Abstract
One of the problems hindering the development of DNA vaccines is the relatively low immunogenicity often seen in humans and large animals compared to that in mice. In the present study, we tried to enhance the immunogenicity of a pcDNA1/faeG19 DNA vaccine in pigs by optimizing the FaeG expression plasmid and by coadministration of the plasmid vectors encoding the A and B subunits of the Escherichia coli thermolabile enterotoxin (LT). The insertion of a Kozak sequence and optimization of vector (cellular localization and expression) and both vector and codon usage were all shown to enhance in vitro FaeG expression compared to that of pcDNA1/faeG19. Subsequently, pcDNA1/faeG19 and the vector-optimized and the vector-codon-optimized construct were tested for their immunogenicity in pigs. In line with the in vitro results, antibody responses were better induced with increasing expression. The LT vectors additionally enhanced the antibody response, although not significantly, and were necessary to induce an F4-specific cellular response. These vectors were also added because LT has been described to direct the systemic response towards a mucosal immunoglobulin A (IgA) response in mice. Here, however, the intradermal FaeG DNA prime-oral F4 boost immunization resulted in a mainly systemic IgG response, with only a marginal but significant reduction in F4+ E. coli fecal excretion when the piglets were primed with pWRGFaeGopt and pWRGFaeGopt with the LT vectors.
Intestinal infections with enterotoxigenic Escherichia coli (ETEC) remain problematic for humans, pigs, and calves. The bacteria possess adhesins which allow them to colonize the small intestine and to produce enterotoxins which act locally on enterocytes. These adhesins bind to specific receptors on the enterocyte brush borders, and the absence of these receptors renders the animal resistant to bacterial colonization and consequently to ETEC-induced diarrhea (27). In pigs, F4 (K88) fimbriae are the best-characterized adhesins (37). They are long proteinaceous appendages composed mainly of several hundreds of identical adhesive subunits called FaeG (22).
Our laboratory developed a challenge model in pigs in which new vaccination strategies against ETEC can be tested. With this model, it was demonstrated that the oral administration of F4 fimbriae to weaned F4R+ pigs led to protection against a subsequent F4+ ETEC infection (35). In practice, a protective mucosal immune response often needs to be induced in the presence of maternal antibodies.
Several studies have shown that DNA vaccines, in contrast to conventional vaccines, can successfully prime immune responses in the presence of maternal antibodies, pointing out their potential use in young animals (3). Our previous pcDNA1/faeG19 DNA vaccine, however, was only marginally immunogenic in pigs (42). This is in agreement with other studies in large animals and humans, suggesting poor immunogenicity of DNA vaccines in these species (20, 39). Several approaches have been explored to enhance the potency of DNA vaccines. These include the optimization of expression (23, 30), the optimization of transfection efficiency (2), the targeting of DNA or the encoded antigen to dendritic cells (8, 29), and the coadministration of adjuvants, such as vector-encoded cytokines (16, 21). Promising results were also observed when DNA vaccines were used in heterologous prime-boost models (13, 32).
In the present study, we evaluated the impact of a Kozak sequence and the optimization of vector usage [intron A, bovine growth hormone poly(A), tissue plasminogen activator (tPA) leader sequence] and codon usage on the in vitro FaeG expression using two porcine, one human, and one nonhuman primate cell line. Subsequently, the influence of expression optimization and that of coadministration of plasmids encoding the LTA and LTB subunits (collectively named the thermolabile enterotoxin [LT] vectors) was analyzed on the immunogenicity of the FaeG DNA vaccine in pigs. Additionally, the priming of an F4-specific intestinal mucosal immune response was assessed in an intradermal (i.d.) DNA prime-oral F4 boost protocol.
MATERIALS AND METHODS
Isolation of F4 fimbriae.
F4 fimbriae were isolated from ETEC strains GIS26 (O149:K91:F4ac; LT+ STa+ STb+) and IMM 01 (O147:F4ac; LT+ STb+) as previously described (35). GIS26 F4 fimbriae were used for the enzyme-linked immunosorbent assay (ELISA), enzyme-linked immunospot, and oral immunizations. IMM01 fimbrial solutions were used in the FaeG-specific lymphocyte proliferation assay as they don't contain flagellin (40).
Plasmids.
pcDNA1/faeG19 and pWRGFaeG were constructed as previously described by cloning the GIS26 faeG sequence into pcDNA1 and pWRG7079, respectively (21, 42). The latter vector contains the tPA signal sequence, which allows extracellular secretion of the vector-encoded protein. The faeGK fragment was derived from pcDNA1/faeG19 by PCR using a forward primer inserting a Kozak sequence (GCC ACC ATG G) around the ATG translation initiation codon (underlined) and was subsequently cloned into pcDNA1, resulting in pcDNA1/faeGK. The faeG sequence was optimized for porcine expression by using the GenScript algorithm, increasing the codon adaptation index (28) of the faeG from 0.542 to 0.811. This codon-optimized sequence was constructed synthetically (GenScript Corp., Edison, NJ) and subsequently cloned into pWRG7079, resulting in pWRGFaeGopt. The pcDNA3-rpGM-CSF plasmid encodes the porcine granulocyte-macrophage colony-stimulating factor. pJV2004 and pJV2005 (1) encode the LTA and LTB subunits, respectively, behind a tPA signal sequence. Each construct was verified by sequencing the insert and its junction site, and large-scale purification was conducted by QIAGEN Endofree plasmid kits (QIAGEN GmbH, Germany).
In vitro expression of recombinant FaeG.
Swine kidney (SK-6 and PK-15), human embryonic kidney (293T), and monkey kidney (COS-7) cells were grown in Dulbecco's modified Eagle's medium (GIBCO BRL, Paisley, Scotland) supplemented with penicillin (100 IU/ml) (GIBCO BRL), streptomycin (100 μg/ml) (GIBCO BRL), l-glutamine (400 mM) (GIBCO BRL), and 10% fetal calf serum. The cells (2 × 105 cells for each transfection) were transfected transiently using Lipofectamine 2000 (Invitrogen, Merelbeke, Belgium) following the manufacturer's protocol. Twenty-four, 48, 72, and 96 h after transfection, culture media (supernatants) were collected and centrifuged to remove cell debris, after which transfected cells were collected, resuspended in phosphate-buffered saline, and lysed by freezing three times (−80°C) and thawing. Each transfection was performed four times. The amount of FaeG was quantified by an F4-specific ELISA as previously described (42) using a serial dilution of a known concentration of purified F4 to establish the standard curve.
Immunization experiment.
Thirty-three F4-seronegative conventionally bred pigs (Belgian Landrace × Piétrain) were weaned at the age of 4 weeks and housed in isolation units. Starting 1 day before weaning, all animals were orally treated during 5 successive days with colistin (150,000 U/kg of body weight; Colivet; Prodivet Pharmaceuticals, Eynatten, Belgium) to prevent E. coli infections during the weaning period.
All pigs were injected i.d. at the left side of the neck with 250 μg of pcDNA3-rpGM-CSF at the age of 6 weeks. Seven and 28 days later (days 0 and 21 postprimary immunization [ppi]), the pigs were i.d. immunized at the pcDNA3-rpGM-CSF injection site with 500 μg of pcDNA1/faeG19 (n = 7; pcDNA1/faeG19 group), pWRGFaeG (n = 7; pWRGFaeG group), pWRGFaeGopt (n = 7; pWRGFaeGopt group), pWRGFaeGopt supplemented with 100 μg pJV2004 and 100 μg pJV2005 (n = 6; pWRGFaeGopt+LT group), or the empty pWRG7079 vector (n = 6; control group). Plasmids were diluted in sterile phosphate-buffered saline and given by multiple injections of a total volume of 1 ml following the sedation of the pigs with azaperone (2 mg/kg; Stressnil, Janssen-Cilag, Beerse, Belgium). All of the pigs were orally boosted with 1 mg F4 fimbriae at days 43, 44, and 45 ppi as described earlier (34). In each group, some animals were euthanized at day 49 or 50 ppi to evaluate antibody responses in the spleen, mesenteric lymph nodes (Mes Lnd; jejunal and ileal), jejunal Peyer's patches (JPP), ileal Peyer's patch (IPP), and lamina propia (LP) (see Fig. 3). The remaining pigs were challenged intragastrically with 1010 F4+ ETEC at days 51 and 52, respectively, as described by Cox et al. (5). Three weeks after challenge, these pigs were euthanized. Euthanasia was performed by intravenous injection of pentobarbital (24 mg/kg; Nembutal, Sanofi Santé Animale, Brussels, Belgium), followed by exsanguination. The experimental procedure was performed following the guidelines of the animal care and ethics committee of our faculty (EC2004/85).
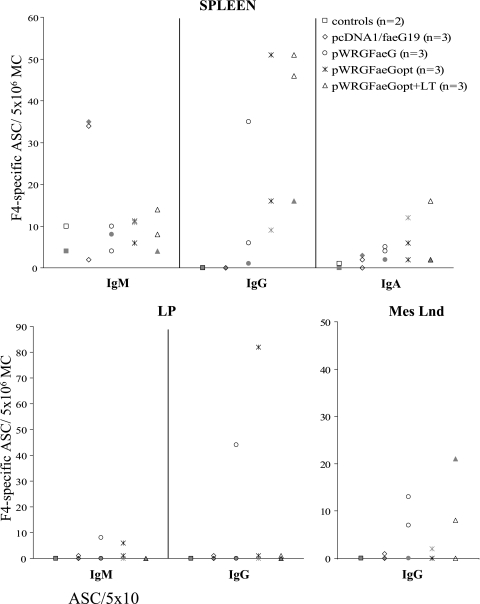
FIG. 3.
F4-specific ASCs per 5 × 106 MCs in the spleen, LP, and Mes Lnd at day 49 or 50 ppi. Pigs were i.d. immunized with DNA and orally boosted with F4 fimbriae as shown in Fig. 1. F4R-negative pigs are represented in gray.
Samples and tests.
Sera were sampled at regular intervals (Fig. 1) and used to determine the F4-specific immunoglobulin G (IgG), IgA, and IgM titers by an indirect ELISA (34). The cutoff values in this ELISA were 0.82, 0.57, and 0.29 for IgM, IgG, and IgA, respectively.
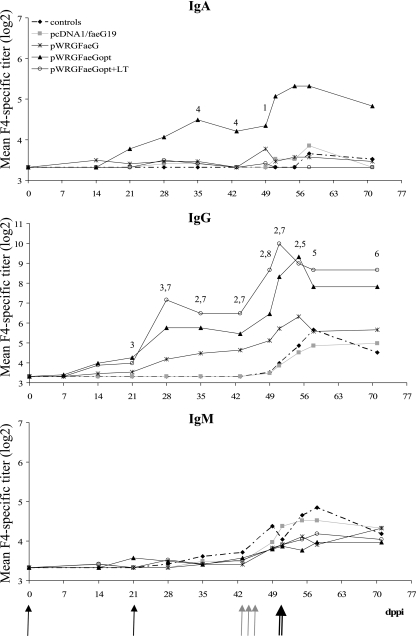
FIG. 1.
Mean F4-specific serum IgA, IgG, and IgM titers following two i.d. immunizations (black arrows) with pcDNA1/faeG19, pWRGFaeG, pWRGFaeGopt, pWRGFaeGopt plus the LT vectors or pWRG7079 (controls), an oral F4 protein boost (all pigs, gray arrows) and an F4+ ETEC challenge (double arrow). Shown are significant differences (P < 0.05) of the pWRGFaeGopt group with the pcDNA1/faeG19 group (1); the control and the pcDNA1/faeG19 group (2); the control, pcDNA1/faeG19 and the pWRGFaeG group (3); all other groups (4) and significant differences of the pWRGFaeGopt+LT group with the pcDNA1/faeG19 group (5); the control and the pcDNA1/faeG19 group (6); the control, pcDNA1/faeG19 and the pWRGFaeG group (7) and all other groups (8). Significant differences between time points are mentioned in the text.
At day 35 ppi, peripheral blood mononuclear cells (PBMCs) were isolated (34) to determine the FaeG-specific lymphocyte proliferation (38).
At day 49 or 50 ppi, MCs were isolated from the spleen, Mes Lnd (jejunal and ileal), JPP, IPP, and LP to enumerate the number of IgG, IgA, and IgM ASCs per 5 × 106 MCs by enzyme-linked immunospot (34). Furthermore, contents were collected from the duodenum, jejunum and ileum (41) and were tested for FaeG-specific IgG and IgA by an indirect ELISA (34). Cutoff values were 0.22 and 0.26 for IgG and IgA, respectively.
After challenge infection, fecal samples were collected daily until day 7 postchallenge and the F4+ E. coli isolates in these samples were enumerated by dot blotting as described previously (35).
The F4 receptor (F4R) status of all pigs was determined by an in vitro villus adhesion assay on small intestinal villi (36).
Statistical analysis.
Statistical analysis was performed using SPSS 12.0 for Windows. Differences in FaeG expression between different constructs within each cell line were tested with a paired-sample t test. Differences in log2 serum antibody titers and in log10 fecal ETEC excretions between groups were analyzed using the general linear model (repeated measures analysis of variance) with the Bonferroni adjustment for multiple comparisons. Differences in log2 serum antibody titers between different time points within groups were tested with a paired-sample t test. Differences in duration of fecal F4+ E. coli excretion between groups were tested using one-way analysis of variance. Differences between groups in lymphocyte proliferation were analyzed with the Kruskal-Wallis test. A P value of <0.05 was considered statistically significant.
RESULTS
In vitro FaeG expression.
Overall, the insertion of a Kozak sequence in pcDNA1/faeG19 significantly increased the FaeG expression. As can be seen in Table 1, pcDNA1/faeGK induced a 1.6- to 4.5-fold increase in expression. The usage of pWRG7079 instead of pcDNA1 further enhanced this expression in 293T and COS-7 cells. However, in only COS-7 cells was this increase significant compared to the expression by pcDNA1/faeGK. The expression in both porcine cell lines was not improved by pWRG7079. In fact, expression became similar again to the expression induced by pcDNA1/faeG19. Codon optimization of FaeG in pWRGFaeG resulted in the highest expression in COS-7 as well as in 293T cells, whereas expression in SK-6 and PK-15 cells increased again towards the level of pcDNA1/faeGK. Only 24 h after the transfection of the porcine cell lines, expression was higher. Taken together, vector optimization improved the expression by up to 1.2-, 1.8-, 5.7-, and 10.0-fold and both vector and codon optimization increased the expression by up to 2.7-, 3.6-, 9.5-, and 11.5-fold in SK-6, PK-15, 293T, and COS-7 cells, respectively, relative to pcDNA1/faeG19 (Table 1).
TABLE 1.
Influence of expression optimization on the in vitro FaeG expression in COS-7, 293T, SK-6, and PK-15 cells
| Cell line | Construct | Total FaeG expression (ng/2 × 105 cells) ata:
|
FaeG in SN (% of total FaeG expression) at:
|
Optimized expression/unoptimized expressionb at:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | ||
| COS-7 | pcDNA1/faeG19 | 1.6 ± 0.6 | 3.8 ± 0.5 | 5.8 ± 1.1 | 6.8 ± 1.4 | 0.0 ± 0.0 | 61.2 ± 13.5 | 61.9 ± 7.8 | 89.4 ± 8.1 | ||||
| pcDNA1/faeGK | 7.7 ± 2.2 (1) | 7.9 ± 1.1 (1) | 14.2 ± 3.1 (1) | 12.7 ± 3.3 | 0.0 ± 0.0 | 29.5 ± 8.3 | 55.0 ± 8.5 | 93.4 ± 2.1 | 4.5 ± 0.1 | 2.1 ± 0.3 | 2.5 ± 0.2 | 2.4 ± 0.6 | |
| pWRGFaeG | 21.0 ± 3.5 (1, 2) | 36.7 ± 4.8 (1, 2) | 47.1 ± 6.8 (1, 2) | 52.3 ± 9.1 (1) | 18.4 ± 8.7 | 31.7 ± 8.5 | 50.6 ± 9.2 | 67.3 ± 7.3 | 9.4 ± 1.4 | 10.0 ± 1.5 | 8.5 ± 1.0 | 8.3 ± 1.9 | |
| pWRGFaeGopt | 22.4 ± 1.4 (1, 2) | 41.8 ± 1.4 (1, 2) | 55.2 ± 4.8 (1, 2) | 54.5 ± 11.6 (2, 3) | 16.1 ± 7.5 | 31.2 ± 4.7 | 56.2 ± 8.1 | 76.8 ± 6.9 | 11.2 ± 1.3 | 11.5 ± 1.1 | 10.4 ± 1.6 | 10.2 ± 2.3 | |
| 293T | pcDNA1/faeG19 | 6.8 ± 2.3 | 33.0 ± 10.9 | 57.5 ± 16.1 | 50.5 ± 10.1 | 0.0 ± 0.0 | 9.0 ± 5.3 | 3.1 ± 1.0 | 8.3 ± 3.0 | ||||
| pcDNA1/faeGK | 17.8 ± 4.0 (1) | 60.9 ± 11.5 | 94.9 ± 20.5 (1) | 93.6 ± 15.7 (1) | 0.0 ± 0.0 | 5.2 ± 1.8 | 4.4 ± 1.8 | 7.5 ± 0.2 | 2.6 ± 0.6 | 1.7 ± 0.2 | 1.8 ± 0.2 | 1.9 ± 0.3 | |
| pWRGFaeG | 38.5 ± 8.7 (1) | 86.0 ± 12.1 (1) | 98.3 ± 20.7 | 112.6 ± 21.5 (1) | 11.5 ± 2.0 | 30.4 ± 11.0 | 31.4 ± 9.2 | 32.8 ± 2.0 | 5.7 ± 0.5 | 2.5 ± 0.4 | 1.9 ± 0.3 | 2.3 ± 0.3 | |
| pWRGFaeGopt | 71.5 ± 14.4 (1, 3) | 137.6 ± 10.6 (1, 2, 3) | 166.6 ± 28.7 (1, 2, 3) | 157.4 ± 26.5 (1, 2) | 7.5 ± 2.5 | 27.8 ± 10.8 | 17.4 ± 2.2 | 33.3 ± 3.2 | 9.5 ± 0.6 | 4.3 ± 1.0 | 3.6 ± 0.9 | 3.2 ± 0.4 | |
| SK-6 | pcDNA1/faeG19 | 14.7 ± 5.4 | 21.5 ± 2.7 | 29.8 ± 0.5 | 32.4 ± 8.1 | 0.0 ± 0.0 | 15.5 ± 5.1 | 38.4 ± 5.9 | 56.7 ± 5.8 | ||||
| pcDNA1/faeGK | 27.4 ± 8.1 (1) | 43.2 ± 3.3 (1, 3) | 47.2 ± 2.9 (1, 3) | 49.1 ± 10.8 (1) | 0.9 ± 0.9 | 15.2 ± 3.0 | 43.1 ± 6.0 | 68.5 ± 7.0 | 2.1 ± 0.3 | 2.1 ± 0.2 | 1.6 ± 0.1 | 1.6 ± 0.1 | |
| pWRGFaeG | 16.1 ± 4.1 | 15.9 ± 4.5 | 26.5 ± 6.9 | 27.0 ± 8.7 | 13.2 ± 10.3 | 40.9 ± 9.8 | 76.9 ± 3.1 | 84.6 ± 5.7 | 1.2 ± 0.1 | 0.9 ± 0.3 | 0.9 ± 0.2 | 0.9 ± 0.2 | |
| pWRGFaeGopt | 35.6 ± 8.1 (1, 3) | 37.7 ± 8.0 | 41.2 ± 9.7 (3) | 42.1 ± 7.8 (3) | 13.6 ± 8.5 | 44.1 ± 5.2 | 72.1 ± 2.7 | 84.9 ± 4.6 | 2.7 ± 0.3 | 1.9 ± 0.6 | 1.4 ± 0.3 | 1.5 ± 0.4 | |
| PK-15 | pcDNA1/faeG19 | 11.7 ± 2.8 | 22.0 ± 7.0 | 24.9 ± 6.2 | 30.1 ± 9.1 | 1.1 ± 1.1 | 16.4 ± 3.4 | 26.8 ± 3.6 | 43.9 ± 3.5 | ||||
| pcDNA1/faeGK | 22.5 ± 7.8 | 37.6 ± 10.5 (1) | 44.5 ± 16.0 | 54.7 ± 18.9 | 0.4 ± 0.4 | 13.0 ± 2.1 | 30.8 ± 4.0 | 54.9 ± 4.3 | 1.7 ± 0.3 | 2.0 ± 0.4 | 1.6 ± 0.3 | 1.6 ± 0.3 | |
| pWRGFaeG | 19.2 ± 4.0 (1) | 17.0 ± 1.9 | 21.9 ± 2.7 | 24.1 ± 7.0 | 9.5 ± 7.0 | 36.9 ± 8.7 | 65.0 ± 4.4 | 81.7 ± 7.1 | 1.8 ± 0.2 | 1.4 ± 0.7 | 1.0 ± 0.2 | 0.9 ± 0.1 | |
| pWRGFaeGopt | 41.1 ± 10.2 (1, 2, 3) | 39.1 ± 7.7 (1, 3) | 43.0 ± 9.8 (1) | 50.3 ± 15.0 (1, 3) | 17.2 ± 7.0 | 43.6 ± 5.6 | 60.9 ± 5.7 | 76.7 ± 9.2 | 3.6 ± 0.2 | 2.5 ± 0.8 | 1.8 ± 0.2 | 1.7 ± 0.1 | |
The calculated total includes FaeG expressed in cell lysates and FaeG expressed in supernatant (SN). The numbers 1, 2, and 3 in parentheses indicate significant increases (P < 0.05) compared to pcDNA1/faeG19, pcDNA1/faeGK, and pWRGFaeG, respectively.
Values are ratios. Optimized expression is the total amount of FaeG expressed after transfection with pcDNA1/faeGK, pWRGFaeG, or pWRGFaeGopt. Unoptimized expression is the total amount of FaeG detected after transfection with pcDNA1/faeG19. Results are the means of four transfections ± SEM.
Following transfection with the secretion constructs pWRGFaeG and pWRGFaeGopt, no correlation was seen between the total amount of FaeG expressed and the amount secreted in the supernatant. The fraction of FaeG in supernatant increased with time, whereas this increase was less pronounced for the total FaeG expression but remained remarkably lower for 293T cells. The transfection with the nonsecretion constructs pcDNA1/faeG19 and pcDNA1/faeGK hardly resulted in FaeG in the supernatant after 24 h, but from then on, the percentage of FaeG in the supernatant also increased with time.
In vivo immunogenicity of the FaeG DNA vaccines and adjuvant effect of the LT vectors.
All pigs were i.d. injected with pcDNA3-rpGM-CSF 1 week before the first DNA vaccination as we previously reported that this injection enhanced the induction of F4-specific immune responses (21). To evaluate the influence of expression optimization on the immunogenicity of the FaeG DNA vaccine in pigs, we opted for pcDNA1/faeG19 and pWRGFaeG because we already used these constructs in an immunization experiment (21, 42) and for pWRGFaeGopt because it gave the highest FaeG expression in porcine cells. In addition, we evaluated the influence of the LT vectors as LT has previously been shown to induce a mucosal immune response when applied to the skin (9, 12).
F4-specific immune responses following intradermal DNA vaccination.
Intradermal DNA vaccination did not induce an F4-specific serum IgM response, whereas a statistically significant F4-specific serum IgA response was detected in only the pWRGFaeGopt group, from 2 weeks after the second immunization onwards. Nevertheless, most groups showed an IgG response of which the height and speed of onset increased in the order pcDNA1/faeG19, pWRGFaeG, pWRGFaeGopt (Fig. 1). Indeed, none of the pigs of the pcDNA1faeG19 and the pWRG7079 group showed F4-specific serum IgG following DNA vaccination, whereas in the pWRGFaeG group, F4-specific IgG was observed in one pig on day 14 ppi and in six of seven pigs 3 weeks after the second immunization. Even more pigs showed F4-specific IgG in the pWRFaeGopt group, namely, four of seven pigs on day 14 ppi and all animals on day 28 ppi. The addition of the LT vectors to pWRGFaeGopt further enhanced this response so that five of six pigs were positive on day 14 ppi and the whole group was positive on day 21 ppi. Consequently, the F4-specific serum IgG titer was significantly higher in the pWRGFaeGopt+LT and the pWRGFaeGopt groups on days 28, 35, and 43 and days 21, 28, and 35 ppi, respectively, than in the other groups.
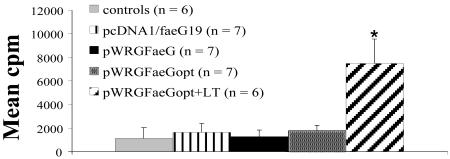
F4-specific proliferation of PBMCs collected at day 35 ppi was observed in only the LT-supplemented group (P < 0.05) (Fig. 2). Medium proliferation was 482 ± 235 (mean counts per minute ± standard error of the mean [SEM]) and not significantly different between the groups.
FIG. 2.
F4-specific proliferation of PBMCs at day 35 ppi. Results are presented as counts per minute. An asterisk indicates significant differences (P < 0.05) between the pWRGFaeGopt+LT group and the other groups. Error bars indicate standard errors of the means.
Oral booster immunization with F4 fimbriae.
To demonstrate priming of the intestinal mucosal immune system by the intradermal DNA vaccination, all pigs received an oral boost with F4. This boost slightly increased the mean F4-specific IgM titer within each group, but no significant differences were observed between the groups. The F4-specific serum IgA titer increased in only one pig of the pWRGFaeGopt group (titer 80), one of the LT group (titer 15), and three of the pWRGFaeG group (titers ranging from 15 to 40). No significant difference was seen between groups. However, a significant increase of the F4-specific serum IgG titer did occur in the pWRGFaeGopt+LT group at day 49 ppi. At that day, a 4.5-fold increase in the mean F4-specific IgG titer was observed in this group, compared to 2-, 1.4-, 1.1-, and 1.1-fold increases in the pWRGFaeGopt, the pWRGFaeG, the pcDNA1/faeG19, and the control groups, respectively. In the latter group, only the F4R+ control pigs showed this increase. Such a difference in response between F4R+ and F4R-negative pigs was not observed for the FaeG DNA-primed pigs.
To localize and quantify the antibody response following the F4 boost, three pigs of each FaeG DNA-vaccinated group and two control pigs were euthanized at day 49 or 50 ppi. One pig in each of these groups was F4R-negative as shown in the in vitro villus adhesion assay. The most important difference between groups was observed in the spleen and, to a lesser extent, in the Mes Lnd (Fig. 3). In the spleen, no IgG antibody-secreting cells (ASCs) could be observed for the control and the pcDNA1/faeG19 groups, while all tested pigs of the three pWRG groups showed IgG ASCs. Furthermore, the number of F4-specific IgG ASCs increased in the order pWRGFaeG, pWRGFaeGopt, pWRGFaeGopt+LT, with medians of 6, 16, and 46 ASCs per 5 × 106 MCs, respectively. Such a difference was not observed for IgM and IgA ASCs, except for the pcDNA1/faeG19 group, which showed a higher IgM response (Fig. 3).
Also in gut-associated lymphoid tissue, F4-specific IgG ASCs could be detected in only the FaeG DNA-primed animals. In the LP, high numbers of F4-specific IgG ASCs and low numbers of F4-specific IgM ASCs were observed in one pig of the pWRGFaeG group (44 and 8 ASCs per 5 × 106 MCs, respectively) and one pig of the pWRGFaeGopt group (82 and 6 ASCs per 5 × 106 MCs, respectively) (Fig. 3). These pigs were both F4R+. In the Mes Lnd, a low F4-specific IgG response could be detected in two pWRGFaeG (7 and 13 ASCs per 5 × 106 MCs) and two pWRGFaeGopt+LT-primed pigs (8 and 21 ASCs per 5 × 106 MCs), irrespective of the F4R status. IgG ASCs were nearly absent (less than five ASCs per 5 × 106 MCs) in the Mes Lnd of the other DNA-primed pigs (Fig. 3) and in the IPP and JPP of all pigs (data not shown). Low numbers of F4-specific IgM (0 to 12 ASCs per 5 × 106 MCs) and IgA ASCs (0 to 14 ASCs per 5 × 106 MCs) could be detected in the IPP, JPP, and Mes Lnd and in all gut-associated lymphoid tissue, respectively, independent of DNA priming (data not shown).
In the intestinal contents, F4-specific IgA and IgG antibodies could be detected in the duodenum of the F4R+ pWRGFaeG pig that showed F4-specific IgG ASCs in its LP and throughout the small intestine of the two F4R+ pigs of the pWRGFaeGopt group (titers of 2 to 16; data not shown).
F4+ ETEC challenge.
To determine whether the induced F4-specific immune response was protective, the remaining pigs were intragastrically challenged with F4+ ETEC at days 51 and 52 ppi. The in vitro villus adhesion assay showed that one pig of the control group and one pig of the pcDNA1/faeG19 group were F4R negative. Data from these pigs were not further considered.
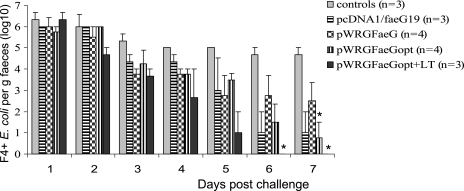
All groups excreted F4+ E. coli after challenge. Compared to that in the control group, excretion was significantly reduced in the pWRGFaeGopt group at day 7 and in the pWRGFaeGopt+LT group at days 6 and 7 postchallenge. Moreover, the duration of the fecal F4+ E. coli excretion was reduced in the pWRGFaeGopt+LT group compared to the control group with at least 2 days (P = 0.081) (Fig. 4).
FIG. 4.
Mean F4+ E. coli excretion per gram of feces (log10) following intragastric challenge with F4+ E. coli at days 51 and 52 ppi. Pigs were i.d. primed with DNA and orally boosted with F4 fimbriae as shown in Fig. 1. An asterisk indicates significant differences (P < 0.05) in excretion with the control group. Error bars indicate standard errors of the means.
After challenge, the serum IgM titer increased 1.7-fold (day 58 ppi) in the control group, whereas almost no increase occurred in the DNA-primed groups (Fig. 1). Furthermore, a significant increase in mean serum IgG titer could be observed in the control group (P = 0.025) at day 55 ppi. Insignificant increases occurred in the DNA-primed groups, except for the pWRGFaeGopt and the pWRGFaeGopt+LT group. Here, the IgG titer remained significantly higher for the pWRGFaeGopt group until day 55 and for the pWRGFaeGopt+LT until day 71 ppi. The serum IgA titer increased slightly in all groups except for the pWRGFaeGopt+LT group (Fig. 1).
DISCUSSION
Introducing a “Kozak” consensus sequence for translation initiation (17) into prokaryotic genes has been suggested to increase their expression levels in eukaryotic cells (11). Hereby, the presence of, especially, a purine (G/A) at the −3 and a G at the +4 position is supposed to be important (18). As pcDNA1/faeG19 contains a T at −3 and an A at the +4 position, it was not surprising that the insertion of a Kozak sequence improved the FaeG expression. The further improvement observed with the pWRG7079 vector in COS-7 and 293T cells may be due to the presence of the intron A, which acts as an enhancer of the cytomegalovirus immediate early promoter (4, 43), and/or to the use of the bovine growth hormone poly(A) instead of the simian virus 40 poly(A) used by pcDNA1 (23, 25). The presence of the tPA coding sequence could also have contributed since its AUG start codon is flanked by an A at −3 and a G at the +4 position. Furthermore, it should be mentioned that the tPA leader sequence directs FaeG to the secretory pathway, making it a target for N glycosylation. Indeed, the FaeG contains three putative N glycosylation sites. Nevertheless, although N glycosylation of the FaeG has been demonstrated to occur in plants, it did not abolish the immunogenic character of the FaeG in that study (15). Furthermore, as pWRGFaeG is far more immunogenic than pcDNA1/faeG19, there is no evidence that N glycosylation would interfere with the immunogenicity of the FaeG in our study. Moreover, antibodies induced following the immunization of pigs with pWRGFaeG were able to block the adhesion of F4+ ETEC to porcine intestinal villi, indicating that they retained their biological activity (unpublished results). A positive influence of the pWRG7079 vector on expression was not observed for the PK-15 and the SK-6 cell lines. Possibly, by improving transcription initiation and termination, the process of translation might have become rate limiting. Indeed, codon optimization significantly increased the FaeG expression in these cell lines and in 293T cells but not in COS-7 cells. Furthermore, COS-7 cells showed the highest increase in FaeG expression by using pWRG7079, suggesting that transcription, rather than tRNA availability and/or mRNA stability, was the rate limiting factor in these cells. Moreover, the fact that codon optimization also appeared to be successful in the human cell line was not unexpected since the preferred codon usage in pigs is very similar to that in humans (24).
In accordance with the increasing FaeG expression in vitro, F4-specific antibody responses were better induced in the order pcDNA1/faeG19, pWRGFaeG, and pWRGFaeGopt. The failure of pcDNA1/faeG19 to induce an F4-specific serum antibody response is most likely due to the absence of FaeG secretion, limiting the amount of antigen available for B-cell priming. Furthermore, the possibility that differences in CpG motifs between the vectors as well as due to codon optimization influenced the immunization cannot be ruled out. Indeed, it is well known that CpG added to a vaccine can have an adjuvant effect on the immunization via interaction with Toll-like receptor 9. Changes in the CpG content might thus result in changes in the adjuvant effect of the DNA vaccine. However, it should be mentioned that the potential adjuvant effect of CpG in the vaccine for large animals is still a matter of discussion. While studies in mice demonstrated enhanced humoral responses by inserting CpG motifs in the vector backbone (19), the insertion of up to 88 ruminant-specific CpG motifs did not quantitatively affect the humoral response in cattle (26).
F4-specific lymphocyte proliferation of PBMCs could be demonstrated in only the group that received the LT vectors. A positive effect of LT on cellular responses was also observed by Arrington et al. (1), who evaluated the same vectors for the adjuvant effect in mice. In their study, augmented cellular responses were accompanied by a Th1-modulating effect and were not due to CpG motifs in the vectors, but mainly resulted from the expressed LTA and LTB subunits. In our study, the addition of the LT vectors enhanced the serum IgG response but abolished the serum IgA response observed following immunization with pWRGFaeGopt. These results were confirmed by an IgA capture ELISA (data not shown), eliminating the possibility that the higher IgG levels in the pWRGFaeGopt+LT group competed with binding of IgA in ELISA. Although cytokine expression profiles and IgG subclass ratios remain to be tested, a likely explanation could be the induction of a Th1-like response by the addition of the LT vectors as observed in mice. Indeed, Th1 is less favorable for IgA responses. Intradermal injection of the LT vectors was well tolerated by the pigs, as a mild local reaction could be demonstrated in only three out of six pigs. This reaction resolved by day 7 (data not shown).
The oral F4 administration induced a secondary serum IgG response in the pWRGFaeGopt and especially in the pWRGFaeGopt+LT group. This response was independent of the F4R status, which was not completely unexpected since oral F4 administration has previously been shown to prime systemic responses in both F4R+ and F4R-negative pigs (33). Studying the ASCs in different tissues revealed that the antibody response was mainly induced systemically. Only a few pigs showed an intestinal mucosal immune response with antibodies which were mainly IgG instead of IgA. Normally serum IgA provides the primary defense against intestinal pathogens like ETEC. Nevertheless, priming with pWRGFaeGopt significantly reduced the fecal F4+ E. coli excretion, whereas the coadministration of the LT vectors additionally reduced the duration of fecal excretion. The reason for this partial protection is not clear, but the IgG response might play a role. Indeed, a study by Yu et al. (44) demonstrated the protection of mice against an oral LT challenge by a passive infusion of serum-derived anti-LT IgG. It has been demonstrated that IgG can be transported towards the intestinal lumen via the bidirectional IgG transporting neonatal Fc receptor (FcRn) (7). The expression of the FcRn receptor has been demonstrated on the intestinal epithelium of young and of adult humans and pigs (14, 31). FcRn-mediated IgG transport might explain the presence of F4-specific IgG in the intestinal contents of some DNA-primed pigs. Once in the intestinal lumen, IgG could contribute to protection by neutralizing adhesins so inhibiting bacterial colonization. Indeed, oral administration of milk-derived anti-colonization factor antigen I IgG has protected human volunteers against a colonization factor antigen I-positive ETEC infection (10).
In conclusion, our results showed that the optimization of the expression construct and coadministration of the LT vectors strongly enhanced the immunogenicity of our FaeG DNA vaccine in pigs. Nevertheless, the i.d. FaeG DNA prime-oral F4 boost immunization induced a mainly systemic IgG response, failing to prevent F4+ E. coli excretion upon challenge. For the development of a vaccination strategy which can prevent postweaning diarrhea, further optimizations are required. The inclusion of immunomodulators, such as vitamin D3, which can modulate a systemic response towards a mucosal one (6) or strategies which enable the delivery of the DNA vaccine to the intestinal mucosa might result in the induction of the local intestinal mucosal response that is needed.
Acknowledgments
This work was supported by the Belgian Federal Ministry of Public Health, Federal Environment Division, the Research Found of Ghent University (BOF) and the “FWO-Vlaanderen.”
We thank Powderject Vaccines, Inc. for providing pWRG7079, pJV2004, and pJV2005 and Pirbright Laboratory, IAH, for providing pcDNA3-rpGM-CSF. We also wish to acknowledge Griet De Smet, Denise Slos, and Rudy Cooman for their technical assistance.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Arrington, J., R. P. Braun, L. Dong, D. H. Fuller, M. D. Macklin, S. W. Umlauf, S. J. Wagner, M. S. Wu, L. G. Payne, and J. R. Haynes. 2002. Plasmid vectors encoding cholera toxin or the heat-labile enterotoxin from Escherichia coli are strong adjuvants for DNA vaccines. J. Virol. 76:4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiuk, L. A., R. Pontarollo, S. Babiuk, B. Loehr, and S. van Drunen Littel-van den Hurk. 2003. Induction of immune responses by DNA vaccines in large animals. Vaccine 21:649-658. [DOI] [PubMed] [Google Scholar]
- 3.Bot, A., and C. Bona. 2002. Genetic immunization of neonates. Microbes Infect. 4:511-520. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, B. S., R. M. Thayer, K. A. Vincent, and N. L. Haigwood. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 19:3979-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, E., E. Schrauwen, V. Cools, and A. Houvenaghel. 1991. Experimental induction of diarrhoea in newly-weaned piglets. Zentbl. Vetmed. Reihe A 38:418-426. [DOI] [PubMed] [Google Scholar]
- 6.Cox, E., F. Verdonck, D. Vanrompay, and B. Goddeeris. 2006. Adjuvants modulating mucosal immune responses or directing systemic responses towards the mucosa. Vet. Res. 37:511-539. [DOI] [PubMed] [Google Scholar]
- 7.Dickinson, B. L., K. Badizadegan, Z. Wu, J. C. Ahouse, X. Zhu, N. E. Simister, R. S. Blumberg, and W. I. Lencer. 1999. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J. Clin. Investig. 104:903-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drew, D. R., J. S. Boyle, A. M. Lew, M. W. Lightowlers, P. J. Chaplin, and R. A. Strugnell. 2001. The comparative efficacy of CTLA-4 and L-selectin targeted DNA vaccines in mice and sheep. Vaccine 19:4417-4428. [DOI] [PubMed] [Google Scholar]
- 9.Enioutina, E. Y., D. Visic, and R. A. Daynes. 2000. The induction of systemic and mucosal immune responses to antigen-adjuvant compositions administered into the skin: alterations in the migratory properties of dendritic cells appears to be important for stimulating mucosal immunity. Vaccine 18:2753-2767. [DOI] [PubMed] [Google Scholar]
- 10.Freedman, D. J., C. O. Tacket, A. Delehanty, D. R. Maneval, J. Nataro, and J. H. Crabb. 1998. Milk immunoglobulin with specific activity against purified colonization factor antigens can protect against oral challenge with enterotoxigenic Escherichia coli. J. Infect. Dis. 177:662-667. [DOI] [PubMed] [Google Scholar]
- 11.Garmory, H. S., K. A. Brown, and R. W. Titball. 2003. DNA vaccines: improving expression of antigens. Genet. Vaccines Ther. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glenn, G. M., D. N. Taylor, X. Li, S. Frankel, A. Montemarano, and C. R. Alving. 2000. Transcutaneous immunization: a human vaccine delivery strategy using a patch. Nat. Med. 6:1403-1406. [DOI] [PubMed] [Google Scholar]
- 13.Guo, Y. J., S. H. Sun, Y. Zhang, Z. H. Chen, K. Y. Wang, L. Huang, S. Zhang, H. Y. Zhang, Q. M. Wang, D. Wu, and W. J. Zhu. 2004. Protection of pigs against Taenia solium cysticercosis using recombinant antigen or in combination with DNA vaccine. Vaccine 22:3841-3847. [DOI] [PubMed] [Google Scholar]
- 14.Israel, E. J., S. Taylor, Z. Wu, E. Mizoguchi, R. S. Blumberg, A. Bhan, and N. E. Simister. 1997. Expression of the neonatal Fc receptor, FcRn, on human intestinal epithelial cells. Immunology 92:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joensuu, J. J., M. Kotiaho, T. H. Teeri, L. Valmu, A. M. Nuutila, K. M. Oksman-Caldentey, and V. Niklander-Teeri. 2006. Glycosylated F4 (K88) fimbrial adhesin FaeG expressed in barley endosperm induces ETEC-neutralizing antibodies in mice. Transgenic Res. 15:359-373. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J. J., J. S. Yang, K. H. Manson, and D. B. Weiner. 2001. Modulation of antigen-specific cellular immune responses to DNA vaccination in rhesus macaques through the use of IL-2, IFN-γ, or IL-4 gene adjuvants. Vaccine 19:2496-2505. [DOI] [PubMed] [Google Scholar]
- 17.Kozak, M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196:947-950. [DOI] [PubMed] [Google Scholar]
- 18.Kozak, M. 1997. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 16:2482-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, X., X. Forns, R. Gutierrez, et al. 2002. DNA-based vaccination against hepatitis C virus (HCV): effect of expressing different forms of HCV E2 protein and use of CpG-optimized vectors in mice. Vaccine 20:3263-3271. [DOI] [PubMed] [Google Scholar]
- 20.MacGregor, R. R., J. D. Boyer, K. E. Ugen, K. E. Lacy, S. J. Gluckman, M. L. Bagarazzi, M. A. Chattergoon, Y. Baine, T. J. Higgins, R. B. Ciccarelli, L. R. Coney, R. S. Ginsberg, and D. B. Weiner. 1998. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J. Infect. Dis. 178:92-100. [DOI] [PubMed] [Google Scholar]
- 21.Melkebeek, V., F. Verdonck, E. Stuyven, B. Goddeeris, and E. Cox. 2006. Plasmid-encoded GM-CSF induces priming of the F4(K88)-specific serum IgA response by FaeG DNA vaccination in pigs. Vaccine 24:4592-4594. [DOI] [PubMed] [Google Scholar]
- 22.Mol, O., and B. Oudega. 1996. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol. Rev. 19:25-52. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery, D. L., J. W. Shiver, K. R. Leander, H. C. Perry, A. Friedman, D. Martinez, J. B. Ulmer, J. J. Donnelly, and M. A. Liu. 1993. Heterologous and homologous protection against influenza A by DNA vaccination: optimization of DNA vectors. DNA Cell Biol. 12:777-783. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norman, J. A., P. Hobart, M. Manthorpe, P. Felgner, and C. Wheeler. 1997. Development of improved vectors for DNA-based immunization and other gene therapy applications. Vaccine 15:801-803. [DOI] [PubMed] [Google Scholar]
- 26.Pontarollo, R. A., L. A. Babiuk, R. Hecker, and S. Van Drunen Littel-Van Den Hurk. 2002. Augmentation of cellular immune responses to bovine herpesvirus-1 glycoprotein D by vaccination with CpG-enhanced plasmid vectors. J. Gen. Virol. 83:2973-2981. [DOI] [PubMed] [Google Scholar]
- 27.Rutter, J. M., M. R. Burrows, R. Sellwood, and R. A. Gibbons. 1975. A genetic basis for resistance to enteric disease caused by E. coli. Nature 257:135-136. [DOI] [PubMed] [Google Scholar]
- 28.Sharp, P. M., and W. H. Li. 1987. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh, M., M. Briones, G. Ott, and D. O'Hagan. 2000. Cationic microparticles: a potent delivery system for DNA vaccines. Proc. Natl. Acad. Sci. USA 97:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberg, T., P. Ohlschlager, P. Sehr, W. Osen, and L. Gissmann. 2005. Modification of HPV 16 E7 genes: correlation between the level of protein expression and CTL response after immunization of C57BL/6 mice. Vaccine 23:1149-1157. [DOI] [PubMed] [Google Scholar]
- 31.Stirling, C. M., B. Charleston, H. Takamatsu, S. Claypool, W. Lencer, R. S. Blumberg, and T. E. Wileman. 2005. Characterization of the porcine neonatal Fc receptor—potential use for trans-epithelial protein delivery. Immunology 114:542-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toussaint, J. F., C. Letellier, D. Paquet, M. Dispas, and P. Kerkhofs. 2005. Prime-boost strategies combining DNA and inactivated vaccines confer high immunity and protection in cattle against bovine herpesvirus-1. Vaccine 23:5073-5081. [DOI] [PubMed] [Google Scholar]
- 33.Van den Broeck, W., H. Bouchaut, E. Cox, and B. M. Goddeeris. 2002. F4 receptor-independent priming of the systemic immune system of pigs by low oral doses of F4 fimbriae. Vet. Immunol. Immunopathol. 85:171-178. [DOI] [PubMed] [Google Scholar]
- 34.Van den Broeck, W., E. Cox, and B. M. Goddeeris. 1999. Induction of immune responses in pigs following oral administration of purified F4 fimbriae. Vaccine 17:2020-2029. [DOI] [PubMed] [Google Scholar]
- 35.Van den Broeck, W., E. Cox, and B. M. Goddeeris. 1999. Receptor-dependent immune responses in pigs after oral immunization with F4 fimbriae. Infect. Immun. 67:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van den Broeck, W., E. Cox, and B. M. Goddeeris. 1999. Receptor-specific binding of purified F4 to isolated villi. Vet. Microbiol. 68:255-263. [DOI] [PubMed] [Google Scholar]
- 37.Van den Broeck, W., E. Cox, B. Oudega, and B. M. Goddeeris. 2000. The F4 fimbrial antigen of Escherichia coli and its receptors. Vet. Microbiol. 71:223-244. [DOI] [PubMed] [Google Scholar]
- 38.Van der Stede, Y., E. Cox, F. Verdonck, S. Vancaeneghem, and B. M. Goddeeris. 2003. Reduced faecal excretion of F4+-E coli by the intramuscular immunisation of suckling piglets by the addition of 1α,25-dihydroxyvitamin D3 or CpG-oligodeoxynucleotides. Vaccine 21:1023-1032. [DOI] [PubMed] [Google Scholar]
- 39.van Rooij, E. M., B. L. Haagmans, Y. E. de Visser, M. G. de Bruin, W. Boersma, and A. T. Bianchi. 1998. Effect of vaccination route and composition of DNA vaccine on the induction of protective immunity against pseudorabies infection in pigs. Vet. Immunol. Immunopathol. 66:113-126. [DOI] [PubMed] [Google Scholar]
- 40.Verdonck, F., E. Cox, E. Schepers, H. Imberechts, J. Joensuu, and B. M. Goddeeris. 2004. Conserved regions in the sequence of the F4 (K88) fimbrial adhesin FaeG suggest a donor strand mechanism in F4 assembly. Vet. Microbiol. 102:215-225. [DOI] [PubMed] [Google Scholar]
- 41.Verdonck, F., E. Cox, K. van Gog, Y. Van der Stede, L. Duchateau, P. Deprez, and B. M. Goddeeris. 2002. Different kinetic of antibody responses following infection of newly weaned pigs with an F4 enterotoxigenic Escherichia coli strain or an F18 verotoxigenic Escherichia coli strain. Vaccine 20:2995-3004. [DOI] [PubMed] [Google Scholar]
- 42.Verfaillie, T., V. Melkebeek, V. Snoek, S. Douterlungne, E. Cox, F. Verdonck, D. Vanrompay, B. Goddeeris, and E. Cox. 2004. Priming of piglets against enterotoxigenic E. coli F4 fimbriae by immunization with FAEG DNA. Vaccine 22:1640-1647. [DOI] [PubMed] [Google Scholar]
- 43.Wang, S., D. Farfan-Arribas, S. Shen, T. H. Chou, A. Hirsch, F. He, and S. Lu. 2006. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine 24:4531-4540. [DOI] [PubMed] [Google Scholar]
- 44.Yu, J., F. Cassels, T. Scharton-Kersten, S. A. Hammond, A. Hartman, E. Angov, B. Corthesy, C. Alving, and G. Glenn. 2002. Transcutaneous immunization using colonization factor and heat-labile enterotoxin induces correlates of protective immunity for enterotoxigenic Escherichia coli. Infect. Immun. 70:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]