysj.105.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freites JA, Tobias DJ, von Heijne G, White SH. Proc Natl Acad Sci USA. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treptow W, Tarek M. Biophys J. 2006;90:L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sands Z, Grottesi A, Sansom MSP. Biophys J. 2005;90:1598–1606. doi: 10.1529/biophysj.105.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrink SJ, de Vries AH, Mark AE. J Phys Chem B. 2004;108:750–760. [Google Scholar]
- 30.Murtola T, Falck E, Patra M, Karttunen M, Vattulainen I. J Chem Phys. 2004;121:9156–9165. doi: 10.1063/1.1803537. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen SO, Lopez CF, Srinivas G, Klein ML. J Phys Condens Matter. 2004;16:R481–R512. [Google Scholar]
- 32.Tozzini V. Curr Opin Struct Biol. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Shih AY, Arkhipov A, Freddolino PL, Schulten K. J Phys Chem B. 2006;110:3674–3684. doi: 10.1021/jp0550816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Klein ML. J Phys Chem B. 2001;105:4464–4470. [Google Scholar]
- 35.Nielsen SO, Lopez CF, Ivanov I, Moore PB, Shelley JC, Klein ML. Biophys J. 2004;87:2107–2115. doi: 10.1529/biophysj.104.040311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venturoli M, Smit B, Sperotto MM. Biophys J. 2005;88:1778–1798. doi: 10.1529/biophysj.104.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bond PJ, Sansom MSP. J Am Chem Soc. 2006;128:2697–2704. doi: 10.1021/ja0569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond PJ, Holyoake J, Ivetac A, Khalid S, Sansom MSP. J Struct Biol. 2007 doi: 10.1016/j.jsb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Shental-Bechor D, Fleishman SJ, Ben-Tal N. Trends Biochem Sci. 2006;31:192–196. doi: 10.1016/j.tibs.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 40.van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 41.Mishra V, Palgunachari M, Segrest J, Anantharamaiah G. J Biol Chem. 1994;269:7185–7191. [PubMed] [Google Scholar]
- 42.Strandberg E, Killian JA. FEBS Lett. 2003;544:69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 43.Halsall A, Dempsey CE. J Mol Biol. 1999;293:901–915. doi: 10.1006/jmbi.1999.3194. [DOI] [PubMed] [Google Scholar]
- 44.Tatulian SA, Qin S, Pande AH, He X. J Mol Biol. 2005;351:939–947. doi: 10.1016/j.jmb.2005.06.080. [DOI] [PubMed] [Google Scholar]
- 45.Wee CL, Bemporad D, Sands ZA, Gavaghan D, Sansom MSP. Biophys J. 2007;92:L07–L09. doi: 10.1529/biophysj.106.098681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuello LG, Cortes DM, Perozo E. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 47.Guy HR, Conti F. Trends Neurosci. 1990;13:201–206. doi: 10.1016/0166-2236(90)90160-c. [DOI] [PubMed] [Google Scholar]
- 48.Silverman WR, Roux B, Papazian DM. Proc Natl Acad Sci USA. 2003;100:2935–2940. doi: 10.1073/pnas.0636603100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 50.Norman KE, Nymeyer H. Biophys J. 2006;91:2046–2054. doi: 10.1529/biophysj.105.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berendsen HJC, van der Spoel D, van Drunen R. Comp Phys Comm. 1995;95:43–56. [Google Scholar]
- 52.Lindahl E, Hess B, van der Spoel D. J Mol Model. 2001;7:306–317. [Google Scholar]
- 53.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 54.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keskin O, Jernigan RL, Bahar I. Biophys J. 2000;78:2093–2106. doi: 10.1016/S0006-3495(00)76756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.