Abstract
Pumilio family (PUF) proteins affect specific genes by binding to, and inhibiting the translation or stability of, their transcripts. The PUF domain is required and sufficient for this function. One Saccharomyces cerevisiae PUF protein, Mpt5p (also called Puf5p or Uth4p), promotes stress tolerance and replicative life span (the maximum number of doublings a mother cell can undergo before entering into senescence) by an unknown mechanism thought to partly overlap with, but to be independent of, the cell wall integrity (CWI) pathway. Here, we found that mpt5Δ mutants also display a short chronological life span (the time cells stay alive in saturated cultures in synthetic medium), a defect that is suppressed by activation of CWI signaling. We found that Mpt5p is an upstream activator of the CWI pathway: mpt5Δ mutants display the appropriate phenotypes and genetic interactions, display low basal activity of the pathway, and are defective in activation of the pathway upon thermal stress. A set of mRNAs that specifically bind to Mpt5p was recently reported. One such putative target, LRG1, encodes a GTPase-activating protein for Rho1p that directly links Mpt5p to CWI signaling: Lrg1p inhibits CWI signaling, LRG1 mRNA contains a consensus Mpt5p-binding site in its putative 3′ untranslated region, loss of Lrg1p suppresses the temperature sensitivity and CWI signaling defects of mpt5Δ mutants, and LRG1 mRNA abundance is inhibited by Mpt5p. We conclude that Mpt5p is required for normal replicative and chronological life spans and that the CWI pathway is a key and direct downstream target of this PUF protein.
The Pumilio family (PUF) proteins are conserved among the eukaryotes (42). They bind to specific sequences in the 3′ untranslated region (3′UTR) of target transcripts via their conserved and characteristic PUF domain and thereby inhibit the stability or translatability of these target mRNAs (32, 50). Indeed, the PUF domain appears sufficient for PUF proteins to affect their target transcripts (32, 50). Five PUF proteins, Puf1p to Puf5p, were thought to exist in the budding yeast Saccharomyces cerevisiae (37, 49). A sixth, Puf6p, has recently been reported (9). None are essential (9, 37, 49). One of the yeast PUF proteins, Mpt5p, also known as Htr1p (23), Puf5p (37), or Uth4p (20), promotes replicative life span (3, 20, 21), the number of generations a virgin daughter cell can undergo before becoming senescent. Mpt5p is a robust regulator of ageing, since it also affects life span in a long-lived genetic background (17).
In addition to displaying a short replicative life span, mutants lacking MPT5 are sensitive to multiple stresses (20, 23); for example, they cannot proliferate at high temperatures (23), and haploid mutants are unable to recover from mating pheromone arrest (4, 23). Only one direct target transcript for Mpt5p, the HO transcript, has been unequivocally identified to date (43), although this target does not account for the stress tolerance and life span defects of mpt5Δ mutants. Recently, Mpt5p was found to bind over 200 cellular transcripts, a large fraction of which encode proteins involved in chromatin modification and transcriptional control (6). A unique consensus Mpt5p-binding sequence was identified in many of the putative targets (most commonly in their 3′UTRs), and Mpt5p was experimentally confirmed to bind to this consensus sequence (6). It is thus likely that many of the transcripts that bind to Mpt5p are bone fide in vivo targets of the PUF protein. A smaller and overlapping set of putative target transcripts has also been identified recently by three-hybrid analysis (41).
Like all PUF proteins, Mpt5p acts to inhibit its targets transcripts. Mpt5p stimulates the degradation of many if not all of its target transcripts (41, 43), at least in part by Mpt5p-triggered deadenylation. This effect may be direct, since Mpt5p binds to the Pop2p-Ccr4p deadenylase complex (7).
How does Mpt5p promote stress tolerance and long replicative life span? The answer is not yet known. It may do so by affecting one key target transcript or by a cumulative effect on multiple targets. It is always possible that a unique function of Mpt5p regulates life span/stress tolerance, a function that is distinct from the ability to bind to and regulate mRNA.
Intriguingly, MPT5 displays some genetic interactions (11, 16, 36) with genes encoding components of the stress response signaling pathway involving a yeast protein kinase C, Pkc1p (28). This cell wall integrity (CWI) pathway, like Mpt5p, also affects stress tolerance (27). Indeed, the overexpression of PKC1 or genes encoding other upstream components suppresses the temperature sensitivity (16, 36) and partly suppresses the replicative life span defect (16) of mpt5Δ mutants. Intriguingly, Mpt5p, like the CWI pathway, appears to promote cell integrity. mpt5Δ mutants are hypersensitive to cell wall damaging agents, and their temperature-sensitive growth defect is suppressed by osmotic stabilization (15). Although the CWI pathway is thus thought to act in parallel with Mpt5p to affect these functions (15, 16), the relationship is not firmly established.
The CWI pathway is activated by stresses to the cell surface (e.g., those caused by heat shock) that are sensed predominantly by a pair of sensors at the plasma membrane, Slg1p (also called Hcs77p or Wsc1p) (8, 13, 45) and Mid2p (22, 38). These sensors stimulate formation of the GTP-bound form of the small Rho1p GTPase that, in turn, directly binds to and activates Pkc1p (19, 34) and other targets (27). The CWI pathway is also regulated by additional inputs and independently of the cell surface sensors. It is responsive to starvation (24, 44), to actin cytoskeleton defects (10), and to Tor2p function (12, 40). The upstream signaling in the CWI pathway is thus complex (27).
The signaling downstream of Pkc1p is no less complex in that the kinase appears to directly or indirectly regulate multiple distinct targets and to thereby affect a diverse range of cellular functions (27). The pathway acts to stimulate expression of cell wall genes, e.g., via the transcription factors Swi4p (30) and Rlm1p (48), to modulate ribosome synthesis (33), to regulate actin polarity (5), and to affect gene silencing (1, 39). A protein kinase cascade culminating in the mitogen-activated protein (MAP) kinase Slt2p, also known as Mpk1p (25, 26), is the best characterized downstream branch.
In this study, we isolated MPT5 as a dosage suppressor of the vegetative defect of mutants lacking the Slg1p upstream sensor of the Pkc1p branch of the CWI pathway. We set out to reexamine the relationships between Mpt5p, Pkc1p, stress tolerance, and cellular life span.
MATERIALS AND METHODS
Chemicals, reagents, and growth media.
All chemicals were supplied by Sigma-Aldrich Chemical Corp. (St. Louis, MO) unless stated otherwise. Components of growth media were from Becton Dickinson (Sparks, MD). Liquid media were prepared as previously described (35). Solid media contained an additional 2% agar, except for tetrad dissection, for which 4% agar was used.
Strains, plasmids, and genetic manipulations.
Yeast strains used in this study were as follows: for the W303-1a strain background (ura3 leu2 his3 trp1 ade2 can1), the MATa wild type (JVG161), MATa mpt5Δ::TRP1 (JVG2110), MATa slt2Δ::TRP1 (JVG1154), MATa lrg1Δ::KanMX (JVG2976), MATa mpt5Δ::TRP1 lrg1Δ::KanMX (JVG2989), MATa GAL psi+ (wild type; gift of Kenji Irie [43]), and MATa GAL psi+ kanMX6::GAL1p-MPT5 (TTC75; gift of Kenji Irie [43]); and for the S288c strain background (ura3 leu2 his3 trp1 ade2 lys2), MATa/MATα TRP1/TRP1 slg1Δ::LEU2/slg1Δ::LEU2 (JVG1081 [8]).
Plasmids used in this study include pmpt5Δ::URA3 (pYK663; provided kindly by Y. Kikuchi [23]), pura3Δ::TRP1 (pJO122 [35]), pYK601 (a 2μ plasmid containing MPT5 and additional downstream genomic sequences [23]), YEp24 (2μ vector), pPKC1 (a URA3-marked 2μ plasmid containing PKC1; pDL289 [46]), pHCS77 (a URA3-marked 2μ plasmid containing HCS77; pJO36 [8]), pMPT5 (a URA3-marked 2μ plasmid containing MPT5; pG19 [this study]), pRlm1-lacZ (a URA3-marked plasmid containing two Rlm1p-binding sites in the promoter region upstream of the lacZ reporter gene [14]), pMS602 (a plasmid derived from pYK601 lacking 1,200-bp downstream genomic sequences [this study]) and pMPT5ΔPUF (a plasmid derived from pYK601 lacking the majority of the Mpt5p PUF domain [this study; see below]).
The chromosomal copy of MPT5 gene was disrupted by direct gene disruption in wild-type haploid MATa and MATα cells in the W303-1a and S288c strain backgrounds by using pYK663 as previously described (23). The URA3 marker in the resulting mpt5Δ::URA3 strains was subsequently converted to TRP1 by direct gene replacement by using pJO122 as previously described (35).
The chromosomal copy of LRG1 was replaced with the KanMX marker by direct gene replacement in haploid wild-type W303-1a as previously described (2).
Double-mutant combinations were generated by mating single-disruption haploid strains of the opposite mating type and selecting for heterozygous diploids. The subsequent sporulation of the diploids and tetrad dissection using a Zeiss Axioskop Tetrad Dissection system allowed the generation of double-mutant haploid progeny.
Screening for multicopy suppressors of the vegetative growth defect of slg1Δ/slg1Δ mutants.
The slg1Δ/slg1Δ mutant (JVG1081; S288c strain background) displays a profound inability to survive/proliferate at temperatures in excess of 30°C (8). The strain was transformed with a genomic library carried in YEp24, a multicopy vector marked with URA3 (kind gift of J. P. Ogas). Transformants were plated on plates containing synthetic dropout medium lacking uracil (SD-URA) and incubated at room temperature for 4 h prior to a shift to 35°C and incubation for 3 days. Ten transformants that formed colonies at 35°C were identified. The corresponding plasmids were isolated for the transformants, and those that robustly conferred the ability to proliferate at 35°C to the slg1Δ/slg1Δ mutant (JVG1081) were identified by retransformation. Restriction mapping of the confirmed plasmids demonstrated that seven contained SLG1. Three novel plasmids were also identified. The ends of the inserts on the three plasmids were sequenced. All inserts shared only one gene, MPT5. One of these MPT5-containing plasmids, pMPT5 (pG19), which was the most robust suppressor of the slg1Δ/slg1Δ defect, was used in subsequent analyses.
An independently derived high-copy plasmid expressing MPT5 (a kind gift of Y. Kikuchi [23]), pYK601, was also found to efficiently suppress the slg1Δ/slg1Δ growth defect. To test if overexpression of MPT5 is sufficient for this suppression, the sequence beyond 1,200 bp downstream of MPT5 in pYK601 was deleted by the excision of a ZhoI-to-SalI fragment; subsequent recircularization of the plasmid yielded pMS602 by standard methodology. MPT5 is the only gene remaining within the insert in this plasmid. Transformation with pMS602 efficiently suppressed the slg1Δ/slg1Δ growth defect. The overexpression of MPT5 alone is thus sufficient for suppression of slg1Δ/slg1Δ mutants.
Construction of pMPT5ΔPUF.
The PUF domain in Mpt5p lies between residues 196 and 499. In the MPT5-containing plasmid pYK601, the only BamHI site lies in the sequence that corresponds to residue 260 of the Mpt5p protein. A unique XhoI site lies approximately 1,200 bp downstream of the MPT5 open reading frame. Most of the region of MPT5 corresponding to the conserved PUF domain was deleted from pYK601 as follows. The plasmid was used as a template for a PCR amplification with primers “BamH1-Forward” (5′-CATTACTGGAGGATCCTAATAACAATGGGGGTGCT-3′) and “Downstream-Reverse” (5′-CGAAATCCCTTGTATCTT-3′). The amplified fragment contained the 3′ end of the MPT5 gene plus approximately 1,200 bp of downstream flanking sequence (containing the XhoI site) but with a BamHI site engineered immediately upstream of the sequence corresponding to residue 474 of Mpt5p. This PCR product was digested with BamHI and XhoI and ligated into linear pYK601, from which its BamHI-to-XhoI fragment had been excised. The ligated product (pMPT5ΔPUF) expressed a version of Mpt5p (Mpt5pΔPUF) lacking only residues 261 to 473 (inclusive) and expressing a protein that lacks most (70%) of its PUF domain.
Determination of cell viability.
The viability of cells in culture was determined as previously described (24) using the viability dye methylene blue. Live (colorless) and dead (dark blue) cells were visualized using a Leitz Orthoplan fluorescent microscope in nonfluorescence mode. A minimum of 200 cells were counted for each sample.
Determination of cell lysis.
Cell lysis was determined for aliquots of cell cultures as previously described (24) using propidium iodide staining. A minimum of 200 cells were counted for each sample.
Determination of acquired thermotolerance.
Acquisition of thermotolerance was measured essentially as previously described (18).
Determination of Slt2p activity.
Slt2p activity was inferred from its phosphorylation state as determined by Western blot analysis using anti-phospho-p44/42 MAP kinase monoclonal antibody (Cell Signaling Technology, Beverly, MD) as previously described (24). The amount of Slt2p was determined using a goat anti-Slt2p antibody from Santa Cruz Biotechnology (Santa Cruz, CA). A loading control antibody (rabbit anti-Rpd3p) was supplied by Upstate Biotechnology (Lake Placid, NY).
Rlm1p is a transcription factor that is a direct target of Slt2p. Rlm1p activity is a faithful reporter of Slt2p status (14, 27). Therefore, Slt2p activity was also assayed using a plasmid (pRlm1-lacZ) containing a lacZ reporter gene under the control of Rlm1p (kind gift of D. E. Levin) as previously described (14).
Determination of chronological life span.
Duplicate 50-ml cultures of transformants were grown in synthetic medium lacking uracil at 26°C with agitation. At daily intervals after inoculation, 1-ml aliquots were collected and sonicated for 5 s. The optical density at 600 nm (OD600) was measured, and samples were diluted into distilled water prior to plating onto yeast extract-peptone-dextrose (YPD) plates. Each sample was plated in triplicate. Plates were incubated at 30°C for 4 days prior to colony counting. Time zero for the chronological ageing experiment was determined as the first point at which the OD600 reached a plateau, i.e., when the cultures had saturated.
RNA preparation, Northern blot analysis, and quantification.
A 5-ml overnight culture was diluted 1:10 and allowed to grow to an OD600 of 0.3 to 0.4. For the mpt5Δ cells and the corresponding wild type, the cells were grown in YPD medium; for the GALp-MPT5 cells and the corresponding wild type, the cells were grown initially in YP medium supplemented with raffinose (YP-raffinose) and then diluted into either YP-raffinose or YP-galactose and grown for 6 h. RNA preparation, gel electrophoresis, Northern blot analysis, and quantification were performed as previously described (41) with the following exceptions: RNA was precipitated with 3 volumes of 100% ethanol, and the phosphorimages were processed using a Fujifilm BAS-1500 phosphorimager and analyzed using Fuji Image Reader and Fuji Image Gauge software. The LRG1 mRNA level in each sample was calculated relative to the ACT1 mRNA level for the same sample. The resulting ratios were used to calculate the relative expression levels of LRG1 mRNA referenced to the ratio in untreated wild-type control samples. The following primers (Sigma-Genosys, Suffolk, United Kingdom) were used for PCR amplification of the LRG1 probe from Saccharomyces cerevisiae genomic DNA (Invitrogen, Paisley, United Kingdom): LRG1F (5′-TTCTGCTGGTTATCGATCGCT-3′) and LRG1R (5′-TTTCACCTTGTATGACCGTTG-3′). The following primers were used for PCR amplification of the ACT1 gene: ACT1A (5′-CTCCTTTTTGAAATATTTTTGG-3′) and ACT1D (5′-ATAAAACTGAAAAGCGATGAAGAGA-3′).
RESULTS
mpt5Δ mutants display a short chronological life span, a defect suppressed by overexpression of PKC1.
Many mutations that affect replicative life span also affect chronological life span—the time that cells grown to saturation in synthetic medium retain the ability to proliferate when returned to fresh medium (for a review, see reference 3). Mutants lacking components of the Pkc1p branch of the CWI pathway display a short chronological life span (24). Is the same true for mpt5 mutants? We cultured congenic wild-type and mpt5Δ mutants, each transformed with a vector, to stationary phase in selective synthetic medium and monitored CFU as a function of time. As shown in Fig. 1A, mpt5Δ mutants displayed a severely reduced chronological life span compared with congenic wild-type cells. We conclude that Mpt5p is a positive regulator of both replicative and chronological life span.
FIG. 1.
mpt5Δ mutants display a short chronological life span, a defect suppressed by overexpression of PKC1. Wild-type (WT; JVG161) and mpt5Δ (JVG2110) cells were transformed with a (A) vector or (B) pPKC1 (pDL289). The transformants were grown to stationary phase in synthetic dropout medium at 30°C. Numbers of CFU were determined as a function of time after saturation by plating dilutions onto YPD plates.
We found that the overexpression of PKC1 also efficiently suppressed the chronological ageing defect of mpt5Δ mutants (Fig. 1B). Consistent with its effects on replicative life span (21), the overexpression of PKC1 did not extend the chronological life span of wild-type cells; indeed, it may have shortened it (time for 50% loss of CFU was reduced from 11 days to 10 days). We conclude that the overexpression of PKC1 is sufficient to suppress the chronological life span defect of mpt5Δ mutants but not to extend the life span of otherwise wild-type cells. The functional overlap between MPT5 and PKC1 is thus extensive, and it covers stress tolerance, replicative life span, and chronological life span.
mpt5Δ mutants display phenotypes characteristic of partial loss of Pkc1p activity.
Mutants lacking MPT5 in the W303-1a strain background have previously been reported to display a temperature-sensitive proliferation defect (23) that is suppressed by osmotic stabilization (15). Such osmotic remediality is a characteristic of, although it is not necessarily unique to, mutants lacking components of the Pkc1p branch of the CWI pathway.
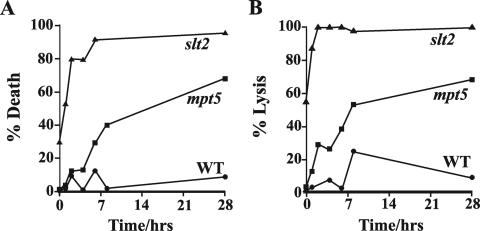
Another key characteristic of mutants that are partly defective in Pkc1p signaling is death by cell lysis at high temperatures (27). Do mpt5Δ mutants also display this phenotype? We grew congenic cultures of haploid mpt5Δ mutants, slt2Δ mutants (as a positive control), and congenic wild-type cells (as a negative control), all in the W303-1a strain background, in liquid rich medium to mid-logarithmic phase at 23°C. Cultures were shifted to 37°C, and the cell viability and degree of cell lysis were determined as a function of time after the temperature upshift. As shown in Fig. 2A, mpt5Δ cells, like slt2Δ cells, lost viability significantly more than did wild-type cells over the time course of the experiment. Furthermore, as shown in Fig. 2B, mpt5Δ mutant cells were prone to lyse at high temperature, concomitant with the loss of cell viability. We conclude that mpt5Δ mutants, like those defective in Pkc1p signaling, die by cell lysis at high temperatures.
FIG. 2.
mpt5Δ mutants die by lysis at high temperatures. Wild-type (WT; JVG161), mpt5Δ (JVG2110), and slt2Δ (JVG1154) cells were grown to mid-logarithmic phase at 23°C and shifted to 37°C. Cell viability (A) and cell lysis (B) were monitored as a function of time after the temperature upshift. At least 200 cells were counted per time point. Representative data are shown.
Overexpression of MPT5 suppresses the temperature-sensitive proliferation defect of mutants lacking Slg1p, a cell surface sensor that acts upstream of Pkc1p.
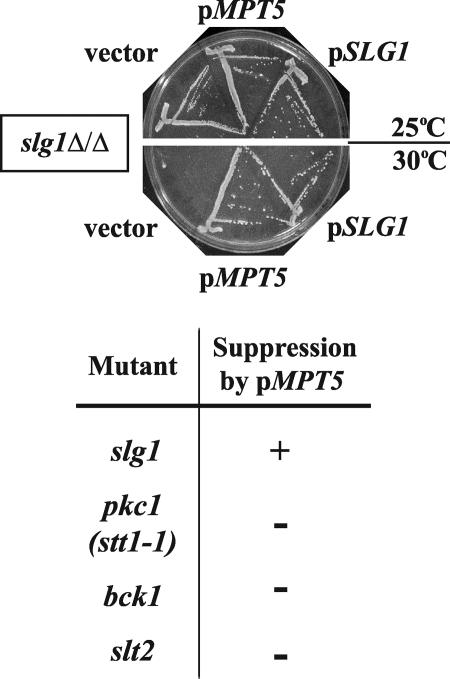
In an independent attempt to identify novel components of the Pkc1p pathway, we screened a genomic library for genes that, when overexpressed, suppressed the temperature-sensitive growth defect of homozygous diploid mutants lacking SLG1, the gene encoding a key cell surface sensor of the CWI pathway. Such high-copy suppressors may encode positive activators of Pkc1p function, since the overexpression of PKC1 itself suppresses the vegetative defect of these slg1Δ/slg1Δ mutants (8). In addition to finding plasmids containing SGL1 itself or PKC1, we isolated three independent plasmids sharing only MPT5 (Fig. 3 and data not shown). We showed that expression of MPT5 was sufficient for this suppression (data not shown). MPT5 thus shares with PKC1 the ability to potently suppress the vegetative growth defect of slg1 mutants.
FIG. 3.
Overexpression of MPT5 suppresses the growth defect of slg1Δ mutants but not mutants affecting downstream signaling in the CWI pathway. slg1Δ/slg1Δ (JVG1081) mutants were transformed with a vector (YEp24), pMPT5 (pG19), or pSLG1 (pJO36). Transformants were selected at 25°C, streaked onto fresh plates, and incubated at 25°C or 30°C for 3 days prior to being photographed. Similar experiments were performed with other mutant strains containing either pkc1-1, bck1Δ, or slt2Δ (data not shown). The results from all these analyses are tabulated here, where “+” indicates suppression of the temperature-sensitive growth defect of each strain by overexpression of MPT5 (pG19) and “−” indicates no such suppression.
Genetic interactions between MPT5 and genes encoding the Pkc1p pathway components.
If Mpt5p acts in the CWI pathway and in the same branch as the Slg1p sensor, we would expect mpt5Δ slg1Δ double mutants to be viable and to display a phenotype no worse than that of the single mutants. We constructed double-mutant haploid progeny by standard genetic manipulation. These progeny proliferated on YPD medium at 25°C, and no synthetic enhancement of phenotype was noted at any temperature tested (data not shown). We found the same for mpt5Δ slt2Δ double mutants (data not shown). MPT5 thus shows no synthetic genetic interaction with SLG1 or SLT2, consistent with the proteins acting in the same rather than in parallel pathways to maintain cell integrity.
If Mpt5p acts in the CWI pathway, then we should be able to place it at a point within the pathway by epistatic analysis. Given that the growth defects of mpt5Δ mutants are suppressed by the overexpression of PKC1 (16, 36; data not shown), we would expect Pkc1p to act downstream of Mpt5p in the pathway. Consistent with this model, we found that overexpression of MPT5 did not suppress the growth/proliferation defects of pkc1ts mutants or of mutants defective in the downstream MAP kinase branch, e.g., bck1Δ or slt2Δ mutants (Fig. 3). Genetic analyses together point to Mpt5p as being a component of the CWI pathway and acting between Slg1p and Pkc1p.
Mpt5p is required for thermal activation of the Pkc1p pathway.
One assayable target of Pkc1p is the downstream MAP kinase Slt2p. This protein kinase is activated and dual phosphorylated on tyrosine and threonine in response to a variety of stresses, including heat shock (31). Heat shock-triggered activation results from thermal stress to the cell surface being detected by the Slg1p and Mid2p cell surface sensors (8, 31). To determine if mutants lacking MPT5 are defective in Pkc1p signaling, we subjected cultures of congenic wild-type and mpt5Δ mutant cells to a rapid heat shock from 23°C to 39°C. Samples were taken up to two hours after the temperature upshift, by which time there had been no appreciable cell death (data not shown). The samples were subjected to Western blot analysis using an anti-dual-phospho-Slt2p antibody. As shown in Fig. 4A, Slt2p was strongly dual phosphorylated by 30 min after heat shock of wild-type cells. In contrast, Slt2p was only poorly dual phosphorylated upon heat shock of mpt5Δ mutants, and this residual activation occurred more slowly than in wild-type cells. Mpt5p is thus required for optimal activation of the Pkc1p-MAP kinase pathway upon heat shock, a stress sensed by the Slg1p and Mid2p cell surface sensors.
FIG. 4.
Mpt5p is required for heat shock-triggered activation, but not latruculin-stimulated activation, of the CWI/Pkc1p pathway. (A) Wild-type (WT; JVG161) and mpt5Δ (JVG2110) cells were grown to mid-logarithmic phase at 23°C and then rapidly shifted to 30°C at time zero. The phosphorylation status of Slt2p was monitored as a function of time after the temperature shift by Western blot analysis. (B) Wild-type (JVG161) and mpt5Δ (JVG2110) cells were grown to mid-logarithmic phase at 23°C and incubated for 1 h at 37°C prior to a rapid shift to 50°C. Cultures were diluted and plated on YPD plates and incubated at 25°C for 3 days. CFU were calculated as a function of time at 50°C. (C) Wild-type (JVG161) and mpt5Δ (JVG2110) cells were grown to mid-logarithmic phase at 23°C and treated with 40 μg/μl latrunculin B or drug vehicle only (ethanol) or left untreated for a further 2 h. The phosphorylation status of Slt2p was determined by Western blot analysis using anti-dual phospho-Slt2p antiserum.
An independent and quantitative output of Slt2p activation upon thermal stress is acquired thermotolerance, the phenomenon whereby prior exposure of cells to a high but sublethal temperature confers increased tolerance to subsequent transfer to a lethal temperature (18). Slt2p is both required and sufficient for acquired thermotolerance in yeast (18). Cultures of congenic wild-type and mpt5Δ mutants were subjected to a lethal temperature (50°C) with and without prior incubation at 37°C. Preincubation of wild-type cells at 37°C largely prevented rapid cell death, and the protection persisted, in part at least, for up to 20 min at 50°C (Fig. 4B). In contrast, mpt5Δ mutants acquired only partial protection from preincubation at 37°C (Fig. 4B). This observation is consistent with mpt5Δ mutants being defective in thermal activation of CWI signaling.
mpt5Δ mutants are not defective in latrunculin-triggered activation of Slt2p.
Depolymerization of the actin cytoskeleton strongly activates the Pkc1p-MAP kinase branch but independently of the cell surface sensors (10). Is Mpt5p also required for this activation? We treated proliferating wild-type and mpt5Δ cells with the actin-depolymerizing drug latrunculin B or with its vehicle alone or left the cells untreated. Samples were harvested two hours later, and the phosphorylation (and thus activation) status of Slt2p was determined as described above. As shown in Fig. 4C, Slt2p was strongly phosphorylated in both wild-type and mpt5Δ mutants in response to treatment with latrunculin B. Thus, Mpt5p is not required for activation of the Pkc1p-MAP kinase branch in response to actin cytoskeletal defects. These data confirm that Pkc1p-to-Slt2p signaling is not inherently compromised by the absence of Mpt5p, nor is actin-to-Pkc1p signaling adversely affected. Rather, we conclude that Mpt5p acts selectively if not specifically on the upstream branch that transmits cell surface state, as sensed by the Slg1p sensor, to Pkc1p.
The PUF domain of Mpt5p is required for its function in the CWI pathway.
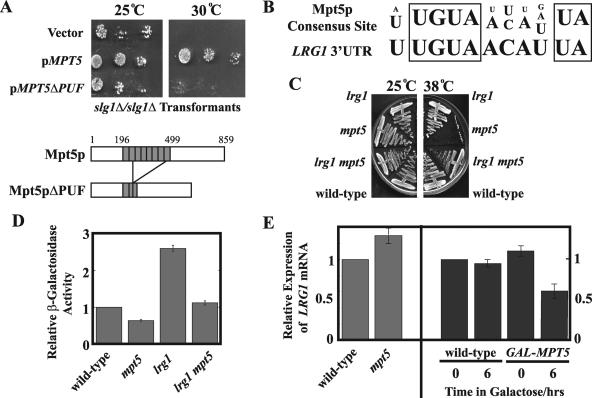
Because Mpt5p is a PUF protein, it is tempting to hypothesize that it affects CWI signaling via its ability to bind to and regulate mRNAs (32, 42, 50). In this scenario, we would expect the PUF domain of Mpt5p to be essential for its function in the Pkc1p pathway. To test this hypothesis, we constructed a plasmid expressing an engineered form of Mpt5p, Mpt5pΔPUF, corresponding to an in-frame fusion of the N- and C-terminal domains of the wild-type protein and lacking most of the PUF domain (Fig. 5A) (see Materials and Methods). This construct, in contrast to a version expressing full-length MPT5, failed to complement the temperature sensitivity of mpt5Δ mutants (data not shown) and failed to suppress the temperature sensitivity of slg1Δ/slgΔ mutants (Fig. 5A). Mpt5p may thus regulate CWI signaling by affecting one or more target transcripts.
FIG. 5.
Mpt5p destabilizes the LRG1 transcript to regulate CWI signaling. (A) The PUF domain of Mpt5p is required for function in the CWI pathway. slg1Δ/slg1Δ (JVG1081) mutants were transformed with a vector (YEp24), pMPT5 (pYK601), or pMPT5ΔPUF. After selection on the appropriate medium, the transformants were grown in selective liquid medium and diluted to an OD600 of 0.02. This dilution was spotted along with two further 1:100 dilutions onto two selective plates, one grown at 25°C and the other at 30°C. The plates were incubated for 3 days. (B) The Mpt5p consensus binding sequence (6) matches the putative Mpt5p-binding sequence in the LRG1 3′UTR. (C) Loss of LRG1 suppresses mpt5 mutants. Wild-type (JVG161), mpt5Δ (JVG2110), lrg1Δ (JVG2976), and mpt5Δ lrg1Δ (JVG2989) cells were plated on rich medium plates and grown at either 25°C or 38°C for 3 days. (D) Loss of LRG1 suppresses the Pkc1p signaling defect of mpt5 mutants. Wild-type (JVG161), mpt5Δ (JVG2110), lrg1Δ (JVG2976), and mpt5Δ lrg1Δ (JVG2989) cells were transformed by conventional methods with the pRlm1-lacZ reporter construct, which faithfully reports on Slt2p activity in vivo. Reporter activity was quantified for each transformant grown to mid-logarithmic phase in synthetic medium at a permissive temperature. (E) Mpt5p regulates the LRG1 mRNA level. Wild-type (JVG161) and mpt5Δ (JVG2110) cells were cultured to mid-logarithmic phase in YPD medium at room temperature and collected, and total RNA was prepared. A GAL-MPT5 strain (TTC75) and its congenic wild type were cultured to mid-logarithmic phase in YP-raffinose and transferred to YP-raffinose or YP-galactose and cultured for 6 h. Samples (t = 0 and t = 6 h) were collected, and total RNA was prepared. All RNA samples were Northern blotted. Transcripts were detected by phosphorimaging. The LRG1 transcript level was quantified relative to the ACT1 transcript control for each sample, and relative expression values between samples were calculated relative to untreated wild-type control samples.
LRG1 is a candidate target transcript linking Mpt5p to the CWI pathway.
Based on the mode of action of PUF domains (42), we predicted that Mpt5p binds to and inhibits a target transcript that encodes an upstream inhibitor of Pkc1p signaling. We examined the 3′UTRs of predicted transcripts encoding known or suspected upstream inhibitors of the CWI pathway (27): the multiple Rho-GTPase activating proteins (Rho-GAPs) and the yeast Rho GDP-dissociation inhibitor. We identified a putative PUF-binding site in the predicted 3′UTR of the predicted LRG1 mRNA (29, 47), encoding a Rho-GAP (29, 47), that matches the consensus Mpt5p-binding sequence (6) (Fig. 5B). The LRG1 transcript was independently found to physically bind to Mpt5p and not to the other yeast PUF proteins tested (6). The LRG1 transcript may thus be an Mpt5p-specific target that links this PUF protein to the CWI pathway.
Lrg1p is an inhibitor of Pkc1p signaling.
Not all Rho-GAPs act on Rho1p, and those that do can selectively affect the activity of only a subset of their targets, only one of which is Pkc1p (27). If the LRG1 transcript is a relevant target of Mpt5p, then it should encode a negative regulator of the Pkc1p signaling. To test this possibility, we constructed an lrg1Δ mutant in the W303 strain background and assayed the basal activity of the Pkc1p pathway by exploiting an Rlm1p-dependent lacZ reporter construct whose expression is dependent on Slt2p activity (and therefore Pkc1p activity) (14). As shown in Fig. 5D, we found that basal reporter activity was low in mpt5Δ mutants (demonstrating that Mpt5p is also required for full basal activity of the CWI pathway) but, in contrast, was significantly elevated in the lrg1Δ mutants. We conclude that Lrg1p is indeed a negative regulator of Pkc1p signaling.
Loss of LRG1 suppresses mpt5Δ defects.
If LRG1 is a key target of Mpt5p, then deletion of LRG1 should suppress the phenotypes of mpt5Δ mutants. To test this possibility, we constructed lrgΔ mpt5Δ double mutants by standard genetic manipulations. As shown in Fig. 5C, we found that deletion of LRG1 efficiently suppressed the temperature-sensitive growth defect of mpt5Δ mutants. As shown in Fig. 5D, we also found that loss of LRG1 suppressed the Pkc1p-signaling defect of mpt5Δ mutants. However, it is clear that LRG1 is not the only relevant target of Mpt5p, since reporter activity was lower in lrg1Δ mpt5Δ mutants than it was in lrg1Δ mutants, i.e., Mpt5p affects Slt2p signaling even in the absence of LRG1.
LRG1 transcript level is dependent on Mpt5p.
Is LRG1 a target of Mpt5p? If so, then Mpt5p should regulate the steady-state abundance of the LRG1 transcript. To test this, we determined the relative abundance of LRG1 mRNA in the presence and absence of Mpt5p and upon galactose-induced overexpression of MPT5 in otherwise wild-type cells. As shown in Fig. 5E, we found that the loss of Mpt5p elevated the abundance of LRG1 mRNA (by approximately a third) and, conversely, that the overexpression of MPT5 repressed its abundance (by approximately 50%). We conclude that the LRG1 transcript is a target of Mpt5p that directly links this PUF protein to the CWI pathway.
DISCUSSION
The CWI pathway is a key downstream target of Mpt5p.
It was proposed that Mpt5p and the Pkc1p/CWI pathway act in parallel to maintain cell integrity and to promote replicative life span (15, 16). Multiple lines of evidence coalesce here to support a revised model in which Mpt5p acts in the CWI pathway (Fig. 6). First, mpt5Δ mutants display the phenotypes characteristic of mutants that are partially defective in the Pkc1p branch of the pathway, e.g., death by lysis at high temperatures (Fig. 2A and B). Second, genetic and epistatic analyses place Mpt5p in the CWI pathway and upstream of Pkc1p (Fig. 3 and data not shown). Third, by multiple independent assays, we find that mpt5Δ mutants are compromised in basal activity (Fig. 5D) as well as in heat-shock activation of the Pkc1p pathway (Fig. 4A and B). Finally, we show that a direct Mpt5p target transcript, the LRG1 mRNA, encodes a relevant inhibitor of Pkc1p signaling (Fig. 5), thereby establishing a direct link between this PUF protein and the CWI pathway.
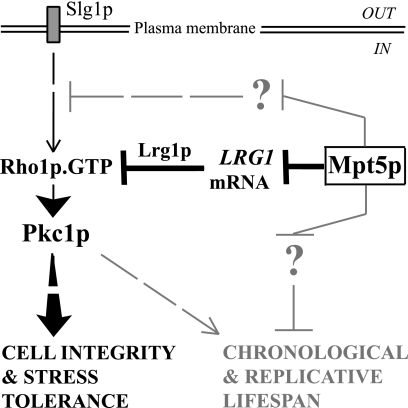
FIG. 6.
Proposed model for Mpt5p action. Mpt5p affects cell integrity and stress tolerance predominantly, if not exclusively, by modulating CWI-Pkc1p activity. Mpt5p affects Pkc1p activity in part by directly binding to and inhibiting the stability of the LRG1 transcript. The reduction in Lrg1p levels, a Rho1-GAP, results in increased amounts of active Rho1p-GTP complex. This complex binds to and activates Pkc1p, which then promotes cell integrity and stress tolerance (27). Mpt5p must also affect the CWI-Pkc1p pathway independently of LRG1, by binding to one or more additional and unidentified transcripts whose products act directly or indirectly in the pathway. The effects of Mpt5p on chronological and replicative life span are only partly dependent on CWI-Pkc1p signaling. Other, as-yet-unknown target transcripts likely regulate other distinct pathways that, with the CWI pathway, cooperate to regulate cellular life span.
A synthetic genetic interaction has been noted (16) between mpt5Δ and swi4Δ, the latter lacking a transcription factor that acts partly in the CWI pathway downstream of Slt2p (27). This interaction may, in contrast to our analysis, point to this PUF protein as acting in parallel to the CWI pathway (16). However, mutations in other components of the CWI pathway, including in SLT2 and SLG1, also show synthetic lethal interactions with swi4Δ mutations (8). This surprising behavior is due to Swi4p also having a second, partly redundant function, to drive expression of genes at the G1-S transition of the cell cycle (35), but independently of the CWI pathway (reviewed in reference 27). A synthetic genetic interaction with swi4Δ mutations is thus a characteristic of mutations in genes encoding CWI components (8), including MPT5.
The Pkc1p branch of the CWI pathway appears to be a critical target of Mpt5p action, since PKC1 suppresses the stress tolerance (16, 36), replicative life span defect (16), and chronological life span defect (Fig. 1A and B) of mpt5 mutants. However, it may not be the sole key target for all the functions of this PUF protein.
It is likely that Mpt5p acts predominantly if not exclusively through Pkc1p to support cell integrity and stress tolerance in proliferating cells. In particular, the lack of any additive or synthetic consequence when mpt5Δ mutations are combined with either the slg1Δ or slt2Δ mutation argues strongly that Mpt5p acts solely in the CWI pathway under the conditions tested, during vegetative proliferation and at elevated temperatures (data not shown).
Mpt5p is thought to extend replicative life span by inhibiting the formation of extrachromosomal rDNA circles (ERCs). These ERCs are derived from recombination at the repetitive rDNA locus. Mpt5p is thought to modulate gene silencing at the rDNA locus via Sir2p (21). Intriguingly, the Pkc1p pathway itself can act to inhibit ERC formation (39), consistent with it mediating, at least in part, the effects of Mpt5p on replicative life span. Paradoxically, the Pkc1p pathway can also act to shorten replicative life span, in this case by phosphorylating the Sir3p silencing protein (39). It is possible that Mpt5p selectively activates the life span-extending function(s) of Pkc1p over its life span-limiting function(s). However, all is not simple. The overexpression of PKC1 does not fully phenocopy high-level overexpression of MPT5; the latter extends replicative and chronological life spans of otherwise wild-type strains, whereas the former does not (16) (Fig. 1B). This disparity could exist for technical reasons (levels of expression, toxicity of ectopically expressed genes) but most likely points to the existence of one or more additional key effectors of this PUF protein in regulating cellular life span (see below).
How does Mpt5p modulate CWI signaling?
Mpt5p appears to selectively if not specifically act on the upstream branch of the CWI pathway that responds to cell surface state. Activation of the Pkc1p-MAP kinase branch in response to heat shock is partly dependent on Mpt5p (Fig. 4A to C); the response to actin depolarization is not affected (Fig. 4D). Heat stress is sensed by the cell surface sensors Slg1p and Mid2p and acts through the Rho1p GTPase (8, 31). The sensing of actin defects appears to be largely independent of these components (10).
It is likely that Mpt5p acts via its PUF domain, since this domain is required for complementation of the temperature-sensitive growth of mpt5Δ strains (data not shown) and for the overexpression of MPT5 to suppress the growth defect of slg1Δ mutants (Fig. 5A). Because PUF domains are known in other cases to be both necessary and sufficient for binding to and inhibiting target transcripts (32, 50), it is likely that Mpt5p also acts by inhibiting one or more target transcripts. The recent identification of transcripts that bind to the Mpt5p PUF domain (6) supports this notion.
The LRG1 transcript is likely to be a relevant target that directly links Mpt5p into the CWI pathway (Fig. 6). First, LRG1 mRNA was found to physically associate with Mpt5p in vitro (6). In an independent study, LRG1 mRNA was confirmed to bind Mpt5p by in vivo three-hybrid analysis (41). LRG1 mRNA has not been found to associate with any of the other yeast PUF proteins tested (6), consistent with it being a specific target of Mpt5p, the only yeast PUF protein to be directly implicated in regulating stress tolerance and cellular life span. Second, the LRG1 mRNA contains a putative Mpt5p-binding site in its 3′UTR that conforms with the unique consensus binding site for this protein (Fig. 5B) (6). This consensus sequence is sufficient for interaction with Mpt5p in vivo (6). Third, Mpt5p acts to inhibit the stability (41) of its target transcripts. It is thus likely that Mpt5p inhibits the expression of an inhibitor of CWI signaling. Here, we show that Lrg1p is such an inhibitor of Pkc1p/CWI signaling (Fig. 5D). Fourth, we show that inactivation of LRG1 is sufficient to elevate Pkc1p/CWI activity in mpt5Δ mutants (Fig. 5D) and to restore their ability to proliferate at high temperatures (Fig. 5C). Finally, we show that LRG1 mRNA abundance is affected by, and is inversely proportional to the levels of, Mpt5p (Fig. 5E).
However, LRG1 is unlikely to be the only relevant target of Mpt5p, since loss of Mpt5p activity can affect Pkc1p signaling even in the absence of LRG1 (Fig. 5D). At least one other target of this PUF protein must directly or indirectly inhibit upstream CWI signaling (Fig. 6). Of the other known direct inhibitors of the CWI pathway, the mRNA for only one, Sac7p, was also found to physically associate with Mpt5p (6). However, this association may not be physiologically relevant, since the SAC7 transcript does not contain a putative PUF-binding site, nor is the association detected between it and Mpt5p an avid one (6). More research is needed.
Any model attributing the effects of Mpt5p to one or to a few direct target transcripts is likely to be overly simplistic. First, PUF proteins, in yeast at least, appear to modulate the expression of their target transcripts rather than to act as on/off switches (41, 42, 43) (Fig. 5E). Mpt5p is therefore likely to affect expression/translation of any specific target to a modest degree, an effect that is likely to have moderate phenotypic consequences on its own. Second, over 200 putative target transcripts of Mpt5p have been identified (6, 41). It would be surprising if the major effects noted for Mpt5p were mediated by its effects on one or on a small number of these targets. Finally, the large set of putative Mpt5p targets is highly enriched for genes encoding proteins that affect gene silencing and chromatin remodeling, functions directly related to replicative ageing (6) and only partly overlapping the known roles for the CWI pathway (27).
The most parsimonious model is that Mpt5p affects stress tolerance and cellular life span by a cumulative effect on many if not most of its target transcripts, a subset of which affect the CWI pathway and thereby stress tolerance and cell integrity and a subset (or most) of which act independently of, but synergistically with, CWI signaling to affect cellular life span (Fig. 6). In this model, Mpt5p acts by the cumulative effect of many individually small changes in gene expression. This complexity notwithstanding, our data point to the CWI pathway as being a key and direct downstream actor in Mpt5p function.
Acknowledgments
We thank Y. Kikuchi, K. Irie, and D. Levin for the gifts of strains or plasmids.
M.S.S. was supported by a 4-year Ph.D. studentship from the Wellcome Trust. The research was also supported by funds from the Research Committee of the Faculty of Biomedical and Life Sciences and by a research grant (C20144) from the Biotechnology and Biological Sciences Research Council to J.V.G.
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Ai, W., P. G. Bertram, C. K. Tsang, T. F. Chan, and X. F. Zheng. 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10:1295-1305. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bitterman, K. J., O. Medvedik, and D. A. Sinclair. 2003. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 67:376-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, T., and J. Kurjan. 1997. Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol. Cell. Biol. 17:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delley, P. A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber, A. P., D. Herschlag, and P. O. Brown. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PloS Biol. 2:342-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstrohm, A. C., B. A. Hook, D. J. Seay, and M. Wickens. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13:533-539. [DOI] [PubMed] [Google Scholar]
- 8.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu, W., Y. Deng, D. Zenklusen, and R. H. Singer. 2004. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18:1452-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison, J. C., E. S. Bardes, Y. Ohya, and D. J. Lew. 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3:417-420. [DOI] [PubMed] [Google Scholar]
- 11.Hata, H., H. Mitsui, H. Liu, Y. Bai, C. L. Denis, Y. Shimizu, and A. Sakai. 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helliwell, S. B., A. Schmidt, Y. Ohya, and M. N. Hall. 1998. The Rho1 effector Pkc1, but not Bni1, mediates signaling from Tor2 to the actin cytoskeleton. Curr. Biol. 8:1211-1214. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby, J. J., S. M. Nilius, and J. J. Heinisch. 1998. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 258:148-155. [DOI] [PubMed] [Google Scholar]
- 14.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781-789. [DOI] [PubMed] [Google Scholar]
- 15.Kaeberlein, M., A. A. Andalis, G. B. Liszt, G. R. Fink, and L. Guarente. 2004. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p-independent mechanism. Genetics 166:1661-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaeberlein, M., and L. Guarente. 2002. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160:83-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaeberlein, M., K. T. Kirkland, S. Fields, and B. K. Kennedy. 2005. Genes determining yeast replicative life span in a long-lived genetic background. Mech. Ageing Dev. 126:491-504. [DOI] [PubMed] [Google Scholar]
- 18.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 19.Kamada, Y., H. Qadota, C. P. Python, Y. Anraku, Y. Ohya, and D. E. Levin. 1996. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 271:9193-9196. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, B. K., N. R. Austriaco, J. Zhang, and L. Guarente. 1995. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80:485-496. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy, B. K., M. Gotta, D. A. Sinclair, K. Mills, D. S. McNabb, M. Murthy, S. M. Pak, T. Laroche, S. M. Gasser, and L. Guarente. 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89:381-391. [DOI] [PubMed] [Google Scholar]
- 22.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi, Y., Y. Oka, M. Kobayashi, Y. Uesono, A. Toh-e, and A. Kikuchi. 1994. A new yeast gene, HTR1, required for growth at high temperature, is needed for recovery from mating pheromone-induced G1 arrest. Mol. Gen. Genet. 245:107-116. [DOI] [PubMed] [Google Scholar]
- 24.Krause, S. A., and J. V. Gray. 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12:588-593. [DOI] [PubMed] [Google Scholar]
- 25.Lee, K. S., K. Irie, Y. Gotoh, Y. Wantanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 13:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. S., and D. E. Levin. 1992. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin, D. E. 2005. Cell wall integrity in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin, D. E., F. O. Fields, R. Kunisawa, J. M. Bishop, and J. Thorner. 1990. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62:213-224. [DOI] [PubMed] [Google Scholar]
- 29.Lorberg, A., H. P. Schmitz, J. J. Jacoby, and J. J. Heinisch. 2001. Lrg1p functions as a putative GTPase-activating protein in the Pkc1p-mediated cell integrity pathway in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:514-526. [DOI] [PubMed] [Google Scholar]
- 30.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 31.Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:1511-1519. [DOI] [PubMed] [Google Scholar]
- 32.Murata, Y., and R. P. Wharton. 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80:747-756. [DOI] [PubMed] [Google Scholar]
- 33.Nierras, C. R., and J. R. Warner. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274:13235-13241. [DOI] [PubMed] [Google Scholar]
- 34.Nonaka, H., K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno, M. Umikawa, A. Mino, and Y. Takai. 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14:5931-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogas, J., B. J. Andrews, and I. Herskowitz. 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66:1015-1026. [DOI] [PubMed] [Google Scholar]
- 36.Ohkuni, K., Y. Kikuchi, K. Hara, T. Taneda, N. Hayashi, and A. Kikuchi. 2006. Suppressor analysis of the mpt5/htr1/uth4/puf5 deletion in Saccharomyces cerevisiae. Mol. Genet. Genomics 275:81-88. [DOI] [PubMed] [Google Scholar]
- 37.Olivas, W., and R. Parker. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19:6602-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajavel, M., B. Philip, B. M. Buehrer, B. Errede, and D. E. Levin. 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3969-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray, A., R. E. Hector, N. Roy, J. H. Song, K. L. Berkner, and K. W. Runge. 2003. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 33:522-526. (Erratum, 34:113.) [DOI] [PubMed] [Google Scholar]
- 40.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88:531-542. [DOI] [PubMed] [Google Scholar]
- 41.Seay, D., B. Hook, K. Evans, and M. Wickens. 2006. A three-hybrid screen identifies mRNAs controlled by a regulatory protein. RNA 12:1594-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spassov, D. S., and R. Jurescic. 2003. The PUF family of RNA binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55:359-366. [DOI] [PubMed] [Google Scholar]
- 43.Tadauchi, T., K. Matsumoto, I. Herskowitz, and K. Irie. 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20:552-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres, J., C. J. Di Como, E. Herrero, and M. A. De La Torre-Ruiz. 2002. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 277:43495-43504. [DOI] [PubMed] [Google Scholar]
- 45.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, B., C.-Y. Chen, and D. E. Levin. 1994. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J. Biol. Chem. 269:16829-16836. [PubMed] [Google Scholar]
- 47.Watanabe, D., M. Abe, and Y. Ohya. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 18:943-951. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickens, M., D. S. Bernstein, J. Kimble, and R. Parker. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18:150-157. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, B., M. Gallegos, Z. Puoti, E. Durkin, S. Fields, J. Kimble, and M. P. Wickens. 1997. A conserved RNA-binding protein that regulates sexual fates in C. elegans hermaphrodite germ line. Nature 390:477-480. [DOI] [PubMed] [Google Scholar]