Abstract
The interaction between Candida albicans and cells of the innate immune system is a key determinant of disease progression. Transcriptional profiling has revealed that C. albicans has a complex response to phagocytosis, much of which is similar to carbon starvation. This suggests that nutrient limitation is a significant stress in vivo, and we have shown that glyoxylate cycle mutants are less virulent in mice. To examine whether other aspects of carbon metabolism are important in vivo during an infection, we have constructed strains lacking FOX2 and FBP1, which encode key components of fatty acid β-oxidation and gluconeogenesis, respectively. As expected, fox2Δ mutants failed to utilize several fatty acids as carbon sources. Surprisingly, however, these mutants also failed to grow in the presence of several other carbon sources, whose assimilation is independent of β-oxidation, including ethanol and citric acid. Mutants lacking the glyoxylate enzyme ICL1 also had more severe carbon utilization phenotypes than were expected. These results suggest that the regulation of alternative carbon metabolism in C. albicans is significantly different from that in other fungi. In vivo, fox2Δ mutants show a moderate but significant reduction in virulence in a mouse model of disseminated candidiasis, while disruption of the glyoxylate cycle or gluconeogenesis confers a severe attenuation in this model. These data indicate that C. albicans often encounters carbon-poor conditions during growth in the host and that the ability to efficiently utilize multiple nonfermentable carbon sources is a virulence determinant. Consistent with this in vivo requirement, C. albicans uniquely regulates carbon metabolism in a more integrated manner than in Saccharomyces cerevisiae, such that defects in one part of the machinery have wider impacts than expected. These aspects of alternative carbon metabolism may then be useful as targets for therapeutic intervention.
Candida albicans is the most important fungal pathogen of humans, particularly affecting individuals with compromised immune systems or implanted medical devices. Disseminated hematogenous candididasis, the most severe manifestation, is the fourth most common hospital-acquired infection and has a mortality rate of about 40% (50). Oropharyngeal thrush and vaginitis, nonlethal infections of mucosal surfaces, are the most frequent forms of the disease, but C. albicans can infect essentially any body site (17, 27). More often, however, C. albicans is a benign component of the mammalian microbiota, living in the gut, mouth, vagina, and on the skin as its primary ecological niche. Despite increasing interest, relatively little is known about the basic biology of C. albicans in either the commensal or pathogenic states. Developing such insight is instrumental in devising novel strategies for reducing the incidence and severity of these infections.
The status of the host's innate immune system is the primary determinant of infection. In vitro, the interaction of C. albicans with phagocytes is very dynamic—phagocytosis by macrophages, for instance, induces hyphal growth within the immune cell as a means of escape. The complexity of this interaction has been confirmed through genomic expression profiling of C. albicans cells phagocytosed by macrophages or neutrophils (12, 18, 32). Over 500 genes are differentially regulated following macrophage phagocytosis (18), and a similarly massive response accompanies neutrophil phagocytosis (12, 32); surprisingly, there are substantial differences between the two programs, suggesting that this fungal pathogen is able to distinguish the two cell types. The macrophage program is similar to that seen after nutrient starvation, including repression of translation and glycolysis and, concurrently, activation of metabolic pathways required to use less favored carbon sources, including the glyoxylate cycle, β-oxidation, and gluconeogenesis. Differential display technology has revealed a similar pattern of gene expression in phagocytosed cells (29). These three interconnected pathways ultimately convert lipids to acetate to glucose (18).
The relevance of these findings has been confirmed by demonstration that the glyoxylate cycle is necessary for full virulence in C. albicans (19). The primary role of this microbe-specific pathway is to assimilate two-carbon compounds (reviewed in reference 20). A similar role for the glyoxylate cycle has been shown in Mycobacterium tuberculosis (22, 23) as well as several fungal and bacterial pathogens of plants, including Magnaporthe grisea, Leptosphaeria maculans, Stagonospora nodorum, and Rhodococcus equi (15, 41, 48, 49). The key glyoxylate enzyme isocitrate lyase is induced in vivo in C. albicans and Cryptococcus neoformans, though it is not required for virulence in the latter species (4, 33).
These results indicate that diverse pathogens metabolize or are exposed to nonsugar carbon sources in vivo. However, beyond the glyoxylate cycle, little is known about the role of metabolic pathways required to utilize such nutrients during infections. Gluconeogenesis and the glyoxylate cycle have been reported in conflicting reports to be either required or dispensable for virulence in Salmonella (2, 10, 45). Gluconeogenesis-defective mutants of the plant pathogen Xanthomonas campestris are avirulent (44), as are peroxisome biogenesis and fatty acid degradation mutants in the plant fungal pathogen Colletotrichum lagenarum (16). It was recently reported that peroxosome function was not required for pathogenicity in C. albicans (28). Deletion of the C. albicans gluconeogenic gene PCK1 confers a moderate reduction in virulence (4). Curiously, disabling both gluconeogenesis and glycolysis via disruption of fructose-1,6-bisaldolase (FBA1) has only a modest impact on infectivity in C. albicans (31).
Some aspects of carbon metabolism present intriguing candidates for drug discovery. The glyoxylate cycle does not exist in mammals, and β-oxidation of fatty acids is highly divergent between mammals and fungi. To better understand the importance of these pathways, we have examined the physiology and virulence of C. albicans strains deficient in β-oxidation, the glyoxylate cycle, and gluconeogenesis, through deletions of genes encoding key enzymes in each pathway, the β-oxidation multifunctional protein (FOX2), isocitrate lyase (ICL1), and fructose-1,6-bisphosphatase (FBP1). Each of these mutants is attenuated to some degree in a mouse model of disseminated candidiasis, confirming that alternative carbon sources are relevant nutrients in vivo, and, in fact, C. albicans likely uses multiple carbon sources during infection. Finally, in vitro phenotypes and genomic analysis indicate that the regulatory networks that control alternative carbon metabolism in C. albicans differ significantly from the paradigms developed in Saccharomyces cerevisiae. Our findings confirm the importance of these central metabolic processes in fungal pathogenesis.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used in this study are listed in Table 1 and are based on SC5314 and its Ura− derivative, CAI4-F2. Standard yeast media were used (37), including YPD (1% yeast extract, 2% peptone, 2% dextrose) and YNB (0.17% yeast nitrogen base, 0.5% ammonium sulfate). Standard YNB was supplemented with 2% glucose. In some experiments, the glucose was replaced with other carbon sources, also at 2%. 5-Fluoroorotic acid (5-FOA) medium was YNB plus 2% glucose, supplemented with 0.2 mM uracil, 0.2 mM uridine, and 0.1% 5-fluoroorotic acid. C. albicans transformations were done either using the modified lithium acetate method (7) or via electroporation (30).
TABLE 1.
Genotypes of the fungal strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| C. albicans | ||
| SC5314 | Wild type | 11 |
| CAI4-F2 | ura3::λimm434/ura3::λimm434 | 11 |
| MRC6 | ura3::λimm434/ura3::λimm434 fox2::hisG/ fox2::hisG RPS10/rps10::URA3 | This study |
| MRC5 | ura3::λimm434/ura3::λimm434 fox2::hisG/ fox2::hisG RPS10/rps10::FOX2-URA | This study |
| MRC8 | ura3::λimm434/ura3::λimm434 fox2::hisG/ fox2::hisG RPS10/rps10::URA3 | This study |
| MRC10 | ura3::λimm434/ura3::λimm434 icl1::hisG/ icl1::hisG RPS10/rps10::URA3 | This study |
| MRC11 | ura3::λimm434/ura3::λimm434 icl1::hisG/ icl1::hisG RPS10/rps10::ICL1-URA3 | This study |
| MRC14 | ura3::λimm434/ura3::λimm434 fbp1::hisG/ fbp1::hisG RPS10/rps10::URA3 | This study |
| MRC15 | ura3::λimm434/ura3::λimm434 fbp1::hisG/ fbp1::hisG RPS10/rps10::FBP-URA3 | This study |
| MRC16 | ura3::λimm434/ura3::λimm434 fbp1::hisG/ fbp1::hisG RPS10/rps10::URA3 | This study |
| S. cerevisiae | ||
| MLY40 | ura3-52 MATα | 21 |
| MLY41 | ura3-52 MATa | 21 |
| MRY1 | Δfox2::G418 ura3-52 MATa | This study |
| MRY3 | Δicl1::G418 ura3-52 MATα | This study |
Mutant construction.
All mutants were constructed in CAI4-F2 using the standard URA-blaster method of Fonzi and Irwin, which utilizes a recyclable hisG-URA3-hisG cassette originally adapted from S. cerevisiae (1, 11). To make plasmid-based constructs to precisely replace the open reading frame, the 5′ and 3′ untranslated regions (UTR) were cloned on a single fragment, separated by a BamHI site, using an overlap PCR strategy. The fusion products were cloned into pGEM5 (fox2Δ) or pBSKII+ (fbp1Δ). The hisG-URA3-hisG cassette was obtained by digesting plasmid pCUB-6 (11) with BamHI/BglII/PvuII and ligated into the fusion plasmids cut with BamHI. The resulting disruption plasmids were pML304 (fox2Δ::hisG-URA3-hisG) and pMR8 or pMR9 (fbp1Δ::hisG-URA3-hisG).
For transformation, the disruption construct was liberated from the plasmid backbone using PstI (fox2Δ) or HindIII/SacII (fbp1Δ). These were used to transform CAI4-F2 to uridine prototrophy. Correct heterozygotes were confirmed by PCR. Ura− recombinants were selected on 5-FOA medium (5). The second allele was disrupted in the same manner, and homozygous mutants were confirmed by PCR for the presence of the disruption alleles and the absence of the wild-type allele. For fbp1Δ, the first allele was disrupted with pMR8 and the second was disrupted with pMR9. These constructs differ in the orientation of the hisG-URA3-hisG cassette with respect to the FBP1 open reading frame.
The icl1Δ/icl1Δ mutant strain MLC9 has been described previously (19). This strain expresses URA3 from the ICL1 locus. Uridine auxotrophs of MLC9 were selected on 5-FOA medium to allow complementation using CIp10-based plasmids, as described below.
S. cerevisiae icl1Δ and fox2Δ mutants were constructed by PCR-mediated disruption, using plasmid pFA6-KanMX2 (47) as a template and oligonucleotide primers with homology to the genes to be disrupted. Linear PCR products were used to transform strains MLY40 for icl1Δ and MLY41 for fox2Δ (21) to G418 resistance. Disruptions were confirmed by PCR. pRS316 (38) was used to complement the uracil auxotrophy for the carbon source growth assay.
Complementation.
Complementation plasmids for FOX2, ICL1, and FBP1 were constructed by PCR by amplifying the open reading frame flanked by 500 to 700 bp of 5′ UTR and ∼300 bp of 3′ UTR. PCR products were subsequently cloned into the integrating plasmid CIp10, which targets integration to the RPS10 locus (25). This methodology has been demonstrated to be phenotypically neutral and to eliminate position effects based on the expression levels of URA3 (6). The complementation plasmids (pMR11, FOX2; pMR12, ICL1; and pMR2, FBP1) were introduced to uridine auxotrophic homozygous mutants by transformation. Empty CIp10 was integrated into the mutant strains. Insertion at RPS10 was verified by PCR. URA3 is integrated at the RPS10 locus in all strains used here, with the exception of the wild-type strain, SC5314.
In vitro growth assays.
Growth assays used solid YNB media containing 2% glucose, potassium acetate, ethanol, oleate, citrate, or glycerol as the sole carbon source and incubation at room temperature (∼24°C), 30°C, or 37°C for 3 to 7 days, depending on the carbon source, as indicated in the figure legends. For the spot dilution assays, strains were grown into the mid-log phase in YPD, collected by centrifugation, and washed with water. They were transferred to 96-well plates, at an optical density at 600 nm (OD600) of 0.5, and serially diluted fivefold. Solid media were spotted with these dilutions using a multiprong pin replicator. The toxicity assays used solid YP medium (1% yeast extract, 2% peptone) with 2% glucose, glycerol, potassium acetate, or ethanol.
For growth assays in liquid media, strains were grown in YNB-glucose at 30°C or 37°C overnight. The next day, cells were harvested by centrifugation, washed twice with water, and resuspended at an OD600 of 0.08 in YNB media containing the appropriate carbon source. Growth of the cultures was assessed over a period of 5 to 7 days by measuring OD600.
Northern analysis.
SC5314 cells were grown overnight in YNB media with 2% glucose, harvested by centrifugation, and washed twice with water. Cells were resuspended in YNB media containing 2% glucose, acetate, or oleate and grown for 1 h, pelleted by centrifugation, and frozen on dry ice-ethanol. RNA was isolated using the hot acidic phenol method (3). RNA was also prepared from icl1Δ, fbp1Δ, and fox2Δ mutants grown in acetate for 1 h. A total of 15 ng of RNA was loaded and run on a 1% MOPS (morpholinepropanesulfonic acid)-agarose gel and then transferred to a nylon membrane. Gene-specific probes were amplified by PCR and were labeled with the RadPrime DNA labeling system from Invitrogen. Probes were purified using Roche Quick Spin columns. Blots were incubated in prehybridization solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 5× Denhardt's solution, 0.1% sodium dodecyl sulfate (SDS), and 100 μg/ml single-stranded DNA for 2 h at 42°C followed by hybridization overnight. Images were processed using a Storm PhosphorImager and exposed to film for autoradiography. rRNA was used as a loading control.
In vivo virulence assays.
Mouse virulence assays were performed as described previously (19). Adult, female, outbred ICR mice were obtained from Harlan. Cultures of C. albicans were grown in YPD to mid-log phase and collected by centrifugation. Cells were washed and resuspended in phosphate-buffered saline, and mice were infected via tail vein injection with 106 C. albicans yeast-form cells. The group sizes were 10 to 12 mice per strain. Infected mice were subsequently monitored for signs of infection and euthanized when moribund according to approved protocols. Survival data were analyzed with Prism3 (Graphpad Software) using the log rank test. Statistical significance was defined as a P value of less than 0.05. All animal assays were conducted in accordance with protocols approved by the University of Texas Health Science Center Animal Welfare Committee.
RESULTS
Our previous work showed that C. albicans responds to phagocytosis by macrophages with a metabolic shift that is highly similar to carbon starvation. We further demonstrated that the glyoxylate cycle was necessary for full virulence (19). These results indicated that C. albicans likely encounters glucose-poor conditions during infection and that the ability to utilize alternative carbon sources is a virulence attribute in this species. Other than the role of the glyoxylate cycle, little is known about nutrient availability in different host environments or the importance of carbon metabolic pathways in either commensalism or pathogenicity of C. albicans. In fact, while some nutrients are well known to be scarce in vivo—iron is a classic example—there have been relatively few studies addressing carbon acquisition and/or metabolic activity in pathogens of any kind.
C. albicans, like most fungi, uses sugars, particularly glucose, as the preferred carbon source. However, a wide variety of nonfermentable carbon sources can also satisfy cellular requirements, including but not limited to ethanol, acetate, amino acids, glycerol, and fatty acids. Collectively, these compounds are sometimes called “alternative” or “nonpreferred” carbon sources because fungi do not use them in the presence of sugars. These alternative sources are metabolized by three main pathways: β-oxidation of fatty acids, the glyoxylate cycle, and gluconeogenesis. The main purposes of these pathways are to provide energy, replenish tricarboxylic acid (TCA) cycle intermediates and acetyl-coenzyme A (CoA), and produce glucose.
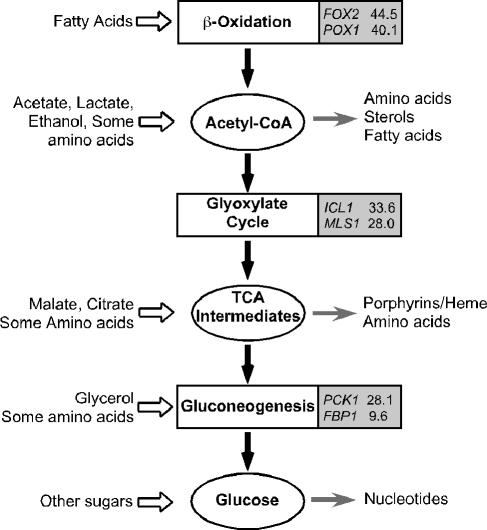
These pathways are shown in schematic form in Fig. 1. Virtually all cellular building blocks are synthesized starting from one of the three intermediates highlighted here: acetyl-CoA, compounds of the TCA cycle, and glucose, which can be produced from various carbon sources through these central pathways. Figure 1 also lists the induction of key genes in each pathway following macrophage phagocytosis (18). Such a coordinated and rapid response strongly implicates these processes in pathogenesis. To understand the role of alternative carbon metabolism in vivo, the carbon sources encountered by C. albicans during infections, and how these systems may differ from those of other species, we used a gene disruption approach to disable these pathways individually.
FIG. 1.
Central carbon metabolism in the absence of glucose. The biochemical pathways are shown in boxes, with the postphagocytosis induction of key genes in the shaded part. Ovals represent the critical intermediates. Inputs are on the left (open arrows); catabolic products are indicated on the right (gray arrows). Induction data are from reference 18.
Construction of alternative carbon metabolism mutants.
Genes chosen for analysis were FOX2, FBP1, and ICL1. FOX2 (orf19.1288) encodes the “multifunctional protein” of β-oxidation, so named because it has both 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase activities, each of which is required for fatty acid degradation. FOX2 was selected because it is essential for β-oxidation in S. cerevisiae (13), is present as a single gene in C. albicans, and is the most highly induced gene of β-oxidation following phagocytosis (44.5-fold; see reference 18). The 923-amino-acid CaFOX2 protein is 52% identical to FOX2 from S. cerevisiae. FOX2 was disrupted using the standard Ura-blaster methodology (1, 11). The disruption construct was made using overlap PCR to precisely remove the open reading frame, from the ATG to the stop codon.
FBP1, encoding fructose-1,6-bisphosphatase, was disrupted in a similar manner. FBP1 (orf19.6178; 331 amino acids) is one of two gluconeogenesis-specific proteins (the other is phosphoenolpyruvate carboxykinase; PCK1), catalyzing the penultimate step in the synthesis of glucose. It, too, is a highly conserved enzyme, with 64% identity to S. cerevisiae FBP1 and 45% identity to the Escherichia coli Fbp. An fbp1 mutant in S. cerevisiae does not grow in the absence of glucose or other hexoses (36).
Both mutants were complemented by cloning the PCR-amplified genes into the CIp10 plasmid (25), which directs integration to the RPS10 locus. This method has been demonstrated to eliminate potential position effects based on altered URA3 expression (6) and to be appropriate for both in vitro and in vivo experiments. In the mutant strains, URA3 was also integrated at RPS10 using an empty CIp10 vector. For both the fox2Δ and fbp1Δ mutants, we tested two independently constructed mutants, one of which was complemented with a wild-type copy of the gene. Our earlier work with ICL1 was done prior to the appreciation of URA3 position effects on virulence (19); for this reason, we reconstructed a pair of strains (mutant and complemented) with CIp10 to compare with the fox2Δ and fbp1Δ strains.
In vitro phenotypes.
Strains lacking FOX2, ICL1, or FBP1 are viable and grow at wild-type rates on media containing glucose as the carbon source, as expected. Using precedents from S. cerevisiae and numerous other species, we established predictions for the carbon utilization phenotypes of these mutant strains: fox2Δ mutants should fail to grow on media containing fatty acids, such as oleic acid, a monounsaturated C-18 lipid; icl1Δ mutants should not grow in the presence of fatty acids, acetate, or ethanol; and fbp1Δ mutants should not utilize any nonfermentable carbon source.
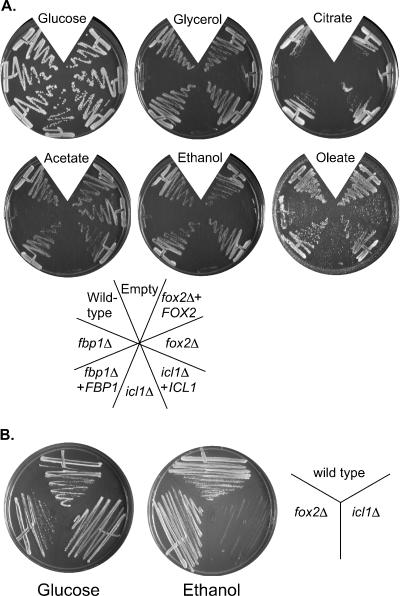
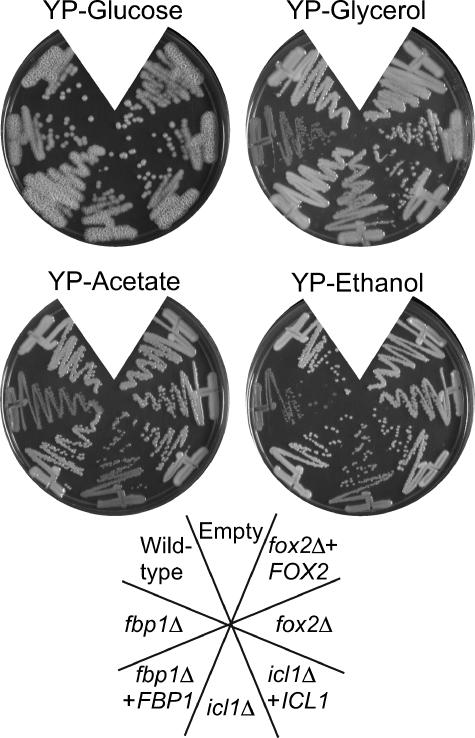
We tested these predictions using a straightforward growth assay on solid media containing different compounds as the carbon source. Included in this panel were glucose, glycerol, potassium acetate, ethanol, citric acid, and oleic acid, each present at 2% in minimal YNB media. As expected, fox2Δ mutant strains were unable to grow when fatty acids such as oleic, palmitic, linoleic, or myristic acids were the sole carbon source (Fig. 2A and data not shown). Surprisingly, the growth defects were not limited to media containing lipids (Fig. 2A). In particular, the fox2Δ mutant was also unable to utilize ethanol as a carbon source and grew poorly on citrate and glycerol. The icl1Δ mutant strain also exhibited more extensive defects than predicted and was entirely unable to grow on citrate and glycerol, in addition to the anticipated phenotypes on fatty acids, ethanol, and oleate. Growth was restored in the complemented strains. We also tested a second, independently constructed, mutant for each gene with identical results (data not shown).
FIG. 2.
Growth phenotypes of the fox2Δ, icl1Δ, and fbp1Δ mutants. (A) C. albicans strains were grown on minimal YNB media containing the indicated carbon source for 3 days (glucose, glycerol, acetate, ethanol) or 6 days (citrate, oleate) at 30°C. Strains were wild type (SC5314), fbp1Δ (MRC14), fbp1Δ + FBP1 (MRC15), icl1Δ (MRC10), icl1Δ + ICL1 (MRC11), fox2Δ (MRC6), and fox2Δ + FOX2 (MRC5). (B) S. cerevisiae wild-type (MLY41), icl1Δ (MRY1), and fox2Δ (MRY2) cells were streaked on minimal YNB media containing 2% glucose or 2% ethanol and grown for 3 days at 30°C.
These observations were surprising for several reasons. FOX2, ICL1, and FBP1 are each structural enzymes in their respective pathways. Their substrate specificity and biochemical function have been thoroughly established, and we did not expect any “cross talk” between the pathways. Mutations in these genes have been studied in a wide variety of systems, and there have been no indications that they have more global regulatory roles. In S. cerevisiae, for instance, the phenotypes of these mutants are clearly confined to growth on their relevant substrate. To test one of these, we constructed S. cerevisiae strains lacking ICL1 or FOX2 and assessed their ability to use ethanol as the sole carbon source (Fig. 2B). The S. cerevisiae fox2Δ strain grows at wild-type rates on this carbon source, in contrast to the C. albicans results.
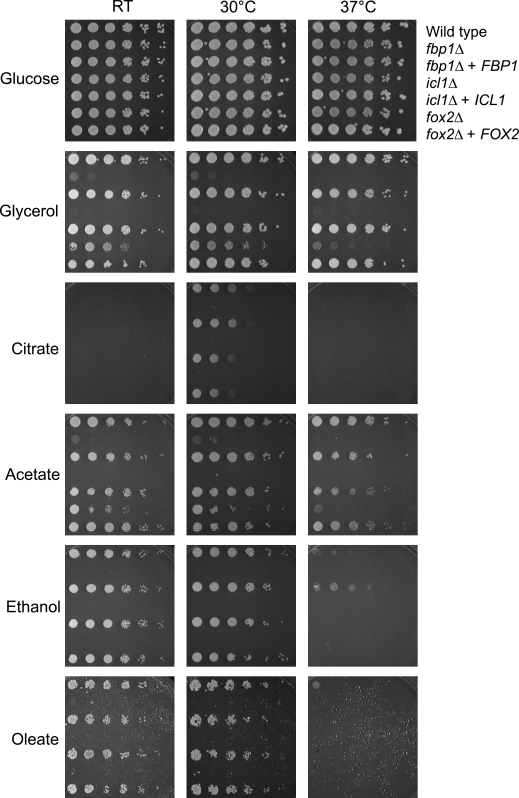
To more thoroughly test these strains on alternate carbon sources, we used a spot dilution assay. In this assay, logarithmically growing cells in rich YPD medium were washed in water and diluted to an OD600 of 0.5 in a 96-well plate. They were then serially diluted 1:5 in water five more times and applied as spots to solid media using a pin replicator. Because initial observations had suggested that the carbon utilization phenotypes may be more severe at higher temperatures, we performed this assay at room temperature (∼24°C), 30°C, and 37°C (Fig. 3).
FIG. 3.
Temperature and carbon source affect alternative carbon utilization. The C. albicans wild-type (SC5314), fbp1Δ (MRC14), fbp1Δ + FBP1 (MRC15), icl1Δ (MRC10), icl1Δ + ICL1 (MRC11), fox2Δ (MRC6), and fox2Δ + FOX2 (MRC5) strains were grown to mid-log phase and serially diluted, 1:5, in a 96-well plate. The dilutions were spotted using a multiprong replicator onto minimal YNB media containing the indicated carbon source at 2%.
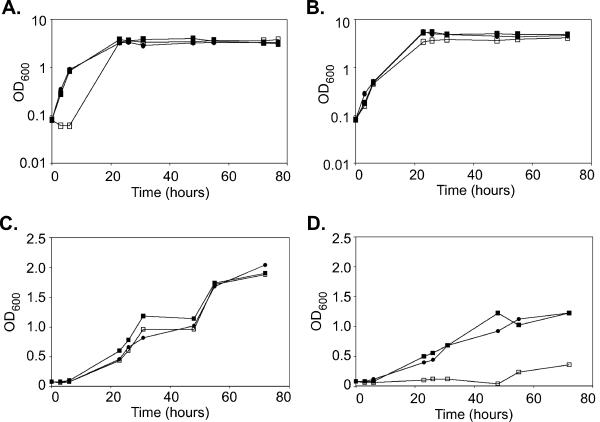
Three observations stand out in these data. First, there are no growth differences in the presence of glucose for any of these mutants (see also Fig. 4A and B). Second, the ability to assimilate nonpreferred carbon sources is impaired at elevated temperatures. C. albicans does not grow as well at 37°C as at 30°C on acetate or glycerol. This defect is most pronounced on media containing citrate, ethanol, and oleate. Citrate utilization, in particular, is very sensitive to temperature, with growth observed only at 30°C. This is somewhat surprising as C. albicans generally grows more rapidly at 37°C, the temperature of its primary niche in the mammalian host, than it does at 30°C. Finally, the temperature effect is even more severe in the mutants. This is particularly visible for the fox2Δ mutant on acetate, which exhibits the wild-type growth rate at 30°C, but does not grow at 37°C (Fig. 4C and D).
FIG. 4.
fox2Δ mutant strains cannot grow in the presence of acetate only at elevated temperature. The wild-type (SC5314), fox2Δ (MRC6), and fox2Δ + FOX2 (MRC5) strains were grown in liquid minimal YNB medium with 2% glucose, washed with water, resuspended in YNB medium containing either glucose or potassium acetate, and incubated in shaking cultures at 30°C or 37°C. Growth was monitored by OD600 at the indicated times. (A) Glucose, 30°C. (B) Glucose, 37°C. (C) Potassium acetate, 30°C. (D) Potassium acetate, 37°C. Note that the glucose growth curves are plotted on a logarithmic scale, while the acetate curves are on a linear scale. Filled circles, wild type (SC5314); open squares, fox2Δ (MRC6); filled squares, fox2Δ + FOX2 (MRC5).
Alternative carbon sources are not toxic.
We considered whether it was possible that perturbations in carbon metabolism may render certain compounds toxic to the cell. For instance, these mutations may lead to the accumulation of usually transient intermediates that are deleterious at high concentrations: acetaldehyde, an intermediate in the conversion of ethanol to acetyl-CoA, inhibits the growth of S. cerevisiae at concentrations above 0.3 g/liter, while ethanol itself is well tolerated even above 100 g/liter (43). To test this hypothesis, we incubated the C. albicans wild-type and mutant strains on the undefined medium YP supplemented with glucose (standard YPD), glycerol, acetate, or ethanol, each present at 2%. In this medium, there are numerous compounds to satisfy carbon requirements, including amino acids, nucleotides, fatty acids, etc. The fbp1Δ, icl1Δ, and fox2Δ mutants grew on each of these media, indicating the growth defects shown in Fig. 2 to 4 on minimal media were not due to toxic effects of these carbon sources. These assays were performed at 37°C, the temperature at which the carbon source-dependent growth effects are most pronounced. As a second test, we incubated these strains on media containing both glucose and an alternative carbon source (e.g., YPDE with 2% glucose and 2% ethanol). The presence of ethanol, glycerol, or acetate, in addition to glucose, did not inhibit growth compared to media containing glucose alone (data not shown), again providing evidence against the toxic effects of these compounds. As can be seen in Fig. 5, the fbp1Δ mutant does grow more slowly in the presence of these alternative carbon sources, presumably because gluconeogenesis is required to use many of the compounds in YP medium.
FIG. 5.
Alternative carbon sources are not toxic. C. albicans fbp1Δ, icl1Δ, and fox2Δ strains (see the legend to Fig. 2) were grown on YP media supplemented with 2% glucose, glycerol, acetate, or ethanol and grown for 2 days at 37°C.
The three mutants were also tested for other phenotypes, including filamentous growth under a variety of inducing conditions and stress sensitivity. In all tests, they behaved identically to the wild type (data not shown).
Regulation of alternative carbon genes.
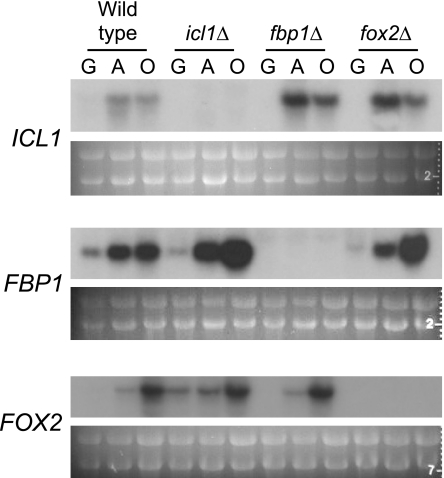
Given the unusual phenotypes, we investigated the regulation of these genes in different carbon sources. In wild-type strains, FBP1, ICL1, and FOX2 are expressed at low levels in glucose-grown cells (Fig. 6). Expression increases markedly in cells incubated in potassium acetate for 1 h. FOX2 is further induced in the presence of oleic acid. These results are similar to the expression profiles of the S. cerevisiae homologs (34, 36, 39).
FIG. 6.
Carbon source-dependent expression of FBP1, ICL1, and FOX2. The wild-type (SC5314), icl1Δ (MRC10), fbp1Δ (MRC14), and fox2Δ (MRC6) strains were grown to mid-log phase in minimal YNB medium with glucose, collected by centrifugation, washed with water, and shifted to media containing glucose (G), potassium acetate (A), or oleate (O) for 1 h. RNA was prepared from these cells, and expression of the three genes was determined by Northern analysis. The absence of signal in the mutant strains also confirms the genotype. The numbers visible at the right side of the rRNA images are from the fluorescent sizing ruler that was inadvertently photographed with the gel.
We also assayed expression of these three genes in the mutant strains to determine whether the unexpected growth phenotypes observed, particularly for the icl1Δ and fox2Δ mutant strains, could be explained by misregulation of other carbon metabolic genes. However, expression levels of FBP1, ICL1, and FOX2 were very similar in the mutants compared to the wild-type strain (Fig. 6), indicating that transcriptional regulation of these genes does not explain the growth phenotypes. Transcripts were not detected in strains lacking the gene used as a probe (e.g., ICL1 message is not seen in the icl1Δ mutant), confirming the mutant genotypes and probe specificity.
Alternative carbon metabolism is required in vivo.
Previous work has shown that icl1Δ mutants are significantly attenuated in a mouse model of hematogenously disseminated candidiasis (19). To determine the extent to which alternative carbon sources are relevant in vivo, we tested these mutants in the standard mouse tail vein injection model. In this model, 106 C. albicans cells (yeast form) are injected into outbred adult female ICR mice. Subsequently, the animals are monitored for signs of clinical infection and euthanized when moribund according to approved protocols.
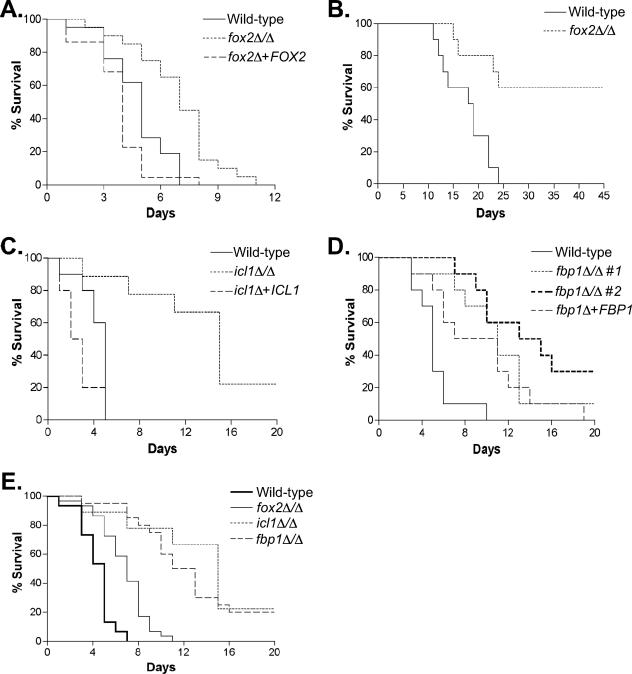
Using this assay, we determined that fox2Δ mutant strains have a mild reduction in virulence (Fig. 7A). The mean time to death (MTD) for the fox2Δ mutant was 6.9 days, compared to 4.8 days for the wild-type strain. Complementation of the mutant with the FOX2 gene fully restored virulence. Given this small difference in virulence, the assay was repeated, giving essentially identical results; the data in Fig. 7A are a combination of both assays (a total of 20 to 22 mice per strain). Statistical analysis of these data is presented in Table 2. In addition, we tested a second fox2Δ mutant, constructed completely independently, which had a similar phenotype (Table 2) (data not shown).
FIG. 7.
Alternative carbon metabolism is required for full virulence. (A) A total of 106 cells of each strain were injected via the tail vein. Strains are wild type (SC5314), fox2Δ (MRC6), and fox2Δ + FOX2 (MRC5). The data are a composite of two separate experiments; see the text and Table 2 for details. (B) A lower inoculum (105 cells per strain) was injected. Strains are wild type (SC5314) and fox2Δ (MRC6). (C) A single strain pair was tested (icl1Δ, MRC10; icl1Δ + ICL1, MRC11). These data are similar to those previously reported; unlike those experiments, URA3 is integrated at RPS10 in these strains; see the text and Table 2 for details. (D) fbp1Δ. Two independent mutant strains (#1, MRC14; #2, MRC16) and one complemented strain were tested (MRC15, isogenic to MRC14). (E) Composite data. Data from the independent mutant strains and replicate experiments, when performed, were pooled to see the overall pattern of virulence.
TABLE 2.
Statistical analysis of virulence data
| Type of strain | No. of mice | MTD in days (% surviving)a | P valueb |
|---|---|---|---|
| fox2 | |||
| Wild type (SC5314) | 21c | 4.8 | |
| fox2Δ (MRC6) | 20c | 6.9 | 0.0007 |
| fox2Δ + FOX2 (MRC5) | 22c | 3.8 | |
| Wild type | 10 | 4.4 | |
| fox2Δ (MRC8) | 10 | 8.4 | 0.008 |
| Wild type (105) | 10 | 17.4 | |
| fox2Δ (MRC6 [105]) | 10 | 45 (60) | 0.0015 |
| icl1 | |||
| Wild type (SC5314) | 10 | 4.2 | |
| icl1Δ (MRC10) | 10 | 20 (20) | 0.002 |
| icl1Δ + ICL1 (MRC11) | 10 | 2.7 | |
| fbp1 | |||
| Wild type (SC5314) | 10 | 5.2 | |
| fbp1Δ (MRC14) | 10 | 20 (10) | 0.0006 |
| fbp1Δ (MRC16) | 10 | 20 (30) | <0.0001 |
| fbp1Δ + FBP1 (MRC15) | 10 | 9.4 |
In several experiments, not all animals died. Values in parentheses represent the percentages of animals surviving at the end of the experiment.
P values are calculated compared to the wild-type strain used in that experiment using the log rank test (Prism3; Graphpad Software).
This experiment was repeated twice. The data are grouped here for convenience.
We also used a smaller inoculum to determine whether the virulence defect would be more pronounced at the lower level. Mice injected with 105 wild-type cells succumbed to the infection with an MTD of 17.4 days, while 60% of the fox2Δ mutant-injected mice survived to 45 days, when the experiment was terminated (Fig. 7B).
To continue our analysis of alternative carbon metabolism during infection, we tested the icl1Δ and fbp1Δ mutant strains as well (Fig. 7C and D). We retested the icl1Δ mutant using strains that control for URA3 position effects that may have complicated our earlier work (19). The reduction in virulence was similar to what we observed previously (19) and what another group using a properly complemented strain pair reported recently (4).
We also tested two independent fbp1Δ mutant strains (Fig. 7B). Both strains behaved similarly in this assay, with either 10% or 30% of the animals surviving to day 20, compared to an MTD of 5.2 days for the wild type. The complemented strain partly restored virulence (MTD = 9.4 days). The wild-type-mutant comparisons were again significant (n = 10, P < 0.05).
Figure 7E shows a composite survival curve to illustrate the overall patterns of virulence seen in mutant strains for these three genes. The fox2Δ mutant is modestly attenuated compared to the wild type. The icl1Δ and fbp1Δ mutants are severely attenuated and are very similar to each other. From these studies, we conclude that C. albicans must acquire and utilize less-preferred carbon sources during an infection.
DISCUSSION
This work demonstrates that several facets of alternative carbon metabolism in C. albicans are important during systemic infection. Deletions of key enzymes in the pathways of β-oxidation of fatty acids (FOX2), the glyoxylate cycle (ICL1), and gluconeogenesis (FBP1) confer virulence defects from moderate to severe. These data strongly support a model whereby C. albicans acquires and assimilates nonfermentable (nonsugar) compounds as primary sources of carbon. The potential nature of these compounds will be discussed below.
Furthermore, we have shown that these mutations alter the utilization of alternative carbon sources in unexpected ways. We find that fox2Δ mutants are unable to efficiently assimilate ethanol or acetate as sole carbon sources, the metabolism of which is independent of the enzymatic steps of β-oxidation. In S. cerevisiae, the fox2Δ mutant grows at wild-type rates in the presence of ethanol, marking an important difference in carbon metabolisms in these two species. Similarly surprising, C. albicans icl1Δ mutants use glycerol and citrate poorly or not at all, in addition to their expected inability to grow on acetate, ethanol, and fatty acids. The growth defects are not caused by accumulation of toxic intermediates resulting from perturbations in the metabolic pathways. Despite these differences, these three key genes are regulated by carbon source in a manner similar to S. cerevisiae; obviously, though, there are many genes whose misregulation in these mutants might cause the observed phenotypes and a more global analysis may be required to find the relevant perturbations in these mutants. Taken together, these data suggest that the pathways controlling alternative carbon metabolism in C. albicans are more integrated than in other fungi. We speculate that C. albicans has evolved this network to meet its specialized needs as a fungal commensal/pathogen. Further work will be required to decipher these pathways.
Why do mutations in carbon utilization pathways confer pleiotropic phenotypes?
FOX2 is an enzyme of the β-oxidation cycle. In several other bacterial and fungal systems, mutation of this “multifunctional protein” confers a single phenotype—the inability to catabolize fatty acids. In C. albicans, uniquely, FOX2 is also required for the efficient utilization of acetate, ethanol, citrate, and lactate, compounds whose assimilation is completely independent of the enzymatic steps of β-oxidation (this work and reference 28). The icl1Δ mutant is similarly pleiotropic. Why might this be? We speculate that these mutations perturb alternative carbon metabolism in one of three ways: by altering cellular physiology, gene regulation, or metabolite balances to disrupt the normal function of these pathways.
Much of alternative carbon metabolism is compartmentalized, occurring in the peroxisomes or mitochondria. Thus, disruption of normal organellar function could inhibit assimilation of nonfermentable carbon sources. Recent work from Distel and colleagues, described below, suggests that peroxisomal dysfunction is not responsible for these phenotypes (28), but does not exclude a role for the mitochondria. In S. cerevisiae, respiration-deficient mitochondrial mutants are unable to grow on nonfermentable carbon sources, but even less dramatic changes can confer carbon source- and temperature-dependent phenotypes. For instance, strains lacking the mitochondrial protein chaperone CPR3 are specifically unable to grow in the presence of lactate at 37°C (9).
Alternatively, fox2Δ and icl1Δ strains may misregulate a gene or genes required for alternative carbon utilization. We show here (Fig. 6) that several key genes are properly regulated at the transcriptional level in the mutant strains, but this was not a comprehensive analysis. Transcriptional repression of genes at key intersections of carbon metabolism—such as acetyl-CoA synthases, carnitine acetyltransferases, or mitochondrial metabolite transporters—could explain these phenotypes, but the possibilities are too numerous for a candidate gene approach and a more global survey will be required. Finally, the mutations described here may instead result in a skewed metabolite profile, such as depletion of coenzyme A or distortion of the NADP/NADPH ratio, that inhibits specific enzymatic processes or disrupts cellular energetics. Most likely, the pleiotropic phenotypes are caused by a combination of effects in which altered levels of metabolites lead to changes in cell physiology that prevent growth. Further studies will be necessary to understand these processes.
What alternative carbon sources are available in vivo?
Our previous work showed that cells lacking the key glyoxylate enzyme isocitrate lyase (ICL1) are significantly impaired during infections (19). This initial finding has been confirmed, both here and by others (4), using reengineered strains that control for possible position effects with the URA3 disruption marker used previously (6, 25). A conclusion from these earlier studies was that C. albicans must use a substrate incorporated via the glyoxylate cycle during infections. However, there are numerous such compounds, including acetate, fatty acids, ethanol, and several amino acids. Because some aspects of alternative carbon metabolism are unique to microorganisms, the identification of relevant carbon sources in vivo may highlight enzymes or pathways as attractive candidates for antifungal drug discovery.
Our data begin to address this question. The mild reduction of virulence seen with the fox2Δ mutant suggests that fatty acids are a relevant carbon compound in vivo. The unexpected carbon source-dependent growth defects of this mutant complicate this analysis, however, and recent work has called into question the importance of β-oxidation. Piekarska et al. reported very recently that disruption of peroxisomal function via mutation of the biogenesis factor PEX5 blocks β-oxidation without any discernible effect on virulence (28). They also showed that the fox2Δ mutant is unable to use ethanol, acetate, and lactate, similar to our findings, and is attenuated in the mouse model. They concluded from these observations that β-oxidation is not relevant in vivo (28).
In contrast, it is apparent that some nonfermentable compounds are important during infection. The similarly severe virulence phenotypes of fbp1Δ and icl1Δ mutants strongly indicated that substrates of the glyoxylate cycle and gluconeogenesis are found and used in at least some in vivo niches. So what is the spectrum of potential compounds? It includes any compound for which acetyl-CoA is a catabolic intermediate: acetate, ethanol, fatty acids (which we have shown to be a minor component), and 10 of the amino acids. The remaining amino acids are converted to TCA cycle intermediates (which are also potential nutrients). Several amino acid auxotrophs are fully virulent, including those unable to synthesize arginine, histidine, and leucine (26), implying that these compounds are present at sufficient levels in tissue or blood to support growth of C. albicans. Alternatively, the secreted aspartyl proteases allow C. albicans to grow in the presence of protein as a nutrient source (14, 42). The secreted aspartyl proteases are widely considered to be virulence factors, and nutrient acquisition may be one of their functions. Degradation of lipids by the host, which occurs partly in the lysosome, may provide a pool of acetate or acetyl-CoA available to a phagocytosed cell. Very recently, Distel and colleagues proposed that lactate is the relevant carbon source (28). This is certainly a physiologically accessible compound, and the data are consistent with this suggestion, but as discussed above, numerous other compounds also meet these criteria.
We believe it is likely that C. albicans finds and uses multiple carbon sources during infection. These would include substrates of the glyoxylate cycle and gluconeogenesis, in addition to glucose. C. albicans could use several compounds simultaneously at the same infection site; this is not generally favored in S. cerevisiae, but our work has illustrated key differences between these species. Alternatively, different body sites could offer diverse nutrients and our virulence data are an amalgamation of these effects. Further work will be required to dissect these responses, perhaps in a manner similar to the green fluorescent protein-promoter fusions used by Brown and colleagues to examine ICL1 expression in vivo (4).
Regulation of alternative carbon metabolism.
Northern analysis has shown that FBP1, ICL1, and FOX2 are expressed at low levels in the presence of glucose and are induced markedly after a shift to media containing acetate or oleate. Particularly highly induced is FOX2 when oleate is the sole carbon source. This carbon source regulation is similar to that observed in S. cerevisiae (34, 36, 39). However, this regulatory similarity belies the intriguing growth phenotypes of the C. albicans mutants.
To examine the regulatory networks that govern alternative carbon utilization, we have begun to identify homologs of the transcriptional regulators known in S. cerevisiae. In that model eukaryote, a shift to poor carbon conditions alleviates glucose repression mediated by the MIG1 transcriptional regulator (reviewed in references 8 and 35). Transcriptional profiling indicates that the C. albicans MIG1 homolog serves a similar function (24). Specific induction of genes based on carbon source is mediated in S. cerevisiae by the regulators CAT8 (gluconeogenesis, glyoxylate cycle), ADR1 (ethanol, acetate, amino acids), and the OAF1/PIP2 heterodimer (fatty acids, peroxisome biogenesis). Other regulators, such as GAL4, are active in the presence of sugars other than glucose. C. albicans has a clearly recognizable CAT8 homolog (40). There are two marginally conserved homologs of ADR1; in contrast, OAF1 and PIP2 are not readily apparent through sequence analysis (M. Lorenz and M. Ramirez, unpublished observations). This divergence of regulatory proteins is consistent with our suggestion that C. albicans has a more highly integrated network governing carbon metabolism such that defects in discrete aspects of carbon utilization may feed back to globally downregulate the system.
Why might C. albicans have this more integrated regulatory network? In S. cerevisiae, cellular metabolism is tuned to run mostly on glucose. When that preferred source is not available, it activates pathways to utilize small subsets of alternative carbon sources: other sugars, ethanol, amino acids, etc. For a saprobic species, the time required to switch from one source to another may be a small cost in terms of the efficiency of expressing only those genes necessary in a given condition. A pathogen like C. albicans, however, may not be able to tolerate periods of metabolic inactivity while new sets of genes are expressed in the face of immunological pressures. This may have driven the evolutionary divergence in this aspect of metabolism between the two species.
Nutritional stress during infection.
It has long been appreciated that iron limitation is a key stress encountered in the host, and pathogens of all kinds have developed sophisticated means to acquire this scarce nutrient. There has been far less emphasis on limitation for other nutrients, such as carbon and nitrogen, but there is increasing evidence from many systems that carbon starvation, in particular, is a common obstacle in vivo. Many bacterial commensals and pathogens have the ability to utilize compounds such as ethanolamine, fucose, and propanediol, suggesting that these compounds may be routinely encountered in their natural environment. Similarly, the glyoxylate cycle is required for full virulence in C. albicans (19) and Mycobacterium tuberculosis (22, 23), the most common fungal and bacterial pathogens of humans. It is also important in several plant pathogens, both bacterial and fungal (15, 41, 46, 48, 49). Each of these, then, apparently experiences carbon starvation during the infection process. Further work will be required to understand the temporal and spatial requirements for alternative carbon metabolism during an infection and to define the exact compounds available to microbial pathogens in the body.
Acknowledgments
We thank A. Carman for assistance with mouse experiments and K. Morano for help with the Northern analysis. We also appreciate helpful comments on the manuscript from H. Zhou, K. Morano, and D. Garsin.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, J. H., M. Utley, H. van den Bosch, P. Nuijten, M. Witvliet, B. A. McCormick, K. A. Krogfelt, T. R. Licht, D. Brown, M. Mauel, M. P. Leatham, D. C. Laux, and P. S. Cohen. 2000. A functional cra gene is required for Salmonella enterica serovar Typhimurium virulence in BALB/c mice. Infect. Immun. 68:3772-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., B. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Edison, NJ.
- 4.Barelle, C. J., C. L. Priest, D. M. MacCallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Brand, A., D. M. MacCallum, A. J. P. Brown, N. A. R. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 9.Davis, E. S., A. Becker, J. Heitman, M. N. Hall, and M. B. Brennan. 1992. A yeast cyclophilin gene essential for lactate metabolism at high temperature. Proc. Natl. Acad. Sci. USA 89:11169-11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, F. C., S. J. Libby, M. E. Castor, and A. M. Fung. 2005. Isocitrate lyase (AceA) is required for salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 73:2547-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fradin, C., P. De Groot, D. MacCallum, M. Schaller, F. Klis, F. C. Odds, and B. Hube. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397-415. [DOI] [PubMed] [Google Scholar]
- 13.Hiltunen, J. K., B. Wenzel, A. Beyer, R. Erdmann, A. Fossa, and W. H. Kunau. 1992. Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J. Biol. Chem. 267:6646-6653. [PubMed] [Google Scholar]
- 14.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 15.Idnurm, A., and B. J. Howlett. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 1:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura, A., Y. Takano, I. Furusawa, and T. Okuno. 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13:1945-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea & Febiger, Philadelphia, PA.
- 18.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 20.Lorenz, M. C., and G. R. Fink. 2002. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot. Cell 1:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 23.Munoz-Elias, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 25.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 26.Noble, S. M., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odds, F. C. 1988. Candida and candidosis. Bailliere Tindall, Philadelphia, PA.
- 28.Piekarska, K., E. Mol, M. van den Berg, G. Hardy, J. van den Burg, C. van Roermund, D. MacCallum, F. Odds, and B. Distel. 2006. Peroxisomal fatty acid β-oxidation is not essential for virulence of Candida albicans. Eukaryot. Cell 5:1847-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prigneau, O., A. Porta, J. A. Poudrier, S. Colonna-Romano, T. Noel, and B. Maresca. 2003. Genes involved in beta-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast 20:723-730. [DOI] [PubMed] [Google Scholar]
- 30.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 31.Rodaki, A., T. Young, and A. J. Brown. 2006. Effects of depleting the essential central metabolic enzyme fructose-1,6-bisphosphate aldolase on the growth and viability of Candida albicans: implications for antifungal drug target discovery. Eukaryot. Cell 5:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin-Bejerano, I., I. Fraser, P. Grisafi, and G. R. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 100:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rude, T. H., D. L. Toffaletti, G. M. Cox, and J. R. Perfect. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect. Immun. 70:5684-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholer, A., and H. J. Schuller. 1993. Structure and regulation of the isocitrate lyase gene ICL1 from the yeast Saccharomyces cerevisiae. Curr. Genet. 23:375-381. [DOI] [PubMed] [Google Scholar]
- 35.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139-160. [DOI] [PubMed] [Google Scholar]
- 36.Sedivy, J. M., and D. G. Fraenkel. 1985. Fructose bisphosphatase of Saccharomyces cerevisiae. Cloning, disruption and regulation of the FBP1 structural gene. J. Mol. Biol. 186:307-319. [DOI] [PubMed] [Google Scholar]
- 37.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, M. M., P. Pavlik, A. Hartig, M. Binder, H. Ruis, W. J. Cook, C. L. Denis, and B. Schanz. 1995. A C-terminal region of the Saccharomyces cerevisiae transcription factor ADR1 plays an important role in the regulation of peroxisome proliferation by fatty acids. Mol. Gen. Genet. 249:289-296. [DOI] [PubMed] [Google Scholar]
- 40.Soares-Silva, I., S. Paiva, P. Kotter, K. D. Entian, and M. Casal. 2004. The disruption of JEN1 from Candida albicans impairs the transport of lactate. Mol. Membr. Biol. 21:403-411. [DOI] [PubMed] [Google Scholar]
- 41.Solomon, P. S., R. C. Lee, T. J. Wilson, and R. P. Oliver. 2004. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 53:1065-1073. [DOI] [PubMed] [Google Scholar]
- 42.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 43.Stanley, G. A., N. G. Douglas, E. J. Every, T. Tzanatos, and N. B. Pamment. 1993. Inhibition and stimulation of yeast growth by acetaldehyde. Biotechnol. Lett. 15:1199-1204. [Google Scholar]
- 44.Tang, D.-J., Y.-Q. He, J.-X. Feng, B.-R. He, B.-L. Jiang, G.-T. Lu, B. Chen, and J.-L. Tang. 2005. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J. Bacteriol. 187:6231-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchawa Yimga, M., M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74:1130-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vereecke, D., K. Cornelis, W. Temmerman, M. Jaziri, M. Van Montagu, M. Holsters, and K. Goethals. 2002. Chromosomal locus that affects pathogenicity of Rhodococcus fascians. J. Bacteriol. 184:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 48.Wall, D. M., P. S. Duffy, C. Dupont, J. F. Prescott, and W. G. Meijer. 2005. Isocitrate lyase activity is required for virulence of the intracellular pathogen Rhodococcus equi. Infect. Immun. 73:6736-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Z. Y., C. R. Thornton, M. J. Kershaw, L. Debao, and N. J. Talbot. 2003. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 47:1601-1612. [DOI] [PubMed] [Google Scholar]
- 50.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]