Abstract
Since antibiotic resistance usually affords a gain of function, there is an associated biological cost resulting in a loss of fitness of the bacterial host. Considering that antibiotic resistance is most often only transiently advantageous to bacteria, an efficient and elegant way for them to escape the lethal action of drugs is the alteration of resistance gene expression. It appears that expression of bacterial resistance to antibiotics is frequently regulated, which indicates that modulation of gene expression probably reflects a good compromise between energy saving and adjustment to a rapidly evolving environment. Modulation of gene expression can occur at the transcriptional or translational level following mutations or the movement of mobile genetic elements and may involve induction by the antibiotic. In the latter case, the antibiotic can have a triple activity: as an antibacterial agent, as an inducer of resistance to itself, and as an inducer of the dissemination of resistance determinants. We will review certain mechanisms, all reversible, that bacteria have elaborated to achieve antibiotic resistance by the fine-tuning of the expression of genetic information.
INTRODUCTION
Bacteria may use various biochemicalpathways to escape the lethal action of drugs: (i) decreased intracellular accumulation of the antibiotic by an alteration of outer membrane permeability, diminished transport across the inner membrane, or active efflux; (ii) alteration of the target by mutation or enzymatic modification; (iii) enzymatic detoxification of the drug; and (iv) bypass of the drug target. The coexistence of several of these mechanisms in the same host can lead to multidrug resistance (MDR). However, since antibiotic resistance usually affords a gain of function, there is an associated biological cost resulting in the loss of fitness of the bacterial host. Considering that antibiotic resistance is most often only transiently advantageous to bacteria, an efficient and elegant way for them to escape the lethal action of drugs is the alteration of resistance gene expression. It appears that the expression of bacterial resistance to antibiotics is frequently regulated, which indicates that modulation of gene expression probably reflects a good compromise between energy saving and adjustment to a rapidly evolving environment. Modulation of gene expression can occur at the transcriptional or translational level, following mutations or the movement of mobile genetic elements, and may involve induction by the antibiotic. In the latter case, the antibiotic can have a triple activity: as an antibacterial agent, as an inducer of resistance to itself, and, as in the case of tetracycline and gram-positive bacteria harboring conjugative transposons, as an inducer of the dissemination of a resistance determinant. We will review certain mechanisms, all reversible, that bacteria have elaborated to achieve antibiotic resistance by fine-tuning the expression of genetic information.
REGULATION OF RESISTANCE EXPRESSION BY TWO-COMPONENT SYSTEMS IN GRAM-POSITIVE BACTERIA
Two-Component Regulatory Systems
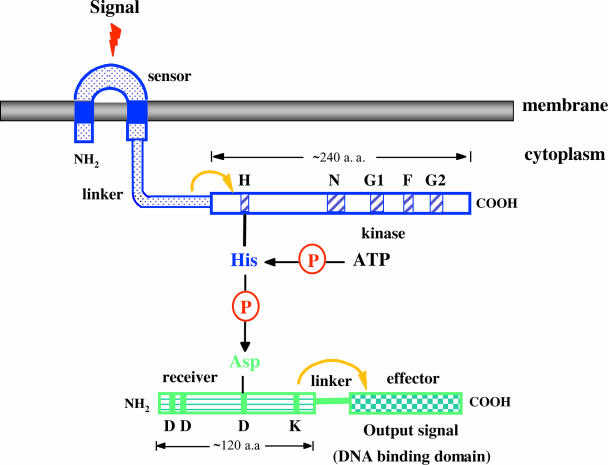
Bacteria live in precarious environments and must constantly adapt to external conditions by adjusting their structure, physiology, and behavior to survive. Many signaling proteins from both gram-positive and gram-negative bacteria are built from modular domains that promote information transfer within and between proteins (242). One such system, designated the “two-component regulatory system,” comprises two proteins: a sensor usually located in the membrane that detects certain environmental signals and a cytoplasmic response regulator that mediates an adaptative response, usually a change in gene expression (Fig. 1) (110, 242). The terms “kinase” and “response regulator” are used since they seem to best represent the essential activities of these proteins. The large majority of histidine kinases are homodimeric proteins with an N-terminal periplasmic sensing domain coupled to a C-terminal cytoplasmic kinase domain (Fig. 1). The sensing domains are variable in sequence, reflecting the many different environmental signals to which histidine kinases are responsive and consequently the numerous specific functions that they regulate. Communication with the cytoplasmic transmitter domain involves the propagation of sensory information across the cytoplasmic membrane, presumably with the induction of conformational changes. The kinase domain that binds ATP and catalyzes the autophosphorylation of a histidine (Fig. 1) is more conserved. It is divided into two subdomains, a variable connecting linker and a second subdomain containing several highly conserved sequences designated H, N, D, F, and G boxes, which may play the role of catalytic center (Fig. 1) (71, 188). The phosphate group of the histidine residue is then transferred to a highly conserved aspartate residue in the receiver domain of the regulator (Fig. 1) (110, 242). Response regulators are characterized by a conserved domain of approximately 125 amino acids usually attached by a linker sequence to a domain with an effector function (Fig. 1) (110, 242). Prominent sequence features of regulators include two aspartate residues near the amino terminus, a lysine close to the carboxyl terminus, and a centrally located aspartate (Fig. 1). The effector domain generally has DNA binding activity, and in that instance, response regulator phosphorylation results in the activation of transcription (Fig. 1). In many instances, the response regulators act as transcriptional activators or repressors.
FIG. 1.
Schematicrepresentation of a two-component regulatory system. Structural features of sensor (top) and regulator (bottom) proteins. H, N, G1, F, and G2 refer to the motifs conserved in histidine protein kinases and are shown as hatched blue boxes. The phosphorylated histidine is nested in a highly conserved sequence termed the H box, close to the N-terminal border of the conserved kinase domain. The G1 and G2 domains are glycine rich and resemble nucleotide binding motifs seen in other proteins. The sequences of the remaining D and F boxes reveal little about their possible functions. In the regulator, the central aspartate is the site of phosphorylation, whereas the amino-terminal pair is probably important for catalysis. The conserved lysine may be involved in effecting the phosphorylation-induced conformational changes that regulate output activity. Asp, aspartate; His, histidine; P, phosphate; dotted blue box, sensor domain; blue box, transmembrane domain; white box, kinase domain; horizontally striped green box, receiver domain; checkerboard green box, effector domain. a.a., amino acids.
Several mechanisms control the rate of dephosphorylation of the phosphorylated response regulators. First, some of the regulators exhibit an autophosphatase activity with half-lives ranging from a few seconds to many minutes. Second, dephosphorylation can be mediated by the corresponding kinase. Finally, auxiliary regulatory proteins can also function as phosphatases to enhance the rate of dephosphorylation of the response regulators.
Resistance to Glycopeptides in Enterococci
The molecular target of glycopeptide antibiotics is the d-alanyl-d-alanine (d-Ala-d-Ala) terminus of intermediates in peptidoglycan synthesis. By binding to this dipeptide, vancomycin and teicoplanin inhibit the transglycosylation and transpeptidation reactions in peptidoglycan assembly (215).
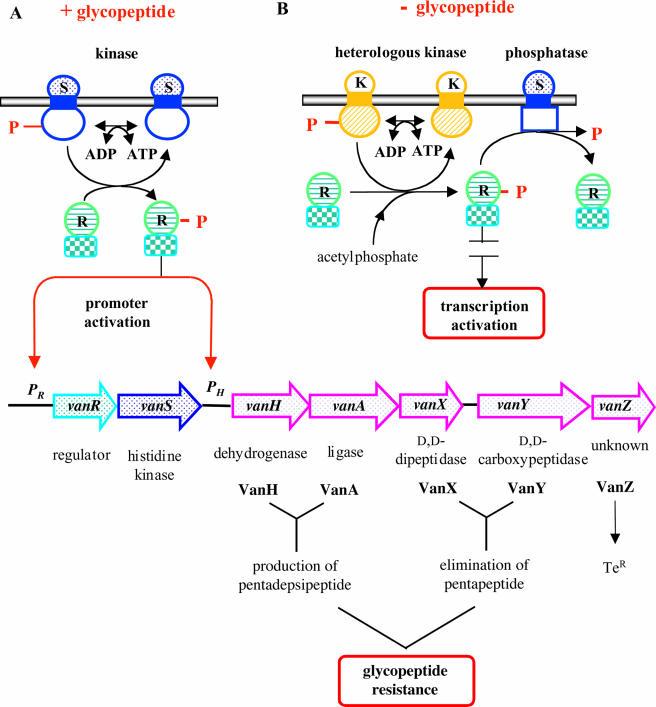
Glycopeptide resistance in enterococci results from the production of modified peptidoglycan precursors ending in d-Ala-d-Lac (VanA, VanB, and VanD) or d-Ala-d-Ser (VanC, VanE, and VanG), to which glycopeptides exhibit low binding affinities, and from the elimination of the high-affinity precursors ending in d-Ala-d-Ala and synthesized by the host Ddl ligase (17, 218). In enterococci with the VanA, VanB, or VanD phenotype, the synthesis of d-Ala-d-Lac requires the presence of a ligase (VanA, VanB, or VanD) of altered specificity compared to the host Ddl ligase and of a dehydrogenase (VanH, VanHB, or VanHD) that converts pyruvate to d-Lac (Fig. 2)(19). In VanC-, VanE-, and VanG-type strains, the ligase genes (vanC, vanE, or vanG) encode a protein catalyzing the synthesis of d-Ala-d-Ser (218), and the production of d-Ser is due to a membrane-bound serine racemase (VanT, VanTE, or VanTG) (Fig. 2) (1, 11, 63).
FIG. 2.
Comparison of the van gene clusters. Open arrows represent coding sequences (red arrows, regulatory genes; purple arrows, genes required for resistance; blue arrows, accessory genes; pink and yellow arrows, genes of unknown function) and indicate the direction of transcription. The percentages of amino acid (aa) identity between the deduced proteins of reference strains BM4147 (VanA) (19), V583 (VanB) (77), BM4339 (VanD) (42), BM4174 (VanC) (10), BM4405 (VanE) (1), and BM4518 (VanG) (63) are indicated under the arrows. The vertical bar in vanYG indicates the frameshift mutation leading to a predicted truncated protein. NA, not applicable.
The interaction of a glycopeptide with its normal target is prevented by the removal of precursors terminating in d-Ala (216). Two enzymes are involved in this process: a cytoplasmic d,d-dipeptidase (VanX, VanXB, or VanXD) that hydrolyzes the dipeptide d-Ala-d-Ala synthesized by the host Ddl ligase and a membrane-bound d,d-carboxypeptidase (VanY, VanYB, or VanYD) that removes the C-terminal d-Ala residue of late peptidoglycan precursors when the elimination of d-Ala-d-Ala by VanX is incomplete (Fig. 2) (12, 217). In VanC-, VanE-, and VanG-type resistance, both activities are encoded by a single gene, vanXYC, vanXYE, or vanXYG.
Classification of glycopeptide resistance is based on the primary sequence of the structural genes for the resistance-mediating ligases. VanA-type strains display high-level inducible resistance to both vancomycin and teicoplanin, whereas VanB-type strains have variable levels of inducible resistance to vancomycin only, since teicoplanin is not an inducer (15, 211). VanD-type strains are characterized by constitutive resistance to moderate levels of both glycopeptides (66, 67). VanC, VanE, and VanG are resistant to low levels of vancomycin but remain susceptible to teicoplanin (63, 80, 135). VanC- and VanE-type strains are inducibly or constitutively resistant (2, 187). In several constitutive strains of these types, various mutations in VanS could, as in VanB-type strains, account for constitutivity (26, 65).
Although all six types of resistance involve genes encoding related enzymatic functions, they can be distinguished by the location of the genes and by the various modes of regulation of gene expression (Fig. 2). The vanA and vanB operons are located on plasmids or in the chromosome (20), whereas the vanD (42, 66, 67), vanG (63), vanE (1), and vanC (10) operons have so far been found exclusively in the chromosome.
Two-component regulatory systems in Van-type enterococci.
Among the ubiquitous two-component systems that constitute one of the largest families of transcriptional regulators in bacteria, the VanS/VanR-type systems are the only ones that control the expression of genes that mediate antibiotic resistance. Expression of VanA-, VanB-, VanD-, VanC-, VanE-, and VanG-type resistance is regulated by a VanS/VanR-type two-component signal transduction system composed of a membrane-bound histidine kinase (VanS, VanSB, VanSD, VanSC, VanSE, or VanSG) and a cytoplasmic response regulator (VanR, VanRB, VanRD, VanRC, VanRE, or VanRG) that acts as a transcriptional activator (Fig. 2) (1, 10, 18, 42, 63, 66, 67, 77). In the vanA, vanB, vanD, and vanG operons, the genes for the two-component regulatory system (vanRS, vanRBSB, vanRDSD, and vanRGSG) are present upstream from the structural genes for the resistance proteins (20, 42, 63, 67), whereas in the vanC and vanE clusters, vanRCSC and vanRESE are located downstream (Fig. 2) (1, 10). The regulatory and resistance genes in the vanA, vanB, and vanD operons are transcribed from distinct promoters, PR, PRB, and PRD and PH, PYB, and PYD, respectively, that are coordinately regulated (13, 14, 43, 65, 77). The vanC and vanE clusters are cotranscribed from a single upstream promoter (Fig. 2) (1, 2, 187).
The vanRG and vanSG genes have the highest homology with vanRD and vanSD, respectively (Fig. 2). Additionally, vanUG encodes a predicted transcriptional activator (63), and a protein of this type has not previously been associated with glycopeptide resistance. Thus, as opposed to the other van gene clusters, the vanG operon contains three genes, vanUG, vanRG, and vanSG, for a putative regulatory system that are cotranscribed constitutively from the PUG promoter, whereas inducible transcription of the vanYG, vanWG, vanG, vanXYG, and vanTG resistance genes is initiated from the PYG promoter (Fig. 2) (63).
Phosphotransfer reactions catalyzed by VanRS and VanRBSB two-component systems.
Despite the fact that the VanS/VanR and VanSB/VanRB two-component systems are only distantly related, they catalyze similar reactions. The two response regulators are 34% identical, whereas the histidine kinases possess only 23% sequence identity, with unrelated amino-terminal sensing domains (Fig. 2). VanS-type sensors comprise an N-terminal sensor domain with two membrane-spanning segments and a C-terminal cytoplasmic kinase domain (Fig. 1) (18, 269). Following a signal related to the presence of a glycopeptide in the culture medium, the cytoplasmic domain of VanS or VanSB catalyzes ATP-dependent autophosphorylation of a specific histidine residue at positions 164 and 233, respectively, and transfers the phosphate group to an aspartate residue at position 53 of VanR or VanRB present in the effector domain (Fig. 3) (13, 18, 269).
FIG. 3.
Model for positive (phosphorylation) and negative (dephosphorylation) control of VanR by VanS and schematic representation of the synthesis of peptidoglycan precursors in VanA- or VanB-type strains. Kinase (A) and phosphatase (B) activities of VanS are depicted. K, heterologous kinase; R, regulator; S, sensor. Dotted blue circle, sensor domain; blue box, transmembrane domain; white circle, kinase domain; horizontally striped green circle, receiver domain; checkerboard green box, effector domain.
Purified VanS and VanSB autophosphorylate in the presence of ATP and act as both a kinase and a phosphatase for VanR and VanRB, respectively (65, 269). VanR and VanRB are phosphorylated following incubation either with the phosphorylated form of VanS or VanSB, respectively, or with acetylphosphate. VanS and VanSB also stimulate the dephosphorylation of VanR and VanRB (65, 269). The VanS and VanSB sensors therefore respectively modulate the levels of phosphorylation of the VanR and VanRB regulators: they act primarily as a phosphatase under noninducing conditions and as a kinase in the presence of glycopeptides, leading to the phosphorylation of the response regulator and the activation of the resistance genes (Fig. 3) (13, 14, 64, 65, 112). The phosphorylation of VanR-type regulators enhances the affinity of the effector portion of the protein for the promoters and stimulates transcription of the regulatory and resistance genes of the van clusters (Fig. 3) (112). In contrast to VanR-VanS, the VanRB-VanSB system mediates the activation of the PYB promoter only in the presence of vancomycin, and the lack of activation by teicoplanin accounts for the susceptibility of VanB-type strains to this antibiotic (15, 65, 77). Spontaneous dephosphorylation of VanR and VanRB is slow in comparison with other response regulators, with half-lives of 10 h and 150 min, respectively, but VanS and VanSB stimulate the reaction (65, 269). The phosphatase activity of VanS and VanSB is required for the negative regulation of resistance genes in the absence of glycopeptides preventing the accumulation of VanR-phosphate (VanR-P) or VanRB-phosphate (VanRB-P) phosphorylated by acetylphosphate or by kinases encoded by the host chromosome (Fig. 3) (13).
In vitro binding of VanR and VanRB to promoter regulatory regions.
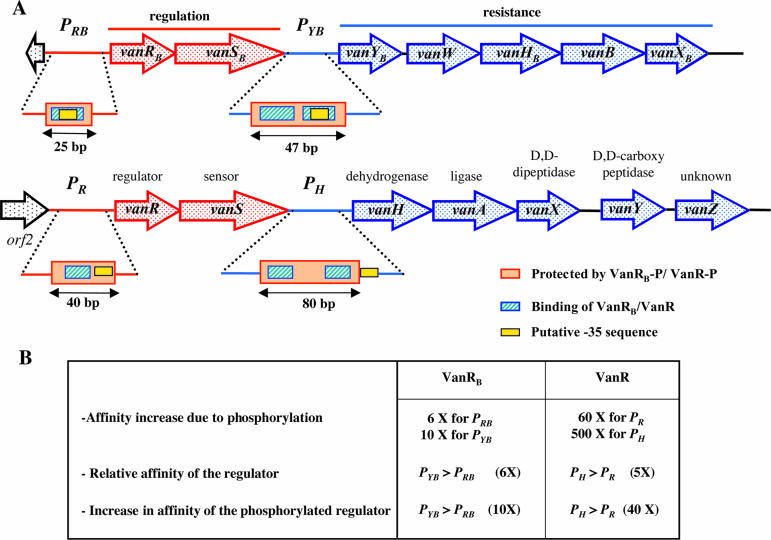
There is sequence similarity between VanR and VanRB and response regulators of the OmpR/PhoB subclass in both the effector and DNA binding domains, with VanR being closer to OmpR (37% similarity) than to PhoB (35%), whereas VanRB is closer to PhoB (32% similarity) than to OmpR (26%). Phosphorylation of VanR and VanRB increases their DNA affinity, but VanR-P (112) appears to be more stable than VanRB-P (64). The promoters in the vanA and vanB operons have common features, with a single binding site in the PR and PRB promoters and two sites in the PH and PYB promoters (64, 112). However, the positionings of these sites in the promoter regions differ: in the case of VanR, the binding site is upstream from the −35 region (112), whereas it overlaps the −35 region for VanRB (Fig. 4) (64). The binding site is centered at −54.5 for VanR in PR and at −32.5 for VanRB in PRB. In the PH and PYB promoter regions, the sites are centered at −53.5 and −86.5 for VanR (112) and at −33.5 and −55.5 for VanRB (64), respectively. The two copies of the binding sites at PH and PYB are 33 bp (112) and 22 bp (64) apart, respectively, suggesting that since these figures differ almost exactly by three or two helical turns of B-DNA (10.5 bp/turn), they both lie on the same face of the DNA helix. VanR and VanRB bind with higher affinity to the corresponding PH and PYB promoters controlling the resistance genes than to the PR and PRB promoters for the regulatory genes (Fig. 4) (64). Phosphorylation increases the affinity for PH by 40-fold but increases the affinity for PYB by only 10-fold, indicating that the cooperativity is higher at PH than at PYB (Fig. 4) (64, 112). A direct relationship between the binding cooperativity of VanRB-P to its sites and the expression of the resistance genes may exist, since the levels of induction of the resistance genes are lower with VanRB than with VanR.
FIG. 4.
(A) Schematic representation of the binding of VanR-type regulators to the vanA and vanB promoters and (B) comparison of affinity of VanR or VanRB and VanR-P or VanRB-P for DNA fragments carrying the PR/PRB and PH/PYB promoters. Open arrows represent coding sequences (red arrows, regulatory genes; purple arrows, genes required for resistance; black arrows, genes of unknown function).
VanR and VanR-P bind to a similar 80-bp stretch of the regulatory region of PH that contains two putative 12-bp binding sites (Fig. 4) (112). The PR promoter contains a single 12-bp binding site, and the phosphorylation of VanR increases the size of the protected region from 20 to 40 bp (Fig. 4) (112), whereas the phosphorylation of VanRB does not increase the size of the protected region in PRB (64). After phosphorylation, VanR generates a more extensive footprint than VanRB (40 bp for PR versus 25 bp for PRB and 80 bp for PH versus 47 bp for PYB) due to higher cooperativity (Fig. 4).
A 21-bp consensus was identified within the binding regions of PRB and PYB, which consists of two and four direct repeats of the CTACAG(G/A)heptanucleotide, respectively (64). A similar organization has been observed in other response regulators such as CtsR (68) and PhoP (74, 270) from Bacillus subtilis and DcuR from Escherichia coli (3). The heptanucleotides, which correspond to the VanRB recognition sequence, are separated by four nucleotides, and at each site, the protected guanines are 10 bp apart and are thus positioned on the same face of the B-DNA helix (64). This tandem symmetry is consistent with the notion that VanRB binds to DNA as a head-to-tail dimer, as reported previously for PhoB (35). The consensus sequence of PRB and PYB is not present in the promoter regions of the other van operons. In contrast, sequence comparison of the PYG promoter, controlling the resistance genes in the vanG operon; the PYD promoter, controlling those of the vanD operon; and the PH promoter revealed a 12-bp consensus sequence, (T/C)CGTAXGAAA(T/A)T, similar to T(T/C)GTA(G/A)GAAA(T/A)T, corresponding to the regions protected by VanR and VanR-P in the vanA operon (112) that is present three times in the PYG region (63) and twice in the PYD region.
VanRB-P recruits the RNA polymerase to the regulatory and resistance gene promoters.
As mentioned above, VanRB and VanRB-P bind specifically to the same regions of the PRB and PYB promoters, and although not essential for binding, phosphorylation of the regulator significantly increases the affinity for the DNA targets (64). Treatment with acetylphosphate converts VanRB from a monomer with low affinity for its binding site into a homodimer with higher DNA affinity (64). Activation of gene expression in vivo most likely requires the phosphorylation and consequently the dimerization of VanRB to raise the binding affinity to physiologically relevant levels. In order to switch on the positive autoregulatory loop that leads to the expression of the vancomycin resistance genes, a VanB-type strain needs to synthesize a minimum number of VanRB and VanSB molecules even in the absence of antibiotic. VanRB-P has a higher affinity for its targets than VanRB and appears to be more efficient than VanRB in promoting an open complex formation with PRB and PYB (64). The RNA polymerase is able to interact with the PRB promoter region in the absence or presence of VanRB but is able to interact with PYB only in the presence of VanRB and in both cases with an increased affinity when VanRB is phosphorylated. In vitro transcription assays showed that VanRB-P activates PYB more strongly than PRB (64). The higher affinity of VanRB for PYB relative to PRB may result from PYB having two heptanucleotide direct repeats, possibly resulting in the cooperative binding of the regulator to the two adjacent sites, which may serve as recognition sites for VanRB and VanRB-P binding. Although the regions protected by VanRB and VanRB-P encompass the −35 regions of the promoters, VanRB-P is able to recruit the RNA polymerase at the promoters and allows efficient open complex formation. Unlike the situation with PhoB, the C-terminal domain of the RNA polymerase α subunit is required for transcription activation from the PRB and PYB promoters, possibly by making direct contact with the activator or by being mandatory for promoter binding (64).
In vivo activation of the PR and PH promoters in VanA-type strains.
In VanA-type strains, the activation of the PR and PH promoters has been studied using various transcriptional fusions with reporter genes (13, 14). Determinations of d,d-dipeptidase activity and of the cytoplasmic pool of peptidoglycan precursors show that the expression of glycopeptide resistance is regulated at the level of transcriptional initiation at these promoters. The PR and PH promoters have similar strengths and are regulated similarly. They are not activated in the absence of VanR and VanS, are induced by glycopeptides when VanR and VanS are present, and are constitutively activated by VanR in the absence of VanS due, presumably, to phosphorylation of VanR by host kinases (13, 14). Consequently, VanR is a transcriptional activator required for initiation at both promoters, whereas VanS is not necessary for the full activation of the promoters since VanR can be phosphorylated independently of its partner sensor. However, VanS is required for negative control of the promoters in the absence of glycopeptides, acting as a phosphatase under noninducing conditions, thus preventing the accumulation of VanR-P. VanR-P binds to the PR promoter and activates the transcription of the vanR and vanS genes. Regulation of the vanA gene cluster therefore involves not only a modulation of the relative amounts of VanR and VanR-P by the kinase and phosphatase activities of VanS but also a modulation of the concentration of the response regulator. An amplification loop results from the binding of VanR-P to the PR promoter with a resultant increased expression of vanR and accumulation of VanR-P following phosphorylation. This may explain the high-level transcription of the resistance genes observed in vanS null mutants, since the amplification loop, in combination with the long half-life of VanR-P, may compensate for the inefficient phosphorylation of the response regulator by the putative host kinase.
Acquisition of teicoplanin resistance by VanB-type enterococci.
As mentioned above, enterococci harboring clusters of the vanB class remain susceptible to teicoplanin since this antibiotic is not an inducer (15). However, mutations in the vanSB sensor gene have been obtained in vitro (26) and in vivo in animal models (21) following selection by teicoplanin, which have resulted in three phenotypic classes (constitutive, teicoplanin-inducible, or heterogeneous expression of the resistance genes) due to three types of alterations of VanSB function. Mutations leading to teicoplanin resistance also confer low-level resistance to the glycopeptide oritavancine (LY333328) (16). Derivatives of VanB-type strains that are resistant to teicoplanin have been isolated from two patients following treatment with vancomycin (103) or teicoplanin (125), but the isolates were not studied further.
(i) Inducible phenotype.
Substitutions in the sensor domain of VanSB lead to inducible expression of resistance by vancomycin and teicoplanin (Fig. 5) (26). A minority of the mutations are located between the two putative transmembrane segments of VanSB. This portion of the sensor is located at the outer surface of the membrane and may therefore interact directly with ligands, such as glycopeptides, which do not penetrate into the cytoplasm. The majority of the substitutions are located in the linker that connects the membrane-associated domain to the cytoplasmic catalytic domain. The N-terminal domain of VanSB is thus involved in signal recognition and is associated with alterations of specificity that allow induction by teicoplanin but not by the nonglycopeptide moenomycin, which also inhibits the transglycosylation reaction (13, 25).
FIG. 5.
Schematic representation of the VanSB sensor and location of amino acid substitutions in teicoplanin-resistant mutants. H, N, G1, F, and G2 refer to the motifs conserved in histidine protein kinases and are shown as hatched boxes. The putative membrane-associated sensor domain (dotted blue) containing transmembrane segments (blue) and the putative cytoplasmic kinase domain (white) are indicated. Het, heterogeneously resistant; R, resistant; S, sensitive; Te, teicoplanin; Vm, vancomycin.
VanS and VanSB may sense the presence of glycopeptides by different mechanisms. VanA-type resistance is inducible by glycopeptides, moenomycin, and other antibiotics that inhibit the transglycosylation reaction but not by drugs that inhibit the reactions preceding (such as ramoplanin) or following (such as bacitracin and penicillin G) transglycosylation (25, 100). This narrow specificity suggests that the accumulation of lipid intermediate II, resulting from the inhibition of transglycosylation, may be the signal recognized by the VanS sensor. This would account for the induction by antibiotics that inhibit the same step of peptidoglycan synthesis but have different structures and modes of action. However, there are conflicting results in relation to antibiotics that can act as inducers, possibly resulting from using some of them at much higher concentrations than those inhibiting cell growth (260). In particular, bacitracin (6, 132) and ramoplanin (90) have been reported to induce vancomycin resistance, and although its mode of action remains somewhat controversial (260), it has recently been proposed that ramoplanin acts at the transglycosylation step (260). In contrast, the VanSB sensor may interact directly with vancomycin, since teicoplanin is not an inducer.
(ii) Constitutive phenotype.
In the VanS-type sensors, five blocks (H, N, G1, F, and G2) of the kinase domain are highly conserved (Fig. 5). The H block is responsible for both autophosphorylation and kinase/phosphatase activities, and G1 and G2 correspond to ATP binding blocks. Mutations responsible for constitutive expression of the vanB cluster result from amino acid substitutions at two specific positions located on either side of the histidine at position 233, which is the putative autophosphorylation site in VanSB (Fig. 5) (26). Constitutive expression of glycopeptide resistance is most probably due to impaired dephosphorylation of VanRB by VanSB, as similar substitutions affecting homologous residues of related sensor kinases impair the phosphatase but not the kinase activity of the proteins (26, 65). These observations confirm that dephosphorylation of VanRB is required to prevent the transcription of the resistance genes (13).
A VanB-type Enterococcus faecium strain that was resistant to vancomycin and susceptible to teicoplanin was isolated from a patient, and 2 weeks later, a derivative that was constitutively resistant to high levels of both glycopeptides was isolated from the same patient (65). Increased resistance in the derivative was shown to be due to the combination of a frameshift mutation leading to the loss of the Ddl ligase activity and the constitutive synthesis of pentadepsipeptide precursors by the loss of VanSB phosphatase activity following a six-amino-acid deletion, which partially overlaps the conserved G2 ATP-binding domain (Fig. 5) (65).
(iii) Heterogeneous phenotype.
The heterogeneously resistant derivatives most probably harbor null alleles of vanSB since the mutations introduce translation termination codons at various positions in the gene (Fig. 5) (27). The antibiotic disk diffusion assay revealed the presence of inhibition zones containing scattered colonies of resistant bacteria that grew predominantly in 48 h (21, 27).
Resistance to Glycopeptides in Staphylococcus aureus
Some of the genes regulated by the VraSR two-component system in S. aureus are associated with cell wall biosynthesis, including murZ, for the production of murein monomer precursors, and pbp2, sgtA, and sgtB, for the polymerization of peptidoglycan (131). The production of VraSR is induced by the exposure of S. aureus to antibiotics that affect cell wall synthesis, such as glycopeptides, β-lactams, bacitracin, and d-cycloserine, suggesting that the VraS sensor kinase responds to damage or the inhibition of cell wall biosynthesis (131). Additionally, the vraSR null mutants derived from methicillin-resistant S. aureus isolates show reduced transcription of murZ and pbp2, which correlates with a significant decrease in resistance to teicoplanin, β-lactams, bacitracin, and fosfomycin but not to d-cycloserine and levofloxacin. Overexpression of the VraR response regulator confers a low level of resistance to vancomycin. These observations indicate that VraSR constitutes a positive regulator of peptidoglycan synthesis that is involved in the expression of resistance to certain cell wall inhibitors in S. aureus.
The overproduction of PBP2 significantly increases resistance to teicoplanin, whereas the reduction in teicoplanin resistance is observed in vraSR null mutants, which agrees well with a loss of PBP2 induction (97). PBP2 possesses transglycosylase activity that catalyzes the elongation of the nascent peptidoglycan chains (195). However, elongation of the chains is not completely abolished after the inactivation of the transglycosylase domain of PBP2, indicating that other transglycosylases also catalyze the elongation reaction. The VraSR system positively regulates the sgtA and sgtB glycosyltransferase genes. The deduced proteins show significant similarity with transglycosylase domains and, consequently, may be involved in glycopeptide resistance in S. aureus (109). It is considered that increased transglycosylase activity contributes to resistance either by competing with glycopeptides for the capture of the membrane-bound murein monomers or by increasing the production of nascent peptidoglycan chains to provide more d-Ala-d-Ala that serves as a false target for vancomycin. High copy numbers of the vraSR genes do not increase the transcription of pbp2 and sgtB and require the presence of cell wall synthesis inhibitors to induce the expression of the genes (131). This indicates that the signal that activates the VraS sensor kinase could be generated by the inhibition of cell wall synthesis.
Resistance to β-Lactams in Enterococcus faecalis
E. faecalis produces a low-affinity penicillin-binding protein (PBP5) that mediates high-level resistance to cephalosporins. A regulatory system, designated CroRS for ceftriaxone resistance, is essential for this intrinsic resistance (56). Deletion of croRS leads to a 4,000-fold reduction in the MIC of expanded-spectrum cephalosporins such as ceftriaxone. The CroS kinase autophosphorylates and transfers its phosphate to the CroR response regulator. The croR and croS genes are cotranscribed from a promoter (croRp) located upstream from croR. CroRS is induced in response to β-lactams and inhibitors of early and late steps of peptidoglycan synthesis, indicating that this system does not respond to the inhibition of a specific biosynthetic step (56). The croRS null mutant produces PBP5, and the expression of an additional copy of pbp5 under the control of a heterologous promoter does not restore ceftriaxone resistance (56). Deletion of croRS is not associated with any defect in the synthesis of the UDP-MurNAc-pentapeptide precursor or of the d-Ala4→l-Ala-l-Ala-Lys3 peptidoglycan cross-bridge. Thus, the CroRS two-component regulatory system is essential for β-lactam resistance mediated by PBP5 in enterococci. However, CroRS is not required for the production of low-affinity PBP5, suggesting that it controls other, as-yet-unidentified, factors essential for the activity of this low-affinity penicillin binding protein.
Recently, to gain a more comprehensive view of the role of two-component signal transduction pathways in the biology of E. faecalis, each of the 18 response regulators previously identified in E. faecalis V583 was targeted by insertion mutagenesis (99). An insertion in croR led to susceptibility to the cephalosporins, bacitracin, and vancomycin despite the presence of a functional vanB operon in strain V583. CroR is thus involved in resistance to a wide range of cell wall-active agents, indicating that this system may have a role in the regulation of cell wall synthesis.
Resistance by Efflux
Drug resistance among gram-negative bacilli such as Escherichia coli and Pseudomonas aeruginosa and gram-positive cocci such as S. aureus, Staphylococcus epidermidis, other coagulase-negative staphylococci, E. faecalis, E. faecium, and Streptococcus pneumoniae complicates the therapy of infections caused by these microorganisms. An important component of this resistance is the activity of membrane-based efflux proteins commonly referred to as “pumps” (205). The function of these efflux pumps is to export molecules through the bacterial envelope, thus limiting the intracellular accumulation of toxic compounds such as antibiotics. This pumping out is energized by ATP hydrolysis or by an ion antiport mechanism (144, 205). Efflux decreases the antibacterial efficacy of structurally unrelated drug classes and has been shown to be responsible for species- or genus-specific intrinsic or “natural” resistance to antibiotics. If the pump is overproduced, it can be responsible for extended cross-resistance, since it confers, by a single mechanism, resistance to various drug classes.
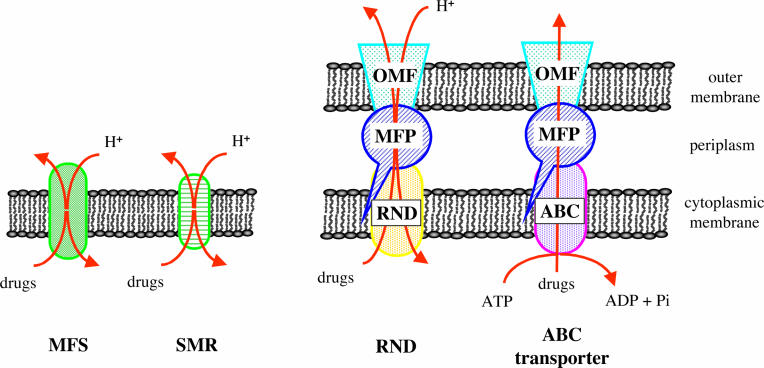
The envelope of gram-negative bacteria comprises two membranes, the inner or cytoplasmic membrane and the outer membrane, which are separated by the periplasmic space, whereas gram-positive bacteria possess a single membrane. The membrane-located transporters can be grouped into the following five families based on sequence homology, mechanisms, and molecular characteristics: the ATP binding cassette (ABC) family, the major facilitator superfamily (MFS), the multidrug and toxin extrusion family, the resistance-nodulation-division (RND) family, and the small multidrug resistance (SMR) family (Fig. 6). In gram-negative bacteria, the efflux machinery is complex, comprising a cytoplasmic membrane-located transporter, a periplasmic membrane adaptor protein, and an outer membrane channel protein. Genomes of gram-negative bacteria usually encode multiple members of each family of multidrug transporters (192). To date, only the ABC, MFS, and SMR families have been described in gram-positive organisms.
FIG. 6.
Schematic representation of the cell membranes with examples of multidrug efflux systems. ABC, ATP binding cassette; MFP, membrane fusion protein; MFS, major facilitator superfamily; OM, outer membrane; OMF, outer membrane factor; RND, resistance nodulation cell division; SMR, small multidrug resistance.
Generally, drug-specific efflux pumps tend to be encoded by plasmids and are thus transmissible, whereas MDR efflux pumps are usually specified by the chromosome (191, 210). The expression of plasmid-borne genes is often sufficient to confer resistance without the need for additional mutations owing to the multicopy state of these genetic elements. However, drug resistance due to chromosomally encoded MDR pump genes most often occurs because of increased gene expression, which can take place as a consequence of substrate-induced transcriptional activation, gene amplification, or the occurrence of regulatory mutations that, in certain instances, confer only low-level resistance to the host (91).
Resistance to quinolones in Staphylococcus aureus.
NorA was the first chromosomally encoded S. aureus pump to be identified. Based on its sequence, the cloned norA gene of a fluoroquinolone-resistant clinical strain was predicted to encode a typical MFS-type protein with 12 membrane-spanning alpha helices. NorA has the highest degree of identity with the Bmr MFS pump of Bacillus subtilis (44%) and only 20 to 25% identity with several tetracycline-specific efflux proteins of gram-negative bacteria (121). Cloning of norA in a plasmid in either S. aureus or E. coli results in fluoroquinolone resistance, particularly to hydrophilic molecules. NorA has a broad substrate specificity, including hydrophilic fluoroquinolones, biocides, and dyes. In addition, the substrates of NorA are typical of those of MDR pumps, namely, amphipathic cations. NorA activity is inhibited by reserpine, a compound known to act as an inhibitor of the function of many MDR efflux proteins. Resistance associated with NorA occurs only when the structural gene for this protein is either amplified or overexpressed as a result of regulatory mutations (121).
Regulation of NorA expression depends on at least two systems, ArlRS and MgrA (formerly NorR) (83, 84, 255). MgrA is composed of 147 residues, has modest similarity with other regulatory proteins such as MarR in E. coli and SarR in S. aureus, and, when overexpressed, causes increased expression of norA. It binds upstream from the norA promoter, and experimental data suggest that repeats of the TTAATT consensus sequence may be involved in the binding of this protein (255). Four such hexamers are located upstream from the −35 motif of the norA promoter. MgrA is not a specific regulator of norA expression but, rather, is a global regulator, since it also regulates autolytic activity and the expression of several virulence factors, including alpha toxin, nuclease, and protein A (153). MgrA is transcribed from two promoters, positively regulates its own expression, and acts at the transcriptional level to enhance the expression of numerous genes. Recently, two novel efflux transporters, NorB and Tet38, that confer resistance to multiple drugs including quinolones and tetracycline, respectively, have been shown to be negatively regulated by MgrA (254).
The ArlR-ArlS two-component regulatory system is involved in adhesion, autolysis, and extracellular proteolytic activity of S. aureus (85). The binding of MgrA to the norA promoter is modified in a strain with a disrupted arlS such that increased norA expression is observed (83, 84, 255). Overexpression of mgrA in a strain producing the ArlS sensor results in increased transcription of norA and reduced susceptibility to various NorA substrates. These data suggest that a mutation in arlS increases the effect of MgrA on the norA promoter and that wild-type levels of MgrA have little effect on norA expression. Highly fluoroquinolone-resistant strains of S. aureus in which norA expression is enhanced in the absence of any modification in arlR-arlS or change in mgrA expression have been reported, indicating that other loci must be involved in the regulation of norA expression.
Resistance to multiple drugs in gram-negative bacteria.
The synthesis of the tripartite efflux systems of gram-negative bacteria (Fig. 6) depends on regulatory genes, implying individual control and thus distinct functions in the cell (180). Two-component systems are not commonly involved in the regulation of drug efflux transporters, although such systems have recently been associated with RND-type pumps, such as AdeABC in Acinetobacter baumannii (154), SmeABC in Stenotrophomonas maltophilia (145), and MdtABC in E. coli (28).
Intrinsic resistance of gram-negative bacteria is due to multidrug efflux by RND pumps that are widely distributed and act in synergy with the outer membrane barrier. The wide substrate range of these transporters often includes β-lactams and aminoglycosides, which are rarely subjected to efflux by other pump classes. RND transporters form a multiprotein complex with members of the outer membrane factor family and of the periplasmic linker membrane fusion protein family. These complexes allow the excretion of drugs directly into the medium. Chromosomally encoded multidrug RND efflux systems appear to be most important for resistance to antimicrobials in P. aeruginosa and other gram-negative pathogens.
(i) Acinetobacter baumannii.
A. baumannii is one of the predominant bacteria associated with outbreaks of nosocomial infections that are often very difficult to treat because of the frequent resistance of this species to multiple antibiotics. Aminoglycosides can be used successfully in combination with a β-lactam, and combinations of a β-lactam with either a fluoroquinolone or rifampin have also been proposed. Partial resistance of A. baumannii to β-lactams is due to the synthesis of a species-specific cephalosporinase (258).
The chromosomally encoded three-component AdeABC pump in A. baumannii is composed of the membrane fusion homolog AdeA, the RND superfamily member AdeB with 12 transmembrane segments, and AdeC an outer membrane protein similar to OprM of P. aeruginosa (154). Insertional inactivation of adeB indicates that the corresponding protein is responsible for resistance not only to aminoglycosides but also to fluoroquinolones, tetracycline, chloramphenicol, erythromycin, and trimethoprim. Thus, this efflux pump recognizes a wide spectrum of substrates including hydrophobic, amphiphilic, and hydrophilic molecules, which can be either positively charged or neutral. When the adeC gene is inactivated, resistance to the various substrates of the AdeABC pump is unaltered (161), suggesting that AdeAB can utilize another outer membrane constituent, as already observed for MexXY from P. aeruginosa (see below).
The expression of multidrug transporters is commonly controlled by specific regulatory proteins. Their structural genes are most often adjacent to those encoding the efflux system. The adeABC genes are cotranscribed and adjacent to the adeS and adeR genes that are transcribed in the opposite direction and encode a sensor and a regulator, respectively (Fig. 7) (161). Inactivation of adeS leads to aminoglycoside susceptibility, indicating that this gene is required for the expression of the adeABC operon. Spontaneous aminoglycoside-resistant derivatives that have mutations in the AdeS sensor or in the AdeR regulator can be obtained in vitro. The T153M substitution in AdeS, downstream from histidine 149, the putative site of autophosphorylation, is presumably responsible for the loss of phosphatase activity of the sensor, as observed for EnvZ (T247R), PhoR (T220N), and VanSB (T237K). In AdeR, the P116L mutation at the first residue of the α5 helix of the receiver domain is involved in interactions that control the output domain of response regulators. These mutations result in the constitutive expression of the AdeABC pump, which is otherwise cryptic in wild-type A. baumannii due to stringent control by AdeRS.
FIG. 7.
Genetic organization of the adeRS-adeABC operon from A. baumannii, the smeRS-smeABC operon from S. maltophilia, and the mexR-mexAB-oprM, mexT-mexEF-oprN, nfxB-mexCD-oprJ, and mexZ-mexXY MDR operons from P. aeruginosa. Purple arrows, structural genes for drug efflux complexes; red arrows, regulatory genes that either repress (−) or activate (+) gene expression (this still has to be confirmed for mexZ).
(ii) Stenotrophomonas maltophilia.
S. maltophilia is an aerobic, nonfermentative, gram-negative bacterium, broadly distributed in nature, that has emerged as an important nosocomial pathogen. This species is characterized by high-level intrinsic resistance to a variety of structurally unrelated antimicrobials, which is partly attributable to limited outer membrane permeability combined with antibiotic efflux (145).
The SmeABC multidrug efflux system, a homolog of the mexAB-oprM efflux operon of P. aeruginosa (see below), is regulated by the SmeSR two-component system (Fig. 7) (145). A strain in which the smeABC genes are overexpressed displays resistance to aminoglycosides, β-lactams, and the fluoroquinolones. Deletions in smeC but not in smeB decrease resistance, suggesting that SmeC only, which possesses its own promoter, contributes to multidrug resistance. Thus, SmeABC does not function as a multidrug efflux system, but it rather appears that SmeC plays a role in antimicrobial resistance independently of SmeAB, possibly as the outer membrane factor component of another unidentified multidrug efflux system (145).
As has been observed for the AdeABC system of A. baumannii, two genes, smeR and smeS, upstream from the smeABC operon and transcribed in the opposite direction, encode a regulatory system composed of a sensor (SmeS) and a regulator (SmeR) (Fig. 7) (145). SmeR positively regulates both smeABC and its own smeSR operon.
(iii) Pseudomonas aeruginosa.
P. aeruginosa is a ubiquitous aerobic gram-negative opportunistic pathogen and one of the most common causes of nosocomial infections. Treatment of P. aeruginosa infections is complicated by the intrinsic resistance of this organism to many antimicrobial agents, which results from the synergistic activity of the outer membrane barrier with that of various broad-substrate-range multidrug efflux systems. In addition to intrinsic resistance, multidrug efflux (Mex) systems promote acquired resistance by overexpression of the structural genes for the pumps following mutational events.
Six RND efflux systems in P. aeruginosa have been characterized (Table 1) (4, 5, 50, 111, 129, 206, 207). The efflux operons each encode an inner membrane RND transporter (MexB, MexD, MexF, MexX, MexK, or MexI), a periplasmic membrane fusion protein (MexA, MexC, MexE, MexY, MexJ, or MexH), and, in certain cases, an outer membrane channel protein (OprM, OprJ, OprN, or OpmD). All these RND operons are similar in their genetic organizations but not with respect to regulation, and the corresponding pumps differ in their substrate specificities (Fig. 7 and Table 1). The antibiotic substrate spectrums of these systems are very wide (Table 1). MexAB-OprM, which exhibits an extraordinarily broad substrate range, is constitutively produced in wild-type bacteria and plays a major role in the intrinsic resistance of P. aeruginosa (Table 1) (128). The MexCD-OprJ, MexEF-OprN, and MexJK-OprM systems are not expressed in wild-type P. aeruginosa (50, 129, 206). Expression of many RND multidrug pumps is controlled by local regulators (Table 1), mostly repressors (Fig. 7). With the exception of MexAB-OprM, the expression of most of these efflux systems is tightly regulated.
TABLE 1.
Substrate profiles and regulatory components of Pseudomonas aeruginosa efflux pumps
| Efflux pump | Regulator(s) | Substratesa | Reference(s) |
|---|---|---|---|
| MexAB-OprM | MexR | β-Lactams (except imipenem), fluoroquinolones, tetracycline, macrolides, chloramphenicol, novobiocin, trimethoprim | 76, 207 |
| MexCD-OprJ | NfxB | β-Lactams (except imipenem), fluoroquinolones, tetracycline, macrolides, chloramphenicol, novobiocin, trimethoprim | 206, 235 |
| MexEF-OprN | MexT | Fluoroquinolones, chloramphenicol, trimethoprim | 127, 129 |
| MexJK-OprM | MexL | Tetracycline, erythromycin | 49, 50 |
| MexXY-OprM | MexZ | Fluoroquinolones, aminoglycosides, tetracycline, macrolides | 5, 111, 165 |
| MexGHI-OpmD | LasR (?), RhlR (?) | Tetracycline, netilmicin, ticarcillin + clavulanic acid | 4 |
The list of substrates is limited to antibiotics.
The mexR and other regulatory genes, nfxB (206, 235), mexZ (166), and mexL (49), encode negative regulators (Table 1) (Fig. 7), and mutations in these genes lead to the overexpression of the mexAB-oprM, mexCD-oprJ, mexXY, and mexJK operons, respectively. MexR (76), NfxB (235), MexZ (166), and MexL (49) have been purified and shown to bind to DNA upstream from mexA, mexC, mexX, and mexJ, respectively. The mexEF-oprN operon is positively regulated by the mexT product, a transcriptional activator of the LysR family (127). Certain clinical isolates can broaden their drug resistance phenotypes by coexpressing MexAB-OprM and MexXY following mutations in multiple regulatory genes (151).
(iv) Escherichia coli.
Certain multidrug efflux pumps in E. coli are regulated by two-component systems. BaeSR is involved in the expression of the RND transporter MdtABCD that pumps out novobiocin and deoxycholate (28, 178). The baeS and baeR genes are immediately downstream from the mdtABCD genes and together probably form an operon.
BaeR and BaeS exhibit in vitro phosphotransfer in the presence of ATP (28), but the nature of the stimulus recognized by the BaeS sensor is not known. The BaeR response regulator binds to the mdtA promoter, and its overexpression strongly stimulates the transcription of the mdtABCD gene cluster, leading to an increase in resistance to novobiocin and deoxycholate. The presence of the BaeS sensor kinase is not required for the full activity of overexpressed BaeR in intact cells. BaeR could be phosphorylated by other sensor kinases present in E. coli, since such cross talk occurs particularly when one of the noncognate partners is present in excess. Cross-regulation has been observed between the various two-component regulatory systems, BaeSR, PhoBR, which is implicated in phosphate metabolism, and CreBC, which is implicated in carbon and energy metabolism (181).
Many of the two-component signal transduction systems in E. coli control the expression of multiple target genes. BaeR modulates the expression of mdtABCD but also that of acrD, which encodes a multidrug exporter system conferring resistance to β-lactams and novobiocin (108).
ROLE OF IS ELEMENTS AND INTEGRONS IN THE MODULATION OF RESISTANCE GENE EXPRESSION
Besides the considerable impact that they have on the mobility and spread of antibiotic resistance genes when they make up composite transposons (31, 81, 146, 219), insertion sequences (ISs) as single elements may also exert noticeable effects on the expression of these genes either directly, by influencing the level of their transcription, or in various ways indirectly, by affecting genes involved in their regulation or in the modulation of resistance levels. Together with the integrons, which are natural expression vectors with the capacity to capture resistance genes (95, 227), they constitute two groups of genetic elements with the potential to contribute much to high-level and multiple-antibiotic resistance in clinical isolates.
Effects of IS Elements on the Expression of Resistance
General characteristics of IS elements.
Insertion sequence elements are small transposable genetic elements, with a size generally between 0.8 and 2.5 kb and encoding only those functions required for their transposition. Currently, approximately 1,000 IS elements have been identified in some 200 gram-negative and gram-positive bacterial species and in archaea and are assigned to 19 families based on theirstructural and functional characteristics (31, 46) (http://www-IS.biotoul.fr).
IS elements may be present in one or several copies and localized on the chromosome, on plasmids, or on both and must reside on conjugative elements for intercellular transfer. Many transpose readily, and others, such as IS200, transpose rarely (32). There is great variability in the distribution of the IS elements of the different families among bacterial species, with some of them restricted to few hosts, such as IS6110, which has been found only in mycobacteria of the tuberculosis complex (156, 250).
IS elements are typically bounded by short repeat sequences of up to ca. 40 bp in an indirect orientation. These inverted repeats are specific for each element, and their presence and integrity are required for transposition, which may or may not be site specific. Upon insertion into the target DNA, a repeat sequence, 2 to 14 bp in length and characteristic for each element, is generated in a direct orientation (Fig. 8). Many elements carry a single, transposase-encoding open reading frame (ORF) covering most of the element, while others carry several ORFs, on a single strand or on both strands, the products of which may also play a role in the regulation of the transposition process. Of particular interest in the present context, IS elements may contain partial or complete promoters, often located at their extremities and in an outward orientation and capable of activating the expression of neighboring genes (Fig. 8) (46, 155).
FIG. 8.
Characteristics of IS elements. DR, direct repeat; IR, inverted repeat; −35/−10 and −35, approximate locations of promoter consensus sequences.
IS-mediated effects on resistance-conferring and resistance-modulating genes.
With respect to IS-mediated effects on antibiotic resistance genes in the strict sense, i.e., genes responsible for drug-specific resistance mechanisms such as antibiotic inactivation, drug target alteration, or specific efflux pump production, gene activation through promoter alteration is the rule. In contrast, insertional inactivation is the predominant effect of IS elements on genes involved in the modulation of resistance levels (which may or may not encode resistance gene repressors), such as ampD, mexR, acrR, nfxB, ompC, ompK36, oprD, and carO in gram-negative bacteria and mecR/I, tcaA, and vanSD in gram-positive cocci (Table 2).
TABLE 2.
IS elements affecting genes conferring or modulating resistance to antibiotics
| Mechanism | Element(s) | Gene(s) affected | Relevant R phenotype(s)a (fold increase)b | Occurrencec
|
Species | Reference(s) | |
|---|---|---|---|---|---|---|---|
| Natl. | Exptl. | ||||||
| Resistance gene activation (hybrid promoter) | IS1 | blaTEM-1 | Amp | + | E. coli | 209 | |
| IS1-like | blaTEM-6 | Caz, Azt (10 P) | + | Enterobacteriaceae | 89 | ||
| IS2 | ampC | Amp (20 P) | + | E. coli | 118 | ||
| acrEF | FQ (∼10) | + | E. coli | 119 | |||
| IS18 | aac(6′)-Ij | Ami | + | Acinetobacter | 229 | ||
| IS26 | aphA7, blaS2A | Kan, Ctx | + | K. pneumoniae | 137 | ||
| blaSHV-2a | Caz, Ctx, Azt | + | P. aeruginosa | 177 | |||
| IS140 (IS26) | aac(3)-IV-aph(4)d | Gen, Hyg | + | Salmonella sp. | 39 | ||
| IS256 | mecA | Met (8-100) | + | + | S. sciuri | 59 | |
| llm | Met (4-16) | + | S. aureus | 158 | |||
| IS257 | dfrA | Tmp | + | S. aureus | 138 | ||
| tetA(K) | Tet | + | S. aureus | 237 | |||
| IS1224 | cepA | Amp | + | B. fragilis | 222 | ||
| Resistance gene activation (complete promoter) | IS257 | tetA(K) | Tet | + | S. aureus | 236 | |
| IS612, IS613, IS614, IS615, IS942, IS943, IS1186, IS1187, IS1188, IS4351 | cfiA | Imi, Mer | + | + | B. fragilis | 124, 197, 198, 199, 239, 262 | |
| IS642, IS1168, IS1169, IS1170 | nimA, nimB, nimC, nimD, nimE | Mtz | + | B. fragilis | 94, 240, 253 | ||
| IS1999 | blaVEB-1 | Caz, Ctx, Azt (1.6 SA) | + | P. aeruginosa | 22 | ||
| oxa-48 | Imi (∼30) | + | K. pneumoniae | 202 | |||
| IS4351 | ermF/S | Ery | + | B. fragilis | 213 | ||
| ISAba1 | ampC | Caz, Tic | + | A. baumannii | 57, 105, 234 | ||
| ISEcp1 | blaCTX-M-15 | Ctx, Atz | + | Enterobacteriaceae | 122 | ||
| blaCTX-M-17 | Ctx, Atz | + | K. pneumoniae | 41 | |||
| ISEcp1B | blaCTX-M-19 | Ctx, Atz | + | K. pneumoniae | 201 | ||
| ISPa12 | blaPER-1 | Caz, Ctx, Azt | + | S. enterica serovar Typhimurium | 200 | ||
| P. aeruginosa | |||||||
| Gene disruption | IS1 | ampD | Pen | + | E. coli | 148 | |
| IS1, IS5, IS26, IS903 | ompK36 | Cfx | + | K. pneumoniae | 107 | ||
| IS5-like, IS102 | ompK36 | Cfx | + | K. pneumoniae | 107 | ||
| IS26 | ompK36 | Imi, Mer | + | K. pneumoniae | 172 | ||
| IS17 | aac(6′)-Ig | AG Sf | + | Acinetobacter | 228 | ||
| IS21 | mexR | Tic, Azt | + | P. aeruginosa | 37 | ||
| IS186 | acrR | FQ (∼30) | + | E. coli | 119 | ||
| IS256 | tcaA | Tei (5-8) | + | S. aureus | 157 | ||
| Van (2) | |||||||
| IS431 | mecI, mecRI | Met (8-32) | + | S. haemolyticus | 123 | ||
| IS1669 | ampD | Caz (64-400) | + | P. aeruginosa | 24 | ||
| IS6110 | pncA | Pyr | + | M. tuberculosis | 139 | ||
| ISAba825, ISAba125 | carO | Imi, Mer (16) | + | A. baumannii | 175 | ||
| ISEfa4 | vanSD | Van constitutive | + | E. faecium | 67 | ||
| ISEfm1/IS19 | ddl | Van constitutive | + | E. faecium | 38, 193 | ||
| ISPa1328, ISPa1635 | oprD | Imi, Mer | + | P. aeruginosa | 268 | ||
| IS NNe | nfxB | Tet, Tig | + | P. aeruginosa | 61 | ||
| IS NN | cmlA, oxa-10 | Cmp, Tic | + | K. pneumoniae | 257 | ||
Abbreviations: R, resistance; Ami, amikacin; Amp, ampicillin; Azt, aztreonam; Caz, ceftazidime; Cfx, cefoxitine; Cmp, chloramphenicol; Ctx, cefotaxime; Ery, erythromycin; FQ, fluoroquinolones; Gen, gentamicin; Hyg, hygromycin; Imi, imipenem; Kan, kanamycin; Mer, meropenem; Met, methicillin; Mtz, metronidazole; Pen, penicillin; Pyr, pyrazinamide; Tei, teicoplanin; Tet, tetracycline; Tic, ticarcillin; Tig, tigecycline; Tmp, trimethoprim; Van, vancomycin.
Numbers in parentheses refer to the increase in MIC (n-fold); numbers followed by “P” indicate changes in promoter strength; numbers followed by “SA” indicate changes in specific β-lactamase activity.
Natl., insertion observed in clinical isolates; Exptl., spontaneous insertion observed under experimental conditions.
The two genes are organized in a transcriptional unit.
NN, IS element not named.
AG S, susceptibility to aminoglycosides.
Altered expression of resistance-conferring or resistance-modulating genes, consisting in some cases of the activation of silent genes, has been described as a consequence of events mediated by over 20 distinct IS elements belonging to at least 10 families (Table 2) (http://www-IS.biotoul.fr). In one way or another, these elements may have a bearing on the efficiency of resistance mechanisms concerning antibiotics of most classes in clinical use, including the β-lactams, aminoglycosides, quinolones, glycopeptides, imidazoles, and tetracyclines, most often affording an increase in resistance levels. Events of this type have been described for members of many groups of bacteria encountered in the clinical setting, including the Enterobacteriaceae, strict aerobic and anaerobic gram-negative bacteria, staphylococci, and enterococci (Table 2).
(i) Activation of resistance genes by promoter alterations.
The molecular mechanisms responsible for altered, IS-mediated expression are not specific for resistance genes. Transcriptional activation may result from IS insertion into a region carrying a weak, an incomplete, or no promoter. Therefore, a hybrid promoter with an alternative or new IS-borne −35 region may be generated, or a complete IS-borne promoter containing both the −35 and the −10 regions may be acquired (Fig. 8). With few exceptions (see below), these two regions conform to the canonical consensus sequences TTGACA and TATAAT, respectively, with a spacing distance of 17 bp for optimal promoter activity as determined for E. coli (149).
(a) Resistance gene activation by IS-mediated formation of hybrid promoters. An IS-mediated rearrangement of the promoter region of the ampC gene of E. coli was shown in an experimental setup (118) only shortly before the observation of similar events affecting resistance genes in clinical isolates. It was found that the insertion of IS2, of which E. coli carries five chromosomal copies, into the −10 region of the artificially plasmid-borne ampC gene resulted in concomitant, ca. 20-fold increases in ampC transcription, β-lactamase production, and ampicillin resistance levels. While the −10 region remained unaltered and the −35 region was changed to a sequence with less homology with the consensus sequence than that of the natural ampC promoter, the critical event was concluded to be the change of the spacer region from 16 to 17 bp. Despite the efficiency of this rearrangement in increasing the resistance level and although IS2 belongs to the family that is most widely distributed among bacterial species (156), this element does not seem to have been involved similarly in clinical isolates. Another IS2 insertion, with the creation of a putative hybrid promoter upstream from the efflux pump-encoding acrEF gene and its increased expression in an E. coli laboratory mutant, facilitated the determination of the substrate profile of the pump (119). Probably the first observation of an IS-mediated formation of a hybrid promoter for an antibiotic resistance gene in a clinical isolate was made by Bräu et al. (39) in Salmonella. They found the plasmid-borne aac(3)-IV and aph(4) genes, coding for gentamicin and hygromycin B resistance, respectively, in an operon-like arrangement downstream from IS140 (IS26), which provided the −35 region.
IS-mediated rearrangements of promoters driving the transcription of genes encoding extended-spectrum β-lactamases belonging to several families of the class A or class D enzymes (117) have been observed (Table 2). The IS26 element has been reported to contribute to the formation of a hybrid promoter for a chromosome-borne SHV-2A gene in P. aeruginosa and for a similar, plasmid-borne gene in a resistance operon (downstream from an aminoglycoside 3′-O-phosphotransferase gene) in Klebsiella pneumoniae, with the new −35 region in each case at the optimal distance of 17 bp from the respective resistance gene-specific −10 region (137, 177). The gene of TEM-6, as identified in a ceftazidime-resistant strain of E. coli, acquired a −35 region after the insertion of an IS1-like element into the spacer region of its “natural” promoter, P3, the strength of which was increased by a factor of 10 (89). It was speculated that this element, which was found to be widespread among β-lactamase-producing and non-β-lactamase-producing Enterobacteriaceae, had been derived from IS1 through a substantial deletion of its central region as well as by point mutations in the remainder, which did not affect the −35 region. In a laboratory mutant, the replacement of the −35 region of the same P3 promoter of the TEM-1 gene carried on plasmid pBR322 by a similar IS1-borne region had previously been shown to result in decreased promoter strength, which was considered to be related to a lesser degree of homology between this region and the −35 consensus sequence (209).
In Acinetobacter species 13, aminoglycosideresistance is conferred by the species-specific 6′-N-acetyltransferase-encoding gene, aac(6′)-Ij, which may be expressed at various levels (133). The activation of silent copies of the aac(6′)-Ij gene in this species by the creation of a putative hybrid promoter with an IS18-borne −35 region appears to occur at a low frequency, at least as judged from the in vitro selection of tobramycin-resistant mutants of a susceptible clinical isolate (229).
The IS256 and IS257 elements have a proven role in the activation of resistance gene transcription in staphylococci. IS256 belongs to a large family with members in gram-negative and gram-positive bacteria (http://www-IS.biotoul.fr). It flanks the composite aminoglycoside resistance transposon Tn4001 and related elements and is involved in their dissemination in staphylococci, enterococci, and streptococci (158). IS256 is infrequently observed in the animal commensal species Staphylococcus sciuri, in which two-thirds of the isolates are susceptible to β-lactam antibiotics including methicillin, although they carry a close homolog of the mecA gene, the primary drug resistance determinant in methicillin-resistant S. aureus (58). Analysis of a heterogeneously methicillin-resistant human clinical isolate of S. sciuri (with MICs of methicillin of between 25 and 800 μg/ml as opposed to between 3 and 6 μg/ml for a mecA-positive, IS256-negative control strain) revealed the insertion of an IS256 copy into the upstream region of mecA with the creation of a powerful hybrid promoter. This led to the speculation that the S. sciuri isolate had acquired IS256 in a clinical environment where the activation of mecA had then been selected under drug pressure (59). In that same study, mecA activation in S. sciuri was obtained in vitro, and the −35 region of the hybrid promoter was the same as that previously identified as being responsible for the transcriptional activation of llm, a gene of S. aureus encoding a putatively membrane-associated protein that contributes to methicillin heteroresistance in this species in an as-yet-unknown manner (159).
A variation on the theme of hybrid promoter formation has been found in S. aureus in connection with the IS257-dependent effects on the levels of trimethoprim resistance resulting from the association of a constant, IS-borne −35 region with variable −10 regions upstream from the resistance gene. Trimethoprim resistance in S. aureus occurs at low or high levels (with MICs of 50 to 300 μg/ml or ≥1,000 μg/ml, respectively) and is mediated by the dihydrofolate reductase gene dfrA, which resides in the center of a three-gene operon carried by Tn4003 (or Tn4003-like elements), a composite transposon flanked by three copies of IS257 (138, 225). The promoter of this operon overlaps the right end of the left copy of the element IS257L, with its −10 sequence located in the central region of the transposon and the −35 sequence in the right terminus of IS257L. Low-level resistance was found to be associated with various deletions that extend ca. 10 to 300 bp away from the right end of IS257L and unmask alternative −10 regions differing in sequence or distance to the −35 box, or both, from the corresponding region in the high-level resistance-conferring form of the transposon. Such deletion variants exist in S. aureus as well as in coagulase-negative staphylococci. It is believed that IS257 itself is involved in the generation of the flanking deletions and that the transposon variants that carry them may have established themselves by imposing less strain on the fitness of their hosts while conferring levels of resistance that are still advantageous (138). IS257 has also been found to affect the level of tetA(K)-dependent efflux-mediated tetracycline resistance in S. aureus (237). Analysis of an IS257-flanked cointegrated copy of a tetA(K)-carrying, pT181-like plasmid in the mec region of a methicillin-resistant strain of S. aureus revealed the replacement of the −35 region of tetA(K), in the nonintegrated form of the plasmid, by the more efficient IS-borne counterpart (the same as in IS257L of Tn4003); in addition, the existence of a complete promoter was detected in the right extremity of IS257, which was, however, less powerful than the hybrid promoter. The combined strength of the complete and the hybrid promoter in the cointegrate, compared to that of the single promoter in the autonomous plasmid, was determined to lead to substantially higher levels of tetracycline resistance as well as relative fitness in the presence of tetracycline at low concentrations (237). Apart from affecting the levels of resistance to trimethoprim and tetracycline, as well as methicillin (see below), IS257 has been found to be associated with genes conferring resistance to antibiotics of five additional classes and is suspected to provide hybrid promoters for the aminoglycoside and mupirocin resistance genes aadA and mupA, respectively. In light of the involvement of IS257 in the capture and expression of resistance genes in staphylococci, its impact on the assembly of multiresistance gene clusters has been likened to that of the integrons in gram-negative bacteria (81).
(b) Resistance gene activation by IS-mediated formation of complete promoters. Many IS elements provide complete promoters for resistance genes (Table 2). The contribution of such a promoter by ISEcp1 to the expression of CTX-M-type extended-spectrum β-lactamase genes has been reported in several instances. The suggested promoter for blaCTX-M-15 on the right end of ISEcp1 (122) as well as the suggested mode of ISEcp1-supported gene mobilization by one-ended transposition (241a) have been validated experimentally for blaCTX-M-17 (41) and for blaCTX-M-19 (201, 203). Considering that ISEcp1 or ISEcp1-like elements are present upstream from genes of multiple other CTX-M- and also CMY-type enzymes in various species of Enterobacteriaceae (see references 36, 73, 150, and 203 and references therein), this element may be among those most largely involved in the expression of extended-spectrum β-lactamase genes.
A complete promoter on the left end of IS1999 was suggested to drive the transcription of oxa-48 in an isolate of K. pneumoniae in which the corresponding extended-spectrum class D enzyme, OXA-48, contributed to carbapenem resistance (202). Also, in P. aeruginosa, this same promoter was present upstream of the experimentally determined site of the initiation of transcription of blaVEB-1 (22). In this case, IS1999 (which was found to coexist with blaVEB-1 frequently in P. aeruginosa but rarely in Enterobacteriaceae) and the adjacent β-lactamase gene were located inside a chromosome-borne integron. The IS-borne promoter, which matches the −35 consensus sequence only poorly (at one out of six positions), was shown to slightly increase the efficiency of the integron-specific promoter Pc (see below) by a factor of 1.6. There was no such increase when a second element, IS2000, was inserted between IS1999 and blaVEB-1, an arrangement observed in some ceftazidime-resistant, VEB-1-producing clinical isolates of P. aeruginosa (22).
Two distinct promoters, one complete and one almost complete and with different spacing, have been found on the left end of ISPa12 upstream from the transcriptional start sites of the extended-spectrum β-lactamase gene blaPER-1 in strains of P. aeruginosa and Salmonella enterica serovar Typhimurium, respectively. In the case of the S. enterica serovar Typhimurium strain, the −10 region overlapped the left inverted repeat and the direct repeat of the element (200). This observation would suggest that, depending upon the nucleotide sequence at the site of its insertion, ISPa12 may have the capacity to promote the expression of resistance genes with variable efficiencies.
The expression of the AmpC gene in Acinetobacter baumannii has been found to vary with the absence or presence of ISAba1, or closely related elements, immediately upstream from the gene (57, 234). An outward-directed promoter was identified after mapping of the transcription initiation site (234), and its strength was determined to be approximately 10-fold greater than that found in the absence of the IS element (105). The ISAba1-borne promoter notably accounted for high-level resistance to ceftazidime (57, 105).
In the gram-negative anaerobe Bacteroides fragilis, the species-specific, endogenous cephalosporinase gene cepA is present in over 90% of the members of the species but is expressed at either low or high levels, with ca. 15- to 100-fold differences in the MICs of ampicillin for the two categories (197, 222). These differences have largely been explained by increased levels of cepA transcription in the highly resistant strains due to a modified promoter structure resulting from the insertion, ca. 50 nucleotides upstream from the translational start codon, of IS1224, an IS21-like element (222). In Bacteroides, the consensus sequences of the two promoter regions (−33, TTTG; −7, TANNTTTG) do not conform to those of the corresponding −35 and −10 regions recognized by typical σ70 factors and do not appear to require as strict a spacing (29), a situation in keeping with the existence of a particular primary sigma factor in the Bacteroidetes phylum (259). In all high-level but not in the low-level ampicillin-resistant B. fragilis strains analyzed, a TTTG sequence was present at the cepA-proximal extremity of a remnant of the IS21-like element and observed with appropriate spacing with respect to the TAccTTTG (c, nonconsensus nucleotide) region, thus contributing to the formation of a hybrid promoter (222). With the exception of cepA and the tet genes, most other resistance genes in B. fragilis are efficiently expressed when transcription is driven by complete, IS-borne promoters. This is the case for cfiA, the nim genes, and ermF/S, conferring resistance to the β-lactam antibiotics including the carbapenems, the nitroimidazoles, and the macrolides, lincosamides, and streptogramins B (MLSB), respectively. The activation of a silent cfiA gene by the Bacteroides-type promoter of IS1186 was first demonstrated in vitro (198) and was also later reported to occur similarly (with an IS942/IS1170-related element) in vivo during imipenem therapy (75). In virtually all B. fragilis strains with MICs of imipenem of ≥16 μg/ml, IS insertions of great diversity have been found in a region of less than 100 bp upstream from cfiA involving over a dozen elements, or isoforms thereof, belonging to at least four families (Table 2) (http://www-IS.biotoul.fr). Curiously, these elements may also carry, next to the Bacteroides-type promoter regions, typical −10, −35 sequences (199), which, in the case of IS942 and IS1187, have been shown to drive reporter gene expression in E. coli (D. Vingadassalom, unpublished data). A similar array of IS elements has been found upstream from the nim genes, and, by analogy but without experimental verification, it is assumed that they also contribute to their expression. IS4351 (which may also activate cfiA) provides the promoter for ermF/S carried by the composite transposon Tn4351, but not all strains with ermF/S-mediated macrolide resistance harbor this IS element (197, 213).
(ii) Disruption of resistance-modulating genes.
There is a variety of examples of insertional inactivation by IS elements of genes encoding proteins that modulate, in one way or another, the efficiency of a given resistance mechanism. These proteins include negative regulators of resistance genes in the strict sense or of multidrug efflux pump genes mediating nonspecific resistance. Other resistance-modulating proteins that may be affected are porins, which condition antibiotic influx across the outer membrane in gram-negative bacteria, or rare proteins without a clearly established function.
In gram-negative bacteria, inducible β-lactam resistance due to the production of the cephalosporinase AmpC is controlled by a complex regulatory circuit involving (next to the transcriptional regulator AmpR and the permease AmpG) AmpD, an amidase affecting the intracellular levels of the muropeptide that conditions the regulatory status of AmpR (116). It had been shown previously that impaired AmpD function leads to the derepression of ampC expression. High semiconstitutive ampC expression resulted from the spontaneous insertion of IS1 into ampD of a strain of E. coli into which the ampR and ampC genes from Citrobacter freundii had been introduced (148). A comparable insertion event occurred in ceftazidime-resistant clinical isolates of P. aeruginosa with stably derepressed AmpC production in which IS1669 had disrupted the AmpD gene (24).
Expression of the mecA gene, encoding the low-affinity PBP2a responsible for methicillin resistance in staphylococci, may be connected to the presence of a regulator region upstream that contains mecR1 and mecI, the divergently transcribed genes of a sensor-transducer and a mecA repressor, respectively (171). IS-mediated rearrangements of the regulator region involving IS1272 or IS431 have resulted in the deletion of mecI and various sections of mecR1. As shown in spontaneous mutants selected in the laboratory, these rearrangements may lead to heterogeneous methicillin resistance (123). The particular deletion configurations characterize three of the five classes of the so-called mecA gene complex, which occur with different frequencies in S. aureus or coagulase-negative staphylococci (123, 126).
In P. aeruginosa, expression of the three-component efflux pumps of the RND family is negatively controlled. Repressor gene disruption leads to resistance phenotypes that depend on the substrate specificity of the corresponding pump. Disruption by IS21 of mexR, which controls the expression of the mexAB-oprM operon (241), was found in a clinical ticarcillin- and aztreonam-resistant P. aeruginosa isolate in which eight- and fourfold-higher MICs of the respective drugs were associated with a threefold-higher level of mexA transcripts in comparison with a control strain containing intact mexR (37). The mexCD-oprJ operon, controlled by nfxB, does not appear to be substantially expressed under normal growth conditions. Its expression was triggered, in a mexB/mexXY-deficient mutant subjected to growth in the presence of tigecycline, by the disruption of nfxB by an unnamed IS element of P. aeruginosa, demonstrating the capability of MexCD-OprJ to pump out the minocycline analog and to afford, in this particular genetic background, a 16- to 32-fold increase in the MIC of the compound (61).
Expression of the multidrug efflux pump AcrAB in E. coli is negatively regulated by acrR and is also controlled by the marRAB locus (184). A discrete, ca. 1.5-fold increase in acrB transcription was accompanied by a similar increase in the MICs of fluoroquinolones and β-lactams in a mar deletion mutant in which an insertion of IS186 into acrR had occurred after exposure to ofloxacin (119).
Bacterial susceptibility to antibiotics, notably to β-lactams, can be altered directly by gene disruption when IS elements insert into the structural genes of porins. This was first shown in K. pneumoniae isolates with a disrupted ompK36, the OmpC gene homolog of the species (106). The IS1, IS5, IS26, or IS903 element was found, in nine randomly in vitro-selected cefoxitin-resistant derivatives, at various positions within the plasmid-borne porin gene that had been introduced into an OmpK36-deficient host, while in a collection of cefoxitin-resistant clinical isolates (some of them from patients undergoing therapy), ompK36 was inactivated by an IS5-like element in three isolates and by IS102 in one isolate (107). A similar gene disruption caused by IS26 in a CTX-M-1-producing K. pneumoniae isolate led to carbapenem resistance (172). Porin gene inactivations were previously described as being responsible for carbapenem resistance in clinical isolates of P. aeruginosa and A. baumannii. In P. aeruginosa, carbapenem diffusion through the outer membrane is facilitated by OprD2 (252). In several multiple-drug-resistant isolates of this species, with MICs of imipenem of 16 to 32 μg/ml, the corresponding gene was disrupted at various sites by ISPa1328 or ISPa1635, leading to the absence of OprD production and also to the down-regulation of oprD transcription (268). In A. baumannii, carbapenem resistance may be associated with the loss of an outer membrane protein termed CarO, which has been suggested to be a functional analog of OprD (147, 175). Among a collection of CarO-deficient isolates, the corresponding gene was found to be disrupted in two isolates by ISAba825 or ISAba125, giving credence to the suggested role of CarO in the diffusion of carbapenems (175).
In strains of Enterococcus faecium and S. aureus, IS elements have been shown to influence glycopeptide resistance. In enterococci, acquired resistance of the VanA, VanB, and VanD type depends on the production of peptidoglycan precursors with a d-Ala-d-Lac instead of the d-Ala-d-Ala terminus, which forms a complex with the glycopeptides in susceptible strains (194). While VanA- and VanB-type resistance is inducible, critically requiring the chromosome-encoded ligase Ddl (for d-Ala-d-Ala synthesis) and the resistance operon-encoded regulatory proteins VanR and VanS, VanD-type resistance to vancomycin and teicoplanin in E. faecium is constitutively expressed. In strains with this resistance phenotype, both ddl and vanS have been found to be mutationally altered, entailing the absence of all d-Ala-d-Ala incorporation into the membrane-associated peptidoglycan precursor along with sustained activation of the resistance genes (vanHDDXD) by VanR, which is only slowly dephosphorylated in the absence of VanS. Either gene has been found to be disrupted by an insertion element: ddl by IS19, also called ISEfm1 (38, 193), and vanSD by ISEfa4 (67).
In intermediately glycopeptide-resistant strains of S. aureus, resistance is not specified by a defined set of acquired genes but, rather, is due to the accumulation of mutations in an array of genes controlling mainly cell wall metabolism and composition (33). One of these genes, tcaA, encodes a putative transmembrane protein that might act as a sensor or a signal transducer. Although up-regulated in the presence of teicoplanin, it is the absence of the gene that causes a decrease in glycopeptide susceptibility. Disruption of tcaA by IS256, accompanied by increased glycopeptide resistance, was found in a spontaneous derivative of a glycopeptide-intermediate S. aureus isolate (157).
IS-mediated gene disruption leading to pyrazinamide resistance in Mycobacterium tuberculosis has been reported in one case. In this species, the susceptibility to pyrazinamide is linked to the production of the pncA-encoded enzyme pyrazinamidase, which transforms the drug into a bactericidal derivative (104). Analysis of 19 pyrazinamide-resistant isolates revealed that the absence of pyrazinamide activity in one of them was due to the insertion of IS6110 into pncA and that this insertion had occurred into the preferential 10-bp target site of the element that is present in the gene (139).
To a large extent, the instances of IS effects on resistance gene expression and on resistance levels reviewed here represent observations of individual cases. Few molecular epidemiological studies seem to have been attempted to determine the frequencies at which the identified elements are involved in the resistance process and to quantify their true impact in the clinical setting [which potentially also includes the abolition of resistance, as observed in an aminoglycoside-susceptible strain of Acinetobacter haemolyticus with its species-specific aac(6′)-Ig gene disrupted by IS17 (228) or as suspected in a strain of K. pneumoniae, with its integron cassette-borne cmlA and oxa-10 genes disrupted by two putative IS elements (257)]. Considering the multiplicity of IS elements and the diversity of resistance-related targets into which they have been found to insert spontaneously, under natural or experimental conditions, a larger involvement than is obvious from the individual descriptions would not be surprising, nor would the future description of known or novel elements as being capable of rendering existing mechanisms of resistance more efficient.
Modulation of Resistance Gene Expression in Class 1 Integrons
General characteristics of integrons.
Integrons are genetic elements that are able to capture genes on small mobile elements, called cassettes, in a process of site-specific recombination (95, 214). These elements comprise, as their characteristic features, a recombinase gene (intI), a recombination site (attI), and a promoter (Pc) that drives the expression of the generally promoterless cassette-associated genes (Fig. 9A) (54, 96, 141). The integrase catalyzes the recombination between attI and the cassette-associated recombination site, called the 59-base element (59-be) or attC, at the recombination point that lies between the G and the first T of the core site sequence GTTRRRY(190, 214, 244).
FIG. 9.
Transcriptional control in class 1 integrons. (A) Schematic representation of the integron platform. 5′ CS and 3′ CS, 5′- and 3′-conserved segments, respectively; Pc and P2, promoter regions (see the text); attI1, recombination site; C1 and C2, gene cassettes; 59 be, 59-base-pair elements with possible stem-loop structures. (B) Relative strength of integron-borne promoter variants. aDetermined relative to the strength of the tac promoter, set at 1 (data are from reference 141). bStreptomycin concentration at which 50% of cells plated formed colonies (data are from reference 54). (C) Effects of cassette order on resistance levels. aResistance is conferred to streptomycin (Sm) by aadA2, to gentamicin (Gm) by aacC1, and to kanamycin (Km) by aacA4 (the genes for which a position effect is observed and the corresponding antibiotic concentrations at which 50% of cells plated formed colonies [IC50s] are shown in boldface type). bIC50 data are from reference 54.
Integrons have been assigned to several classes depending upon their intI sequences and subdivided into two categories, the mobilized integrons and the chromosomal integrons (53, 95, 96, 169, 226). Cassettes that mediate antibiotic resistance are typically found in the mobilized integrons, but chromosomal integrons may also provide or capture such cassettes (53, 226). Integrons of class 1 are the most abundant and exist in a great number of gram-negative genera (82, 168). They reside, although not exclusively, on transposons and conjugative plasmids, accounting for their wide distribution and their significant association with a multiresistance phenotype in Enterobacteriaceae (82, 140). The cassettes in this integron class encode a variety of enzymes, aminoglycoside-modifying enzymes, dihydrofolate reductases, β-lactamases, and chloramphenicol acetyltransferases; they may also encode nonenzymatic chloramphenicol resistance. More recently identified cassettes are associated with genes that mediate resistance to rifampin and quinolones and increasingly also with genes encoding extended-spectrum β-lactamases and carbapenemases (82, 88, 136, 200, 203, 208, 261, 264).
Transcriptional control of resistance gene expression in class 1 integrons.
Transcription of cassette-associated resistance genes is controlled twofold. While the transcription of probably all promoterless genes in a cassette array is driven by the Pc promoter, four variants of which exist, the expression of genes in the second position and further downstream is additionally conditioned by putative 59-be-encoded transcription terminators with probably low efficiency (Fig. 9A) (54).
(i) Impact of the integron-borne promoter region.
The strengths of three variants of Pc (which was initially called P1 or Pant) and of a secondary promoter, P2, located 252 to 223 bp and 133 to 107 bp, respectively, from the recombination point, have been measured relative to the strength of the tac promoter or as reflected in antibiotic resistance levels (54, 141). The three variants of Pc, with the sequences TTGACA/TAAACT, TGGACA/TAAGCT, or TGGACA/TAAACT in their −35/−10 regions (separated by a 17-bp spacer), were categorized as “strong,” “weak,” or “hybrid,” respectively, and vary in strength by a factor of approximately 30 (141). The weak form may be associated with the secondary promoter, P2 (TTGTTA/TAAGCT), which is active when carrying a 3-bp insertion in its otherwise 14-bp spacer. When the transcription of aadA2 (or aadA1) was driven by the strong or the weak form of Pc, or by the combination of Pc[weak] and P2, there was good agreement between the relative promoter strengths and the resistance levels (Fig. 9B). Primer extension mapping of the transcription start sites revealed that when the combination of Pc[weak] and P2 is functional, the majority of the transcripts originates at P2, confirming the finding that, in this configuration, P2 contributes 90% of the total promoter activity (54, 141). The combination of Pc[strong] and P2 promoters has been observed recently, but its strength has not been determined (208).
When a BLAST search was carried out for the class 1 integron sequences that cover the distance between the −35 region of Pc and the inner boundary of the 5′-conserved segment and that are identical with the nucleotide sequences specifying any one of the four promoter variants Pc[strong], Pc[weak], Pc[hybrid], and Pc[weak] plus P2, it appears that among more than 100 retrieved sequences, there are quite similar numbers of each variant. Although this information cannot be taken as true molecular epidemiological data, it would suggest that neither promoter variant may be of singular advantage for resistance gene expression. The same search failed to reveal clearly preferential associations between any of the promoter variants and individual cassette-associated genes inserted at attI1, although there may be a tendency for carbapenemase genes of the blaIMP type to occur more frequently downstream from Pc[strong] or Pc[weak] and of the blaVIM type to occur more frequently downstream from Pc[hybrid].
(ii) Impact of the cassette-borne 59-be.
The expression of promoterless, cassette-associated resistance genes is markedly influenced by their position in a cassette array. Using a series of plasmid constructs in which the transcription of the cassette genes is driven from the same promoter (in this case, Pc[strong]), and as exemplified in Fig. 9C, Collis and Hall showed that the resistance level conferred by a given gene is highest when it is promoter proximal and that this level is reduced, by factors of generally between 2 and 5, when a cassette is present upstream (54). It was suggested that this modulation of resistance gene expression occurs essentially at the level of transcription and that it is linked to properties of the 59-be-containing 3′ ends of the cassettes, since these ends were found to coincide roughly with the 3′ ends of the major mRNA transcripts of the cassettes. The possibility was put forward that the 59-base elements, which generally contain inverted repeats, function as transcription terminators (54). Substantial silencing of a downstream gene, as of aac(6′)-Ib downstream from blaIMP-1, may be observed when there is the potential for the formation of a stable stem-loop structure, although in this case, silencing might alternatively, or in addition, be due to poor translation if one considers the presence of a ribosomal binding site of only three nucleotides (8). Full silencing has also been reported, as for oxa-9 downstream from the cmlA-2 cassette in In40 (196).
(iii) Transcription independent of integron-specific sequences.
The expression of some cassette-associated resistance genes is driven by promoters other than Pc. This was first shown for the cmlA cassette of In4, specifying nonenzymatic chloramphenicol resistance, which contains a cluster of three overlapping promoter sequences (34), and later also for several cmlA cassette variants (176, 196, 204). (The control of cmlA expression at the translational level is described below in the section on translational attenuation.) A promoter region upstream from, but apparently not part of, the resistance gene cassette has been reported for the fused oxa-10-aadA1 cassette of In53 (176). In complex class 1 integrons, various resistance genes are found downstream from the orf513-containing common region, CR1, an atypical class of insertion sequence now called ISCR1 (189, 251). An involvement of this region in gene expression could be suspected, e.g., from the observation that blaCTX-M-2 downstream from ISCR1 confers high levels of β-lactam resistance to Enterobacteriaceae (9), while the closely related, chromosome-encoded class A β-lactamase gene, blaKLUA, from which blaCTX-M-2 is speculated to have been derived, confers only low levels of resistance to its host, Kluyvera ascorbata (115). ISCR1-borne promoter sequences have been identified recently for qnrA, blaCTX-M-2, blaCTX-M-9, and dfrA10 in various gram-negative bacteria (160, 220). A comprehensive compilation of the genes associated with ISCR1 has been published in a recent review (251).
Translational control of gene expression in class 1 integrons.
Efficient gene translation normally requires the presence of a translation initiation region (TIR) consisting of the initiation codon, a Shine-Dalgarno (SD) sequence, and an adequate spacer between them. Typically, the initiation codon ATG (or, less frequently, GTG or, rarely, TTG) is separated by ca. 5 to 15 nucleotides from an SD sequence, made up of any four or more nucleotides (but maybe as few as three) within the sequence AAGGAGG(48, 93, 130). While a canonical TIR is found in the majority of the resistance gene cassettes, examination of published sequences reveals that their presence is not the rule, and in some cases, the mechanism of translation initiation remains obscure.
The absence of a TIR in cassettes inserted at attI of class 1 integrons can be compensated for by the coincidence of two circumstances. The first is the (constant) presence of a short ORF, overlapping the inner boundary of the 5′-conserved segment of the integron, which has an SD sequence (GGAG) eight nucleotides upstream from its initiation codon; the second is the (frequent) occurrence of a stop codon, TAG more often than TAA, at positions 3 to 5 (underlined) in theGTTRRRY core site of the cassette-associated 59-base element. Under these circumstances, insertion of the cassette with the formation of the composite attI1 site (190) results in the placement of a stop codon in phase with the reading frame of the short ORF, which then has a coding capacity for a peptide of 11 amino acids and which, for that reason, has been termed ORF-11 (Fig. 10A) (98).
FIG. 10.
Possible functions of ORF-11 in the translation of cassette-associated genes. SD, Shine-Dalgarno sequence. (A) Translational coupling (a, b, and c) (see the text). The position of the stop codon in the recombination core site is underlined. (B) Fusion. (C) Unknown effect of ORF-11.
A role has been assigned to ORF-11 in the initiation of translation of a TIR-deficient, cassette-associated aac(6′)-Ib-type gene conferring resistance to aminoglycosides (98). It was shown that the deletion of ORF-11, with or without maintenance of the 20-bp region connecting its stop codon to the initiation codon of the resistance gene, reduced the efficiency of translation by over 80%, while the deletion of the connecting region alone had no effect (Fig. 11). Considering also that replacement of either the SD sequence or the initiation codon of ORF-11 with noncanonical sequences reduced the translation efficiency by two-thirds, these results were taken to support the role of ORF-11 as a strong enhancer of the initiation of translation of TIR-deficient genes. Whether the residual translational activity of close to 20% that was observed under experimental conditions in the absence of ORF-11 and of any recognizable TIR feature is of relevance in vivo is unclear. Since there was no evidence for a specific function of the ORF-encoded peptide, the translation of the resistance gene would be dependent on the translation of ORF-11 per se and, as such, should be considered as coupled. However, the precise coupling mechanism that operates in this context has not been determined.
FIG. 11.
Relative ORF-11-mediated translation efficiencies. C, control; WT, configuration depicted in Fig. 10A (wild type); Δa, Δb, and Δc, mutants with deletion of fragments a, b, and c as shown in Fig. 10A (data are from reference 98).
Inspection of recently published sequences supports the assumption made previously that the expression of at least one-quarter, and maybe more, of the cassette-associated genes inserted at attI1 profits from the presence of ORF-11 via translational coupling. This process has been found not to be impeded substantially by an artificial increase in distance to up to ca. 50 nucleotides between the stop codon of ORF-11 and the translation initiation codon of the resistance gene, a distance that is rarely exceeded in naturally occurring cassettes (98).
Cassette integration at attI1 has also resulted in the generation of an alternative ORF with its coding capacity extended to 18 amino acids (ORF-18), as first evidenced in the nucleotide sequence of the oxa-1 cassette (186). In this case, the TIR and the first 11 amino acids are identical to those of ORF-11, but the stop codon, separated from the translation initiation codon of the oxacillinase gene by a stretch of 45 nucleotides, is apparently not provided by attC. Not surprisingly, this configuration is the same in a cassette named oxa-30 (oxa-1 and oxa-30 are identical) (245), in an oxa-31 cassette, and in an aac(6′)-Ib cassette, which encodes an acetyltransferase fused to the N-terminal amino acids of OXA-1 (23, 44, 238). ORF-18 promotes translation of the downstream gene in a way similar to that of ORF-11. Its deletion abolished the translation of oxa-1 fully, as measured in terms of β-lactamase activity and ticarcillin resistance, while the introduction of an SD sequence (GGAG) in the deletion mutant upstream from the oxacillinase gene restored enzyme production and β-lactam resistance to their original levels (B. Berçot, unpublished data).
Several cassettes have borrowed the TIR of ORF-11 directly after an in-frame fusion between the resistance gene and the ORF brought about by small sequence duplications or deletions. Such events have occurred in cassettes with associated aminoglycoside 3-N- or 6′-N-acetyltransferase genes in gram-negative bacteria and dihydropteroate synthase genes in Mycobacterium fortuitum and Corynebacterium striatum (Fig. 10B) (163, 173, 267; M. C. Ploy, unpublished data) (GenBank accession number AJ294721). Whether the common presence of ORF-11 and its TIR has any impact on the efficiency of translation of the genes that have their own canonical SD sequence is not clear (Fig. 10C), but the possibility that it augments the recruitment of ribosomes in the vicinity of the initiation codons of these genes could be imagined.
Since the integrons of classes 1, 2, and 3, i.e., those predominantly bearing resistance gene cassettes, possess similar Pc promoters (there is an 18-bp spacer in class 2) at equivalent positions and since the cassettes are considered to be exchangeable between classes (55, 101), it would seem likely that resistance gene transcription in all classes is similarly subject to the mechanisms of control described for class 1 integrons (54, 141). Whether the potential for enhanced translation of cassette-associated genes in class 1 integrons is a trait that has contributed to their predominance over integrons of the other two classes remains a matter of speculation (53, 98).
POSTTRANSCRIPTIONAL (TRANSLATIONAL) ATTENUATION
Control of mRNA translation is a widespread mechanism of regulation in both prokaryotes and eukaryotes that often complements transcriptional regulation. Posttranscriptional (or translational) attenuation controls the inducible expression of several resistance genes, and this mechanism has been particularly well studied for the erm and cat genes, which are responsible for resistance to macrolides by ribosome methylation and inactivation of chloramphenicol by production of a chloramphenicol acetyltransferase, respectively (72, 263). A similar mechanism regulates the inducible cmlA gene that controls permeability to chloramphenicol and is part of class 1 integrons (see above) (243). The regulation of expression of antimicrobial resistance results from an interplay between the ribosome, a short peptide sequence encoded by the mRNA, and the antibiotic. The following paragraphs will focus on the regulation of macrolide resistance by ribosome methylation because of its major clinical significance.
Inducible Expression of Macrolide Resistance
The erm(C) paradigm.
Resistance to macrolides emerged in staphylococci in 1956 soon after the introduction of erythromycin into therapy (45, 87). Biochemical studies indicated that resistance was due to the methylation of the ribosomal target of the antibiotics, which yielded a broad-spectrum cross-resistance to macrolides, lincosamides, and streptogramins B, the so-called MLSB phenotype. Subsequently, the MLSB phenotype was reported in a large number of microorganisms and was found to be encoded by a variety of erm (erythromycin ribosome methylase) genes. Erm methylases make up a family of highly related proteins that use S-adenosylmethionine as a methyl donor to mono- or dimethylate a single adenine residue (A2058 [E. coli numbering]) in nascent 23S rRNA. The A2058 residue is located within a conserved region of domain V of 23S rRNA and plays a key role in the binding of MLSB antibiotics. As a consequence of 23S rRNA methylation, the binding of erythromycin to its target is impaired. Cross-resistance between all macrolides, lincosamides, and streptogramins B (pristinamycin I and quinupristin) occurs because of overlapping binding sites in 23S rRNA (263).
It rapidly appeared that resistance to macrolides occurred with two major phenotypes. Acquisition of an erm(A) or erm(C) determinant in staphylococci yields either dissociated resistance to macrolides with susceptibility to lincosamides or cross-resistance to macrolides and lincosamides, corresponding to inducible and constitutive expression of MLSB resistance, respectively. Inducible expression yields dissociated resistance to macrolides due to differences in the inducing ability of the antibiotics. The genetic basis for induction has been studied in detail in the case of the erm(C) gene of staphylococcal plasmid pE194 (92, 114). Strains harboring erm(C) are resistant to inducer macrolides such as erythromycin and its derivatives (azithromycin, clarithromycin, dirithromycin, and roxithromycin). In contrast, noninducer macrolides such as spiramycin (a 16-membered macrolide), lincosamides (lincomycin and clindamycin), and streptogramins B (pristinamycin I and quinupristin) remain active.
Early studies in the laboratories of B. Weisblum and D. Dubnau showed that induction arises posttranscriptionally according to the model of translation attenuation (92, 114, 263). erm mRNA is synthesized but in an inactive conformation and becomes active only in the presence of inducer macrolides. The inactivity of the mRNA is due to the structure of its 5′ end, which has a set of four inverted repeats that sequester the initiation sequences (ribosome binding site and initiation codon) for the methylase by base pairing in the absence of erythromycin (Fig. 12, conformation A). Thus, the methylase cannot be produced, since the initiation motifs for the translation of the enzyme are not accessible to the ribosome. Induction is related to the presence of an open reading frame, encoding a short 14-amino-acid peptide, upstream from the erm(C) structural gene. In the presence of low concentrations of erythromycin, the binding of the antibiotic to a ribosome translating the leader peptide causes the ribosome to stall. Ribosome stalling probably induces the destabilization of the two stem-loop structures of configuration A and other conformational rearrangements in the mRNA. In particular, the formation of the alternative stem-loop structure (Fig. 12, conformation B) would unmask the initiation sequences for the methylase, allowing synthesis to proceed by ribosomes that are not complexed with erythromycin or by those that are methylated. Methylation of some ribosomes might occur through transient rearrangements of the stem-loop structures, which would lead to the synthesis of a basal level of methylase. A third alternate mRNA conformation has been predicted, which could occur at the end of the induction process when the concentration of the inducer macrolide has decreased and the majority of ribosomes are methylated (263).
FIG. 12.
Alternative conformations of the mRNA from the inducible erm(C) gene of pE194. Shown is the secondary structure of the mRNA in the absence (A) or presence (B) of erythromycin. RBS, ribosome binding site; LP, leader peptide; ORF, open reading frame; 1, 2, 3, and 4, inverted repeat. Green and red lines indicate the coding sequence.
Three interrelated mechanisms contribute to the regulation of methylase production. The most important mechanism is mRNA stabilization that occurs during the induction process. This stabilization, a consequence of ribosome stalling, protects the transcripts from degradation by RNases (30, 230), leading to a spectacular increase in mRNA half-life that enhances enzyme synthesis. A feedback mechanism negatively regulates methylase synthesis: as the pool of methylated ribosomes increases during induction, fewer ribosomes are able to stall, and therefore, transcripts return to the inactive conformation. Finally, it has been shown in Bacillus subtilis that the Erm(C) methylase binds to its own mRNA at a site with structural similarity to the site of methylation in 23S rRNA. As a consequence, the methylase might block its own production when synthesized in excess (62).
The mechanism responsible for the specificity of induction remains poorly understood. It is not related to the class of erm gene but depends on the structure of the specific attenuator, which controls the expression of the erm gene, and, for a given attenuator, the structure of the MLSB antibiotic determines whether a particular macrolide is an inducer or not (167). Probably, the interactions between the macrolide, the leader peptide, and the ribosome are critical for proper ribosome stalling, which is required for induction. In the erm(C) leader peptide, four amino acids, IFVI, are critical for induction (263). A similar sequence is found in specific small peptides encoded in Escherichia coli 23S rRNA by five-codon minigenes (248). These peptides can render cells resistant to low levels of a variety of macrolide antibiotics (247, 249). A “bottle brush” model of action for these macrolide resistance peptides, in which newly translated peptides interact with the macrolide molecule on the ribosome and actively displace it from its binding site, has been proposed (247). Probably, a similar type of interaction between the leader peptide and macrolides can occur. It seems that the leader peptide could be the selector of the site of ribosome stalling in leader mRNA by cis interference with translation, as previously demonstrated for the leader peptides controlling the inducible expression of cat genes (221).
Considering the importance of the leader peptide sequence for specificity of induction, it is not surprising that certain mutations in this sequence lead to changes in the induction patterns. For instance, changes in the relative activity of erythromycin and lincosamides as inducers of erm(C) have been observed (167). These changes, obtained in the laboratory, are not common in clinical isolates.
Control of expression of other erm genes.
Many other erm genes, including those detected in pathogenic bacteria, are also inducibly expressed. A model of posttranscriptional regulation similar to that described for erm(C) has been proposed for the regulation of erm(A) (mostly found in staphylococci) and erm(B) (mostly found in streptococci and enterococci). However, the regulatory regions of these determinants are more complex than that of erm(C). The attenuator of erm(A) contains two short control peptides, and induction may involve a series of rearrangements of the inverted repeats (174). The subset of the erm(A) class genes previously called erm(TR), which is mostly present in beta-hemolytic streptococci, has a similar attenuator structure. The 5′ end of erm(B) also presents a series of inverted repeats that are responsible for the lack of methylase synthesis in the absence of erythromycin (113). Fourteen pairs of repeats that could form alternative stem-loop structures by base pairing have been identified, and one of them might sequester the ribosome binding site and the initiation codon of the methylase gene. Induction would be related to the presence of sequences coding for a small leader peptide of 36 amino acids upstream from the gene.
The specificity of induction relies, as mentioned above, on the structure of the attenuator and on the precise mode of action of specific MLS compounds. Since the structure of the attenuator differs in each class or subclass of erm gene, different patterns of inducible MLSB resistance are observed. For instance, spiramycin is a common inducer, like erythromycin, of erm(B) expression [whereas it is not an inducer for erm(C) or erm(A)] (51). The genetic background and the bacterial host may also have roles in the specificity of induction, possibly in relation to differences in ribosomal structure or in methylase expression (164).
Again, changes in the sequence of the leader peptide lead to changes in the pattern of inducibility. For instance, a clinical isolate of Enterococcus faecalis that contains an R7C mutation in the putative leader peptide of its erm(B) gene is more strongly induced by tylosin, a 16-membered macrolide, than by erythromycin (183). Similarly, a clinical isolate of S. aureus with an unusual inducible cross-resistance to erythromycin, clindamycin, lincomycin, and quinupristin had mutations in the attenuator of the erm(A) gene (52).
Phenotypes of inducible MLSB resistance.
Due to the diversity of the erm genes and the specificity of the corresponding regulators, inducible expression of these genes gives rise to a large variety of phenotypes. However, in practice, since each class of erm gene is preferentially distributed in certain bacterial species, a few major phenotypes of inducible MLSB resistance are observed in staphylococci and streptococci/enterococci.
In staphylococci, inducible expression of erm(A) or erm(C) leads to a similar dissociated phenotype of resistance. The strains are resistant to 14-membered (clarithromycin, dirithromycin, erythromycin, and roxithromycin) and 15-membered (azithromycin) macrolides, which are inducers. In contrast, the noninducer ketolide (telithromycin), 16-membered macrolides available in certain countries (josamycin, midecamycin, miocamycin, rokitamycin, and spiramycin) or in veterinary practice (tylosin), the lincosamides (lincomycin and clindamycin), and streptogramins B (pristinamycin I and quinupristin) remain active. In disk diffusion tests, the blunting of the clindamycin (or any noninducer macrolide) inhibition zone, as in a D-shaped zone, can be observed, provided that a disk of erythromycin is placed nearby (Fig. 13).
FIG. 13.
S. aureus containing an erm(C) gene that is inducibly expressed. CM, clindamycin; E, erythromycin; L, lincomycin; SP, spiramycin; PI, pristinamycin IA (streptogramin factor B); PII, pristinamycin IIA (streptogramin factor A); PT, pristinamycin; TEL, telithromycin. A D-shaped zone can be observed for the clindamycin (and noninducer macrolides) zone of inhibition on the edge closest to the erythromycin zone of inhibition.
In streptococci, the inducible phenotypes are more diverse and complex. Several members of the MLSB group, including erythromycin and its derivatives, and spiramycin are inducers at various degrees of ErmB methylase production (51). In addition, recent studies have demonstrated that in Streptococcus pyogenes and S. pneumoniae containing an inducible erm(B) gene, the ribosomes are partly methylated in the absence of induction (69). The variety of resistance phenotypes in inducibly resistant streptococci might thus be explained by the complex pattern of inducibility combined with enzyme production at various basal levels, probably in relation to a relaxed control of methylase synthesis by the erm(B) attenuator (223, 224). It has been shown, by induction studies including fusions of attenuators with a reporter gene, that the MLSB phenotype characterized by high-level cross-resistance to macrolides and lincosamides, which is commonly detected in pneumococci, is frequently inducible (223).
Constitutive Expression of erm Genes
In the laboratory, constitutive expression of MLSB resistance can be obtained in initially inducible strains of staphylococci by selection on agar plates containing inhibitory concentrations of a noninducer macrolide, lincosamide, or streptogramin B at frequencies varying between 10−6 and 10−8, depending on the strain and the selector antibiotic. Clinical isolates that are constitutively resistant to erythromycin are widespread, especially in methicillin-resistant staphylococci. Whether in laboratory strains or in clinical isolates, deletion of the entire attenuator yields constitutive resistance; also, point mutations or tandem duplications in the attenuator lead to constitutive resistance by decreasing the stability of the hairpin structure sequestering the initiation sequences for the methylase or by duplicating the initiation signal-containing sequences, which are thus available for translation (231, 232, 233).
Similarly, in vitro selection of constitutive resistance at a frequency of 10−7 with clindamycin has been reported for a clinical isolate of Streptococcus pyogenes UCN1 that is inducibly resistant to erythromycin and that harbors an erm(A) [erm(TR) subset] gene. Clindamycin resistance was associated with deletions of 163 and 6 bp, which is probably explained by illegitimate recombination between different parts of the regulatory region, as well as a tandem duplication of 101 bp in the regulatory sequence of the erm(TR) gene (79).
Deletion of the attenuator has been found in constitutively resistant clinical isolates of Staphylococcus epidermidis and S. aureus containing erm(C) or erm(A) (134, 232, 265) and Enterococcus faecalis, Streptococcus agalactiae, and S. pneumoniae containing erm(B) (162, 224). In addition, point mutations in the attenuators of erm(T) of Lactobacillus reuteri (246) or of erm(A) of S. aureus (265) or tandem duplications in the attenuators of erm(C) of S. aureus, Staphylococcus saprophyticus, and Staphylococcus equorum and of erm(A) of S. aureus (102, 152, 185, 232, 266) have been reported.
Probably, constitutive MLSB-resistant isolates have evolved from the inducibly resistant isolates under selective pressure by noninducer macrolide/lincosamide antibiotics. Constitutive production of a methylase confers a characteristic phenotype with cross-resistance to the MLSB drugs, regardless of the nature of the erm gene (Fig. 14). However, the level of resistance may vary according to the degree of methylation of the ribosome. Although all members of the Erm family methylate the adenine of 23S rRNA located at position A2058, they differ in their capacities to monomethylate or dimethylate the nucleotide at this position. The major Erm methylases detected in pathogens, Erm(A), Erm(B), and Erm(C), generally function as dimethylases that confer high-level cross-resistance to MLSB drugs (including telithromycin). However, it has recently been shown, using mass spectrometry to analyze the methylated DNA, that Erm(B) in a Streptococcus pneumoniae background monomethylates the 23S rRNA, which renders cells resistant to erythromycin and clindamycin but not to telithromycin (69). This explains, at least in part, why telithromycin is active against nearly all S. pneumoniae isolates containing erm(B) but is active against only a few S. pyogenes isolates containing that gene (69, 159).
FIG. 14.
S. aureus containing an erm(C) gene that is constitutively expressed. CM, clindamycin; E, erythromycin; L, lincomycin; SP, spiramycin; PI, pristinamycin IA (streptogramin factor B); PII, pristinamycin IIA (streptogramin factor A); PT, pristinamycin; TEL, telithromycin.
Clinical Implications of Inducible MLSB Resistance
What is the clinical evidence for failure of clindamycin treatment?
As mentioned above, constitutive mutants can be readily selected in vitro from inducibly MLSB-resistant strains in the presence of clindamycin. The frequency of mutation may be as high as 10−6 to 10−7 per parent cell. The notion that bacterial inocula exceeding 107 CFU can be found in mediastinitis and in certain lower respiratory tract infections implies that the use of clindamycin for the treatment of an infection due to an inducibly resistant strain of S. aureus is not devoid of risk. In fact, recent reports indicate that the observation made primarily in the laboratory holds true for clinical isolates (70, 86, 142, 143, 170, 212, 236). However, clinical evidence of the risk of emergence of clindamycin resistance is based on only a few case reports. Also, the limited number of cases may bias the situation, since the reports have been justified by the observation of clinical failures, and successes are usually not reported. Much of the data come from pediatric patients. Infections due to community-acquired methicillin-resistant S. aureus are increasing in this category of patient, and therapeutic options are limited since β-lactams (because of resistance) and quinolones (in children) cannot be used. Therefore, since clindamycin represents an interesting alternative to vancomycin, the proportion of community-acquired methicillin-resistant S. aureus isolates with an inducible MLSB phenotype should be carefully surveyed. In fact, the incidence of isolates that are resistant to erythromycin but susceptible to clindamycin varies worldwide (256). In the United States, the incidence of the inducible MLSB phenotype also varies widely by hospital, by geographical area, and temporally, ranging from 2.6% to 27.9% (7, 40, 47, 182).
For streptococci, similar doubts concerning the activity of clindamycin against isolates susceptible to this antibiotic but with an inducible MLSB phenotype can be raised. Although no clinical failure has been reported, the use of clindamycin does not appear to be safe.
Implications for the clinical microbiology laboratory.
The reporting of clindamycin susceptibility raises problems when the isolate is resistant to erythromycin. In that case, the detection of inducible MLSB resistance (which is possible only by revealing induction of clindamycin resistance) is required. Disk diffusion is an easy method to detect this phenotype, by placing an erythromycin disk in close proximity to a clindamycin disk on an agar plate (78). A D-shaped zone is specific for the inducible MLSB phenotype. The 2004 CLSI (formerly NCCLS) susceptibility testing standards recommend this approach to detect inducible MLSB resistance in staphylococci. When staphylococci are tested using a broth-based method, particularly when using automated instruments, the CLSI recommends placing erythromycin and clindamycin disks 15 mm apart on the blood agar plate that is routinely used to verify the purity of the bacterial inoculum (120).
Isolates displaying a D-shaped zone should be reported as clindamycin resistant by the laboratory (179). However, the CLSI suggests the possibility of including the comment, “This isolate is presumed to be resistant based on detection of inducible clindamycin resistance. Clindamycin may still be effective in some patients.” The final decision, to treat or not to treat the patient with clindamycin, should be based on an analysis of every specific case, and if clindamycin therapy is started, it requires close follow-up of the patient for signs of failure. So far, although it seems reasonable to discourage the use of clindamycin in deep-seated infections or in infections with high bacterial densities that increase the risk of selection of constitutive mutants, there are no criteria to confidently predict the success or the failure of clindamycin therapy in infections due to staphylococci with inducible MLSB resistance. Although the CLSI recommendation is limited to clindamycin, the same reasoning should be applied to telithromycin and 16-membered macrolides in countries where they are available for staphylococci with inducible MLSB resistance.
The inducible MLSB phenotype should be distinguished from another phenotype of dissociated resistance to erythromycin and susceptibility to clindamycin, which is due to the acquisition of msr(A). This gene encodes an inducibly produced efflux pump belonging to the ABC transporters. Erythromycin and related 14- and 15-membered macrolides are inducers and substrates for the pump. In contrast, clindamycin is neither an inducer nor a substrate, and thus, msr(A)-carrying strains are fully susceptible to this compound. Constitutive mutants are resistant to erythromycin but remain fully susceptible to clindamycin. Therefore, the microorganisms that are resistant to erythromycin but susceptible to clindamycin and that do not display a D-shaped zone are presumably resistant to erythromycin by efflux and can be safely reported as being susceptible to clindamycin. The case of telithromycin is different, since this antimicrobial is not an inducer but is a substrate for the MsrA pump and may select for constitutive resistant mutants. However, there have been no reports of clinical failure of telithromycin therapy for patients with infections caused by telithromycin-susceptible, erythromycin-resistant isolates (60).
Although the issue of detection and reporting of inducible MLSB resistance in streptococci has still not been fully addressed, recommendations similar to those for staphylococci should be made for clindamycin and telithromycin. Pratically, this applies to beta-hemolytic streptococci with the erm(A) gene. Isolates that are resistant to erythromycin but susceptible to clindamycin and that do not exhibit a D-shaped zone may be safely reported as being susceptible to clindamycin and telithromycin. In this case, resistance is due to an efflux pump encoded by a mef(A) gene for which neither clindamycin nor telithromycin are substrates. This is in contrast with the MsrA pump for which, as mentioned above, telithromycin is a substrate.
Finally, we need more prospective studies of cases of infections due to staphylococci or hemolytic streptococci treated with clindamycin to more definitively define the place of this antimicrobial compound in the treatment of infections due to microorganisms with various macrolide resistance phenotypes.
CONCLUSION
The plethora of interactions between antibiotics and bacteria testifies to the remarkable adaptability of living organisms to changing and hostile environments. It provides privileged systems for the study of gene regulation and dissemination in that the manifestation of the genetic information, resistance, is easy to trace and that bacteria are subjected to accelerated evolution driven by the massive use of antibiotics. Study of the mode of action of, and of bacterial resistance to, antibiotics in the past decades has helped to elucidate numerous aspects of bacterial physiology and, not least, those of gene expression and its regulation.
Acknowledgments
We gratefully acknowledge P. E. Reynolds for his comments and his critical reading of the manuscript.
REFERENCES
- 1.Abadía, L., P. Courvalin, and B. Périchon. 2002. vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405. J. Bacteriol. 184:6457-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abadia-Patino, L., K. Christiansen, J. Bell, P. Courvalin, and B. Perichon. 2004. VanE-type vancomycin-resistant Enterococcus faecalis clinical isolates from Australia. Antimicrob. Agents Chemother. 48:4882-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abo-Amer, A. E., J. Munn, K. Jackson, M. Aktas, P. Golby, D. J. Kelly, and S. C. Andrews. 2004. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J. Bacteriol. 186:1879-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 5.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen, N. E., and J. N. Hobbs, Jr. 1995. Induction of vancomycin resistance in Enterococcus faecium by non-glycopeptide antibiotics. FEMS Microbiol. Lett. 132:107-114. [DOI] [PubMed] [Google Scholar]
- 7.Almer, L. S., V. D. Shortridge, A. M. Nilius, J. M. Beyer, N. B. Soni, M. H. Bui, G. G. Stone, and R. K. Flamm. 2002. Antimicrobial susceptibility and molecular characterization of community-acquired methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 43:225-232. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arias, C. A., P. Courvalin, and P. E. Reynolds. 2000. vanC cluster of vancomycin-resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 44:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arias, C. A., M. Martin-Martinez, T. L. Blundell, M. Arthur, P. Courvalin, and P. E. Reynolds. 1999. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 31:1653-1664. [DOI] [PubMed] [Google Scholar]
- 12.Arthur, M., F. Depardieu, L. Cabanié, P. Reynolds, and P. Courvalin. 1998. Requirement of the VanY and VanX D,D-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819-830. [DOI] [PubMed] [Google Scholar]
- 13.Arthur, M., F. Depardieu, and P. Courvalin. 1999. Regulated interactions between partner and non-partner sensors and responses regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145:1849-1858. [DOI] [PubMed] [Google Scholar]
- 14.Arthur, M., F. Depardieu, G. Gerbaud, M. Galimand, R. Leclercq, and P. Courvalin. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 179:97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33-44. [DOI] [PubMed] [Google Scholar]
- 16.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1999. Moderate-level resistance to glycopeptide LY333328 mediated by genes of the vanA and vanB clusters in enterococci. Antimicrob. Agents Chemother. 43:1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arthur, M., C. Molinas, T. D. H. Bugg, G. D. Wright, C. T. Walsh, and P. Courvalin. 1992. Evidence for in vivo incorporation of d-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 36:867-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arthur, M., P. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 21.Aslangul, E., M. Baptista, B. Fantin, F. Depardieu, M. Arthur, P. Courvalin, and C. Carbon. 1997. Selection of glycopeptide-resistant mutants of VanB-type Enterococcus faecalis BM4281 in vitro and in experimental endocarditis. J. Infect. Dis. 175:598-605. [DOI] [PubMed] [Google Scholar]
- 22.Aubert, D., T. Naas, and P. Nordmann. 2003. IS1999 increases expression of the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. J. Bacteriol. 185:5314-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubert, D., L. Poirel, J. Chevalier, S. Leotard, J. M. Pages, and P. Nordmann. 2001. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal β-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baptista, M., F. Depardieu, P. Courvalin, and M. Arthur. 1996. Specificity of induction of glycopeptide resistance genes in Enterococcus faecalis. Antimicrob. Agents Chemother. 40:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baptista, M., F. Depardieu, P. Reynolds, P. Courvalin, and M. Arthur. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25:93-105. [DOI] [PubMed] [Google Scholar]
- 27.Baptista, M., P. Rodrigues, F. Depardieu, P. Courvalin, and M. Arthur. 1999. Single-cell analysis of glycopeptide resistance gene expression in teicoplanin-resistant mutants of a VanB-type Enterococcus faecalis. Mol. Microbiol. 32:17-28. [DOI] [PubMed] [Google Scholar]
- 28.Baranova, N., and H. Nikaido. 2002. The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 30.Bechhofer, D. H., and D. Dubnau. 1987. Induced mRNA stability in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 84:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett, P. M. 2004. Genome plasticity: insertion sequence elements, transposons and integrons, and DNA rearrangement. Methods Mol. Biol. 266:71-113. [DOI] [PubMed] [Google Scholar]
- 32.Beuzon, C. R., D. Chessa, and J. Casadesus. 2004. IS200: an old and still bacterial transposon. Int. Microbiol. 7:3-12. [PubMed] [Google Scholar]
- 33.Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bächi. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77-82. [DOI] [PubMed] [Google Scholar]
- 34.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanco, A. G., M. Sola, F. X. Gomis-Ruth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure (Cambridge) 10:701-713. [DOI] [PubMed] [Google Scholar]
- 36.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutoille, D., S. Corvec, N. Caroff, C. Giraudeau, E. Espaze, J. Caillon, P. Plesiat, and A. Reynaud. 2004. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing beta-lactam resistance. FEMS Microbiol. Lett. 230:143-146. [DOI] [PubMed] [Google Scholar]
- 38.Boyd, D. A., J. Conly, H. Dedier, G. Peters, L. Robertson, E. Slater, and M. R. Mulvey. 2000. Molecular characterization of the vanD gene cluster and a novel insertion element in a vancomycin-resistant enterococcus isolated in Canada. J. Clin. Microbiol. 38:2392-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bräu, B., U. Pilz, and W. Piepersberg. 1984. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol. Gen. Genet. 193:179-187. [DOI] [PubMed] [Google Scholar]
- 40.Braun, L., D. Craft, R. Williams, F. Tuamokumo, and M. Ottolini. 2005. Increasing clindamycin resistance among methicillin-resistant Staphylococcus aureus in 57 northeast United States military treatment facilities. Pediatr. Infect. Dis. J. 24:622-626. [DOI] [PubMed] [Google Scholar]
- 41.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadewall, B., and P. Courvalin. 1999. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 181:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casadewall, B., P. E. Reynolds, and P. Courvalin. 2001. Regulation of expression of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 183:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casin, I., B. Hanau-Bercot, I. Podglajen, H. Vahaboglu, and E.Collatz. 2003. Salmonella enterica serovar Typhimurium blaPER-1-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob. Agents Chemother. 47:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chabbert, Y. 1956. Antagonisme in vitro entre l'erythromycine et la spiramycine. Ann. Inst. Pasteur 90:787-790. [PubMed] [Google Scholar]
- 46.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. L. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, DC.
- 47.Chavez-Bueno, S., B. Bozdogan, K. Katz, K. L. Bowlware, N. Cushion, D. Cavuoti, N. Ahmad, G. H. McCracken, Jr., and P. C. Appelbaum. 2005. Inducible clindamycin resistance and molecular epidemiologic trends of pediatric community-acquired methicillin-resistant Staphylococcus aureus in Dallas, Texas. Antimicrob. Agents Chemother. 49:2283-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen, H., M. Bjerknes, R. Kumar, and E. Jay. 1994. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22:4953-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuanchuen, R., J. B. Gaynor, R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. Molecular characterization of MexL, the transcriptional repressor of the mexJK multidrug efflux operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1844-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clarebout, G., and R. Leclercq. 2002. Fluorescence assay for studying the ability of macrolides to induce production of ribosomal methylase. Antimicrob. Agents Chemother. 46:2269-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarebout, G., E. Nativelle, and R. Leclercq. 2001. Unusual inducible cross resistance to macrolides, lincosamides, and streptogramins B by methylase production in clinical isolates of Staphylococcus aureus. Microb. Drug Resist. 7:317-322. [DOI] [PubMed] [Google Scholar]
- 53.Coleman, N. V., and A. J. Holmes. 2005. The native Pseudomonas stutzeri strain Q chromosomal integron can capture and express cassette-associated genes. Microbiology 151:1853-1864. [DOI] [PubMed] [Google Scholar]
- 54.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collis, C. M., M. J. Kim, S. R. Partridge, H. W. Stokes, and R. M. Hall. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 184:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comenge, Y., R. Quintiliani, Jr., L. Li, L. Dubost, J. P. Brouard, J. E. Hugonnet, and M. Arthur. 2003. The CroRS two-component regulatory system is required for intrinsic β-lactam resistance in Enterococcus faecalis. J. Bacteriol. 185:7184-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corvec, S., N. Caroff, E. Espaze, C. Giraudeau, H. Drugeon, and A. Reynaud. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52:629-635. [DOI] [PubMed] [Google Scholar]
- 58.Couto, I., H. de Lencastre, E. Severina, W. Kloos, J. A. Webster, R. J. Hubner, I. S. Sanches, and A. Tomasz. 1996. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 2:377-391. [DOI] [PubMed] [Google Scholar]
- 59.Couto, I., S. W. Wu, A. Tomasz, and H. de Lencastre. 2003. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J. Bacteriol. 185:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis, K. A., S. A. Crawford, K. R. Fiebelkorn, and J. H. Jorgensen. 2005. Induction of telithromycin resistance by erythromycin in isolates of macrolide-resistant Staphylococcus spp. Antimicrob. Agents Chemother. 49:3059-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dean, C. R., M. A. Visalli, S. J. Projan, P. E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denoya, C. D., D. H. Bechhofer, and D. Dubnau. 1986. Translational autoregulation of ermC 23S rRNA methyltransferase expression in Bacillus subtilis. J. Bacteriol. 168:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 64.Depardieu, F., P. Courvalin, and A. Kolb. 2005. Binding sites of VanRB and sigma70 RNA polymerase in the vanB vancomycin resistance operon of Enterococcus faecium BM4524. Mol. Microbiol. 57:550-564. [DOI] [PubMed] [Google Scholar]
- 65.Depardieu, F., P. Courvalin, and T. Msadek. 2003. A six amino acid deletion, partially overlapping the VanSB G2 ATP-binding motif, leads to constitutive glycopeptide resistance in VanB-type Enterococcus faecium. Mol. Microbiol. 50:1069-1083. [DOI] [PubMed] [Google Scholar]
- 66.Depardieu, F., M. Kolbert, H. Pruul, J. Bell, and P. Courvalin. 2004. Characterization of new VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 48:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Depardieu, F., P. E. Reynolds, and P. Courvalin. 2003. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob. Agents Chemother. 47:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 69.Douthwaite, S., J. Jalava, and L. Jakobsen. 2005. Ketolide resistance in Streptococcus pyogenes correlates with the degree of rRNA dimethylation by Erm. Mol. Microbiol. 58:613-622. [DOI] [PubMed] [Google Scholar]
- 70.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. [DOI] [PubMed] [Google Scholar]
- 71.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34:633-640. [DOI] [PubMed] [Google Scholar]
- 72.Duvall, E. J., and P. S. Lovett. 1986. Chloramphenicol induces translation of the mRNA for a chloramphenicol-resistance gene in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 83:3939-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 74.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edwards, R., and P. N. Read. 2000. Expression of the carbapenemase gene (cfiA) in Bacteroides fragilis. J. Antimicrob. Chemother. 46:1009-1012. [DOI] [PubMed] [Google Scholar]
- 76.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evers, S., and P. Courvalin. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J. Bacteriol. 178:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fiebelkorn, K. R., S. A. Crawford, M. L. McElmeel, and J. H. Jorgensen. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fines, M., M. Gueudin, A. Ramon, and R. Leclercq. 2001. In vitro selection of resistance to clindamycin related to alterations in the attenuator of the erm(TR) gene of Streptococcus pyogenes UCN1 inducibly resistant to erythromycin. J. Antimicrob. Chemother. 48:411-416. [DOI] [PubMed] [Google Scholar]
- 80.Fines, M., B. Perichon, P. Reynolds, D. F. Sahm, and P. Courvalin. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Firth, N. 2003. Evolution of antimicrobial multi-resistance in gram-positive bacteria, p. 33-60. In C. F. Amabile-Cuevas (ed.), Multiple drug resistant bacteria. Horizon Scientific Press, Norfolk, United Kingdom.
- 82.Fluit, A. C., and F. J. Schmitz. 2004. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 10:272-288. [DOI] [PubMed] [Google Scholar]
- 83.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 86.Frank, A. I., J. F. Marcinak, P. D. Mangat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530-534. [DOI] [PubMed] [Google Scholar]
- 87.Garrod, L. P. 1957. The erythromycin group of antibiotics. Br. Med. J. 2:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giuliani, F., J. D. Docquier, M. L. Riccio, L. Pagani, and G. M. Rossolini. 2005. OXA-46, a new class D β-lactamase of narrow substrate specificity encoded by a blaVIM-1-containing integron from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 49:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goussard, S., W. Sougakoff, C. Mabilat, A. Bauernfeind, and P. Courvalin. 1991. An IS1-like element is responsible for high-level synthesis of extended-spectrum beta-lactamase TEM-6 in Enterobacteriaceae. J. Gen. Microbiol. 137:2681-2687. [DOI] [PubMed] [Google Scholar]
- 90.Grissom-Arnold, J., W. E. Alborn, Jr., T. I. Nicas, and S. R. Jaskunas. 1997. Induction of VanA vancomycin resistance genes in Enterococcus faecalis: use of a promoter fusion to evaluate glycopeptide and nonglycopeptide induction signals. Microb. Drug Resist. 3:53-64. [DOI] [PubMed] [Google Scholar]
- 91.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gryczan, T. J., G. Grandi, J. Hahn, R. Grandi, and D. Dubnau. 1980. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 8:6081-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gualerzi, C. O., L. Brandi, E. Caserta, A. La Teana, R. Spurio, J. Tomsic, and C. L. Pon. 2000. Translation initiation in bacteria, p. 477-494. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller, (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. American Society for Microbiology, Washington, DC.
- 94.Haggoud, A., G. Reysset, H. Azeddoug, and M. Sebald. 1994. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob. Agents Chemother. 38:1047-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 96.Hall, R. M., and H. W. Stokes. 2004. Integrons or super integrons? Microbiology 150:3-4. [DOI] [PubMed] [Google Scholar]
- 97.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 98.Hanau-Bercot, B., I. Podglajen, I. Casin, and E. Collatz. 2002. An intrinsic control element for translational initiation in class 1 integrons. Mol. Microbiol. 44:119-130. [DOI] [PubMed] [Google Scholar]
- 99.Hancock, L. E., and M. Perego. 2004. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J. Bacteriol. 186:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Handwerger, S., and A. Kolokathis. 1990. Induction of vancomycin resistance in Enterococcus faecium by inhibition of transglycosylation. FEMS Microbiol. Lett. 70:167-170. [DOI] [PubMed] [Google Scholar]
- 101.Hansson, K., L. Sundstrom, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hauschild, T., P. Luthje, and S. Schwarz. 2006. Characterization of a novel type of MLS(B) resistance plasmid from Staphylococcus saprophyticus carrying a constitutively expressed erm(C) gene. Vet. Microbiol. 15:258-263. [DOI] [PubMed] [Google Scholar]
- 103.Hayden, M. K., G. M. Trenholme, J. E. Schultz, and D. F. Sahm. 1993. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J. Infect. Dis. 167:1224-1227. [DOI] [PubMed] [Google Scholar]
- 104.Heifets, L. B., M. A. Flory, and P. J. Lindholm-Levy. 1989. Does pyrazinoic acid as an active moiety of pyrazinamide have specific activity against Mycobacterium tuberculosis? Antimicrob. Agents Chemother. 33:1252-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 106.Hernandez-Alles, S., S. Alberti, D. Alvarez, A. Domenech-Sanchez, L. Martinez-Martinez, J. Gil, J. M. Tomas, and V. J. Benedi. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 107.Hernandez-Alles, S., V. J. Benedi, L. Martinez-Martinez, A. Pascual, A. Aguilar, J. M. Tomas, and S. Alberti. 1999. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob. Agents Chemother. 43:937-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hirakawa, H., K. Nishino, J. Yamada, T. Hirata, and A. Yamaguchi. 2003. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52:576-582. [DOI] [PubMed] [Google Scholar]
- 109.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 110.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, DC.
- 111.Hocquet, D., C. Vogne, F. El Garch, A. Vejux, N. Gotoh, A. Lee, O. Lomovskaya, and P. Plesiat. 2003. MexXY-OprM efflux pump is necessary for a adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Holman, T. R., Z. Wu, B. L. Wanner, and C. T. Walsh. 1994. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry 33:4625-4631. [DOI] [PubMed] [Google Scholar]
- 113.Horinouchi, S., W. H. Byeon, and B. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Horinouchi, S., and B. Weisblum. 1980. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc. Natl. Acad. Sci. USA 77:7079-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jacobs, C., J. M. Frère, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 117.Jacoby, G. A., and L. S. Munoz-Price. 2005. The new beta-lactamases. N. Engl. J. Med. 352:380-391. [DOI] [PubMed] [Google Scholar]
- 118.Jaurin, B., and S. Normark. 1983. Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell 32:809-816. [DOI] [PubMed] [Google Scholar]
- 119.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jorgensen, J. H., S. A. Crawford, M. L. McElmeel, and K. R. Fiebelkorn. 2004. Detection of inducible clindamycin resistance of staphylococci in conjunction with performance of automated broth susceptibility testing. J. Clin. Microbiol. 42:1800-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 123.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kato, N., K. Yamazoe, C. G. Han, and E. Ohtsubo. 2003. New insertion sequence elements in the upstream region of cfiA in imipenem-resistant Bacteroides fragilis strains. Antimicrob. Agents Chemother. 47:979-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kawalec, M., M. Gniadkowski, J. Kedzierska, A. Skotnicki, J. Fiett, and W. Hryniewicz. 2001. Selection of a teicoplanin-resistant Enterococcus faecium mutant during an outbreak caused by vancomycin-resistant enterococci with the VanB phenotype. J. Clin. Microbiol. 39:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kobayashi, N., M. M. Alam, and S. Urasawa. 2001. Genomic rearrangement of the mec regulator region mediated by insertion of IS431 in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45:335-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kohler, T., S. F. Epp, L. K. Curty, and J. C. Pechere. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kohler, T., M. Kok, M. Michea-Hamzehpour, P. Plesiat, N. Gotoh, T. Nishino, L. K. Curty, and J. C. Pechere. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 130.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 131.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 132.Lai, M. H., and D. R. Kirsch. 1996. Induction signals for vancomycin resistance encoded by the vanA gene cluster in Enterococcus faecium. Antimicrob. Agents Chemother. 40:1645-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lambert, T., G. Gerbaud, and P. Courvalin. 1994. Characterization of the chromosomal aac(6′)-Ij gene of Acinetobacter sp. 13 and the aac(6′)-Ih plasmid gene of Acinetobacter baumannii. Antimicrob. Agents Chemother. 38:1883-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lampson, B. C., and J. T. Parisi. 1986. Naturally occurring Staphylococcus epidermidis plasmid expressing constitutive macrolide-lincosamide-streptogramin B resistance contains a deleted attenuator. J. Bacteriol. 166:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Leclercq, R., S. Dutka-Malen, J. Duval, and P. Courvalin. 1992. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob. Agents Chemother. 36:2005-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J.-D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee, K. Y., J. D. Hopkins, and M. Syvanen. 1990. Direct involvement of IS26 in an antibiotic resistance operon. J. Bacteriol. 172:3229-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leelaporn, A., N. Firth, M. E. Byrne, E. Roper, and R. A. Skurray. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob. Agents Chemother. 38:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lemaitre, N., W. Sougakoff, C. Truffot-Pernot, and V. Jarlier. 1999. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob. Agents Chemother. 43:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Leverstein-van Hall, M. A., H. E. Blok, A. R. Donders, A. Paauw, A. C. Fluit, and J. Verhoef. 2003. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 187:251-259. [DOI] [PubMed] [Google Scholar]
- 141.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 42:49-54. [DOI] [PubMed] [Google Scholar]
- 142.Levin, T. P., B. Suh, P. Axelrod, A. L. Truant, and T. Fekete. 2005. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob. Agents Chemother. 49:1222-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lewis, J. S., II, and J. H. Jorgensen. 2005. Inducible clindamycin resistance in staphylococci: should clinicians and microbiologists be concerned? Clin. Infect. Dis. 40:280-285. [DOI] [PubMed] [Google Scholar]
- 144.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 145.Li, X. Z., L. Zhang, and K. Poole. 2002. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 46:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Limansky, A. S., M. A. Mussi, and A. M. Viale. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J. Bacteriol. 169:1923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Literacka, E., J. Empel, A. Baraniak, E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2004. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 48:4136-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lodder, G., C. Werckenthin, S. Schwarz, and K. Dyke. 1997. Molecular analysis of naturally occuring ermC-encoding plasmids in staphylococci isolated from animals with and without previous contact with macrolide/lincosamide antibiotics. FEMS Immunol. Med. Microbiol. 18:7-15. [DOI] [PubMed] [Google Scholar]
- 153.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mahillon, J., C. Leonard, and M. Chandler. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 157.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bächi. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Maki, H., and K. Murakami. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Malhotra-Kumar, S., C. Lammens, S. Chapelle, M. Wijdooghe, J. Piessens, K. Van Herck, and H. Goossens. 2005. Macrolide- and telithromycin-resistant Streptococcus pyogenes, Belgium, 1999-2003. Emerg. Infect. Dis. 11:939-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Martin, B., G. Alloing, V. Mejean, and J. P. Claverys. 1987. Constitutive expression of erythromycin resistance mediated by the ermAM determinant of plasmid pAM beta 1 results from deletion of 5′ leader peptide sequences. Plasmid 18:250-253. [DOI] [PubMed] [Google Scholar]
- 163.Martin, C., J. Timm, J. Rauzier, R. Gomez-Lus, J. Davies, and B. Gicquel. 1990. Transposition of an antibiotic resistance element in mycobacteria. Nature 345:739-743. [DOI] [PubMed] [Google Scholar]
- 164.Martin, P. K., Y. Bao, E. Boyer, K. M. Winterberg, L. McDowell, M. B. Schmid, and J. M. Buysse. 2002. Novel locus required for expression of high-level macrolide-lincosamide-streptogramin B resistance in Staphylococcus aureus. J. Bacteriol. 184:5810-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matsuo, Y., S. Eda, N. Gotoh, E. Yoshihara, and T. Nakae. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 238:23-28. [DOI] [PubMed] [Google Scholar]
- 167.Mayford, M., and B. Weisblum. 1990. The ermC leader peptide: amino acid alterations leading to differential efficiency of induction by macrolide-lincosamide-streptogramin B antibiotics. J. Bacteriol. 172:3772-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 170.McGehee, R. F., F. F. Barrett, and F. Finland. 1968. Resistance of Staphylococcus aureus to lincomycin, clindamycin, and erythromycin. Antimicrob. Agents Chemother. 13:392-397. [PubMed] [Google Scholar]
- 171.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and β-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mena, A., V. Plasencia, L. Garcia, O. Hidalgo, J. I. Ayestaran, S. Alberti, N. Borrell, J. L. Perez, and A. Oliver. 2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mugnier, P., I. Casin, A. T. Bouthors, and E. Collatz. 1998. Novel OXA-10-derived extended-spectrum β-lactamases selected in vivo or in vitro. Antimicrob. Agents Chemother. 42:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Murphy, E. 1985. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J. Bacteriol. 162:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Naas, T., L. Philippon, L. Poirel, E. Ronco, and P. Nordmann. 1999. An SHV-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1281-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing. 14th informational supplement. M100-S14. NCCLS, Wayne, PA.
- 180.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nishino, K., T. Honda, and A. Yamaguchi. 2005. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J. Bacteriol. 187:1763-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ochoa, T. J., J. Mohr, A. Wanger, J. R. Murphy, and G. P. Heresi. 2005. Community-associated methicillin-resistant Staphylococcus aureus in pediatric patients. Emerg. Infect. Dis. 11:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Oh, T. G., A. R. Kwon, and E. C. Choi. 1998. Induction of ermAMR from a clinical strain of Enterococcus faecalis by 16-membered-ring macrolide antibiotics. J. Bacteriol. 180:5788-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Oliveira, S. S., E. Murphy, G. R. Gamon, and M. C. Bastos. 1993. pRJ5: a naturally occurring Staphylococcus aureus plasmid expressing constitutive macrolide-lincosamide-streptogramin B resistance contains a tandem duplication in the leader region of the ermC gene. J. Gen. Microbiol. 139:1461-1467. [DOI] [PubMed] [Google Scholar]
- 186.Ouellette, M., L. Bissonnette, and P. H. Roy. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc. Natl. Acad. Sci. USA 84:7378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Panesso, D., L. Abadia-Patino, N. Vanegas, P. E. Reynolds, P. Courvalin, and C. A. Arias. 2005. Transcriptional analysis of the vanC cluster from Enterococcus gallinarum strains with constitutive and inducible vancomycin resistance. Antimicrob. Agents Chemother. 49:1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 189.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Partridge, S. R., G. D. Recchia, C. Scaramuzzi, C. M. Collis, H. W. Stokes, and R. M. Hall. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855-2864. [DOI] [PubMed] [Google Scholar]
- 191.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Paulsen, I. T., R. A. Skurray, R. Tam, M. H. Saier, Jr., R. J. Turner, J. H. Weiner, E. B. Goldberg, and L. L. Grinius. 1996. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 19:1167-1175. [DOI] [PubMed] [Google Scholar]
- 193.Perichon, B., B. Casadewall, P. Reynolds, and P. Courvalin. 2000. Glycopeptide-resistant Enterococcus faecium BM4416 is a VanD-type strain with an impaired d-alanine:d-alanine ligase. Antimicrob. Agents Chemother. 44:1346-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perichon, B., and P. Courvalin. 2000. Update on vancomycin resistance. Int. J. Clin. Pract. 54:250-254. [PubMed] [Google Scholar]
- 195.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 98:10886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ploy, M. C., P. Courvalin, and T. Lambert. 1998. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob. Agents Chemother. 42:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Podglajen, I., J. Breuil, I. Casin, and E. Collatz. 1995. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J. Bacteriol. 177:5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Podglajen, I., J. Breuil, and E. Collatz. 1994. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105-114. [DOI] [PubMed] [Google Scholar]
- 199.Podglajen, I., J. Breuil, A. Rohaut, C. Monsempes, and E. Collatz. 2001. Multiple mobile promoter regions for the rare carbapenem resistance gene of Bacteroides fragilis. J. Bacteriol. 183:3531-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann. 2005. Genetic environment and expression of the extended-spectrum β-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Poirel, L., J.-W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in the expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Poirel, L., C. Heritier, V. Tolun, and P. Nordmann. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Poole, K. 2004. Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 206.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 207.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pournaras, S., A. Ikonomidis, L. S. Tzouvelekis, D. Tokatlidou, N. Spanakis, A. N. Maniatis, N. J. Legakis, and A. Tsakris. 2005. VIM-12, a novel plasmid-mediated metallo-β-lactamase from Klebsiella pneumoniae that resembles a VIM-1/VIM-2 hybrid. Antimicrob. Agents Chemother. 49:5153-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Prentki, P., B. Teter, M. Chandler, and D. J. Galas. 1986. Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 191:383-393. [DOI] [PubMed] [Google Scholar]
- 210.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Quintiliani, R., Jr., S. Evers, and P. Courvalin. 1993. The vanB gene confers various levels of self-transferable resistance to vancomycin in enterococci. J. Infect. Dis. 167:1220-1223. [DOI] [PubMed] [Google Scholar]
- 212.Rao, G. G. 2000. Should clindamycin be used in treatment of patients with infections caused by erythromycin-resistant staphylococci? J. Antimicrob. Chemother. 45:715-716. [DOI] [PubMed] [Google Scholar]
- 213.Rasmussen, J. L, D. A. Odelson, and F. L. Macrina. 1987. Complete nucleotide sequence of insertion element IS4351 from Bacteroides fragilis. J. Bacteriol. 169:3573-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 215.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 216.Reynolds, P. E. 1998. Control of peptidoglycan synthesis in vancomycin-resistant enterococci: D,D-peptidases and D,D-carboxypeptidases. Cell. Mol. Life Sci. 54:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Reynolds, P. E., F. Depardieu, S. Dutka-Malen, M. Arthur, and P. Courvalin. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol. Microbiol. 13:1065-1070. [DOI] [PubMed] [Google Scholar]
- 218.Reynolds, P. E., H. A. Snaith, A. J. Maguire, S. Dutka-Malen, and P. Courvalin. 1994. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem. J. 301:5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rice, L. B. 2002. Association of different mobile elements to generate novel integrative elements. Cell. Mol. Life Sci. 59:2023-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rodriguez-Martinez, J. M., L. Poirel, R. Canton, and P. Nordmann. 2006. Common region CR1 for expression of antibiotic resistance genes. Antimicrob. Agents Chemother. 50:2544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rogers, E. J., U. J. Kim, N. P. Ambulos, Jr., and P. S. Lovett. 1990. Four codons in the cat-86 leader define a chloramphenicol-sensitive ribosome stall sequence. J. Bacteriol. 172:110-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Rogers, M. B., T. K. Bennett, C. M. Payne, and C. J. Smith. 1994. Insertional activation of cepA leads to high-level β-lactamase expression in Bacteroides fragilis clinical isolates. J. Bacteriol. 176:4376-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Rosato, A., H. Vicarini, A. Bonnefoy, J. F. Chantot, and R. Leclercq. 1998. A new ketolide, HMR 3004, active against streptococci inducibly resistant to erythromycin. Antimicrob. Agents Chemother. 42:1392-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rosato, A., H. Vicarini, and R. Leclercq. 1999. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 43:559-562. [DOI] [PubMed] [Google Scholar]
- 225.Rouch, D. A., L. J. Messerotti, L. S. Loo, C. A. Jackson, and R. A. Skurray. 1989. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol. Microbiol. 3:161-175. [DOI] [PubMed] [Google Scholar]
- 226.Rowe-Magnus, D. A., A. M. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 227.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 228.Rudant, E., P. Courvalin, and T. Lambert. 1997. Loss of intrinsic aminoglycoside resistance in Acinetobacter haemolyticus as a result of three distinct types of alterations in the aac(6′)-Ig gene, including insertion of IS17. Antimicrob. Agents Chemother. 41:2646-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rudant, E., P. Courvalin, and T. Lambert. 1998. Characterization of IS18, an element capable of activating the silent aac(6′)-Ij gene of Acinetobacter sp. 13 strain BM2716 by transposition. Antimicrob. Agents Chemother. 42:2759-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sandler, P., and B. Weisblum. 1989. Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5′-to-3′ nucleolytic cleavage of the ermA transcript. J. Bacteriol. 171:6680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Schmitz, F. J., J. Petridou, N. Astfalk, K. Kohrer, S. Scheuring, and S. Schwarz. 2002. Molecular analysis of constitutively expressed erm(C) genes selected in vitro by incubation in the presence of the noninducers quinupristin, telithromycin, or ABT-773. Microb. Drug Resist. 8:171-177. [DOI] [PubMed] [Google Scholar]
- 232.Schmitz, F. J., J. Petridou, N. Astfalk, S. Scheuring, K. Kohrer, J. Verhoef, A. C. Fluit, and S. Schwarz. 2001. Structural alterations in the translational attenuator of constitutively expressed erm(A) genes in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1603-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Schmitz, F. J., J. Petridou, H. Jagusch, N. Astfalk, S. Scheuring, and S. Schwarz. 2002. Molecular characterization of ketolide-resistant erm(A)-carrying Staphylococcus aureus isolates selected in vitro by telithromycin, ABT-773, quinupristin and clindamycin. J. Antimicrob. Chemother. 49:611-617. [DOI] [PubMed] [Google Scholar]
- 234.Segal, H., E. C. Nelson, and B. G. Elisha. 2004. Genetic environment and transcription of ampC in an Acinetobacter baumannii clinical isolate. Antimicrob. Agents Chemother. 48:612-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shiba, T., K. Ishiguro, N. Takemoto, H. Koibuchi, and K. Sugimoto. 1995. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J. Bacteriol. 177:5872-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257-1260. [DOI] [PubMed] [Google Scholar]
- 237.Simpson, A. E., R. A. Skurray, and N. Firth. 2000. An IS257-derived hybrid promoter directs transcription of a tetA(K) tetracycline resistance gene in the Staphylococcus aureus chromosomal mec region. J. Bacteriol. 182:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Siu, L. K., J. Y. Lo, K. Y. Yuen, P. Y. Chau, M. H. Ng, and P. L. Ho. 2000. β-Lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like β-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Soki, J., E. Fodor, D. W. Hecht, R. Edwards, V. O. Rotimi, I. Kerekes, E. Urban, and E. Nagy. 2004. Molecular characterization of imipenem-resistant, cfiA-positive Bacteroides fragilis isolates from the USA, Hungary and Kuwait. J. Med. Microbiol. 53:413-419. [DOI] [PubMed] [Google Scholar]
- 240.Soki, J., M. Gal, J. S. Brazier, V. O. Rotimi, E. Urban, E. Nagy, and B. I. Duerden. 2006. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J. Antimicrob. Chemother. 57:212-220. [DOI] [PubMed] [Google Scholar]
- 241.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241a.Stapleton, P. D. 1999. Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457.
- 242.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stokes, H. W., and R. M. Hall. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26:10-19. [DOI] [PubMed] [Google Scholar]
- 244.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 245.Sun, T., M. Nukaga, K. Mayama, E. H. Braswell, and J. R. Knox. 2003. Comparison of beta-lactamases of classes A and D: 1.5- Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 12:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thode-Andersen, A. A. Mercer, and T. R. Klaenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid 31:60-71. [DOI] [PubMed] [Google Scholar]
- 247.Tenson, T., A. DeBlasio, and A. S. Mankin. 1996. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl. Acad. Sci. USA 93:5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tenson, T., and A. S. Mankin. 2001. Short peptides conferring resistance to macrolide antibiotics. Peptides 22:1661-1668. [DOI] [PubMed] [Google Scholar]
- 249.Tenson, T., L. Xiong, P. Kloss, and A. S. Mankin. 1997. Erythromycin resistance peptides selected from random peptide libraries. J. Biol. Chem. 272:17425-17430. [DOI] [PubMed] [Google Scholar]
- 250.Thierry, D., M. D. Cave, K. D. Eisenach, J. T. Crawford, J. H. Bates, B. Gicquel, and J. L. Guesdon. 1990. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 18:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Trinh, S., A. Haggoud, G. Reysset, and M. Sebald. 1995. Plasmids pIP419 and pIP421 from Bacteroides: 5-nitroimidazole resistance genes and their upstream insertion sequence elements. Microbiology 141:927-935. [DOI] [PubMed] [Google Scholar]
- 254.Truong-Bolduc, Q. C., P. M. Dunman, J. Strahilevitz, S. J. Projan, and D. C. Hooper. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187:2395-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Truong-Bolduc, Q. C., X. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Verdet, C., Y. Benzerara, V. Gautier, O. Adam, Z. Ould-Hocine, and G. Arlet. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob. Agents Chemother. 50:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Vila, J., A. Marcos, F. Marco, S. Abdalla, Y. Vergara, R. Reig, R. Gomez-Lus, and T. Jimenez de Anta. 1993. In vitro antimicrobial production of β-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferase by and susceptibility of clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 37:138-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Vingadassalom, D., A. Kolb, C. Mayer, T. Rybkine, E. Collatz, and I. Podglajen. 2005. An unusual primary sigma factor in the Bacteroidetes phylum. Mol. Microbiol. 56:888-902. [DOI] [PubMed] [Google Scholar]
- 260.Walker, S., L. Chen, Y. Hu, Y. Rew, D. Shin, and D. L. Boger. 2005. Chemistry and biology of ramoplanin: a lipoglycodepsipeptide with potent antibiotic activity. Chem. Rev. 105:449-476. [DOI] [PubMed] [Google Scholar]
- 261.Walsh, T. R. 2005. The emergence and implications of metallo-beta-lactamases in gram-negative bacteria. Clin. Microbiol. Infect. 11:2-9. [DOI] [PubMed] [Google Scholar]
- 262.Walsh, T. R., A. Onken, B. Haldorsen, M. A. Toleman, and A. Sundsfjord. 2005. Characterization of a carbapenemase-producing clinical isolate of Bacteroides fragilis in Scandinavia: genetic analysis of a unique insertion sequence. Scand. J. Infect. Dis. 37:676-679. [DOI] [PubMed] [Google Scholar]
- 263.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Weldhagen, G. F. 2004. Integrons and beta-lactamases—a novel perspective on resistance. Int. J. Antimicrob. Agents 23:556-562. [DOI] [PubMed] [Google Scholar]
- 265.Werckenthin, C., and S. Schwarz. 2000. Molecular analysis of the translational attenuator of a constitutively expressed erm(A) gene from Staphylococcus intermedius. J. Antimicrob. Chemother. 46:785-788. [DOI] [PubMed] [Google Scholar]
- 266.Werckenthin, C., S. Schwarz, and H. Westh. 1999. Structural alterations in the translational attenuator of constitutively expressed ermC genes. Antimicrob. Agents Chemother. 43:1681-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wohlleben, W., W. Arnold, L. Bissonnette, A. Pelletier, A. Tanguay, P. H. Roy, G. C. Gamboa, G. F. Barry, E. Aubert, J. Davies, et al. 1989. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC(3)-I), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 217:202-208. [DOI] [PubMed] [Google Scholar]
- 268.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2004. Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol. Lett. 236:137-143. [DOI] [PubMed] [Google Scholar]
- 269.Wright, G. D., T. R. Holman, and C. T. Walsh. 1993. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32:5057-5063. [DOI] [PubMed] [Google Scholar]
- 270.Yamamoto, K., H. Ogasawara, N. Fujita, R. Utsumi, and A. Ishihama. 2002. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 45:423-438. [DOI] [PubMed] [Google Scholar]