Abstract
We used a nonredundant microarray of the Salmonella enterica serovar Typhimurium LT2 and Typhi CT18 genomes to assess the genomic content of a diverse set of isolates of serovar Typhi. Comparative genomic hybridization revealed 13 regions of absent or divergent gene content in the eight Typhi strains examined compared to Typhi CT18. In particular, two Typhi CT18 prophage regions, STY1048 to STY1077 and STY2038 to STY2077, as well as a five-gene islet (STY3188 to STY3193) were absent or divergent in all other Typhi strains examined. Seven Typhi strains lacked most or all of the IS1 elements present in strain CT18, and three Typhi strains lacked a P4-like phage (STY4821 to STY4834). One strain was devoid of a 149-gene region (STY4521 to STY4680), which encodes numerous phage genes and the Vi antigen biosynthesis and export gene cluster, a type IV pilus, and numerous phage genes. In Typhi strain 26T25, an amplification of an entire inter-ribosomal region encompassing 31 genes has occurred. Furthermore, a 257-gene region (STY1360 to STY1639) showed an aberrant replication pattern in three Typhi isolates. Overall, these differences in gene content indicate that even within a highly clonal bacterial population the genomic reservoir is unstable.
Salmonella enterica is a facultative intracellular pathogen that is implicated in a wide variety of life-threatening infections ranging from typhoid to gastroenteritis and bacteremia. Salmonella-induced enterocolitis is the leading food-borne illness with a lethal outcome in the United States and causes millions of cases of gastroenteritis each year. Of the seven subspecies (I, II, IIIa, IIIb, IV, VI, and VII) of S. enterica, only subspecies I isolates are routinely associated with humans and warm-blooded animal infections (26). In addition, some serovars of S. enterica subspecies I show remarkable host specificity. For example, serovars Typhi, Dublin, and Gallinarum exclusively infect humans, cattle, and birds, respectively.
S. enterica serovar Typhi is the etiological agent of typhoid fever. Studies have shown that isolates of serovar Typhi are extremely homogeneous (22, 25). Multilocus enzyme electrophoresis (MLEE) analysis measuring chromosomal genotypic diversity and evolutionary relationships among several hundred serovar Typhi isolates of worldwide distribution and collected over several years distinguished only two clones: a predominant clone of global distribution and a second, very similar clone initially confined to West Africa (25).
The sequences of two S. enterica subspecies I genomes, serovar Typhimurium strain LT2 (STM) and serovar Typhi strain CT18 (STY), were completed in 2001 (16, 18). STY CT18 is a multiple-drug-resistant isolate recovered from a typhoid fever patient from Vietnam in 1993 (18). During completion of this manuscript, the genome sequence of a second S. enterica serovar Typhi strain (Ty2) was decoded (8), facilitating the first genome-wide comparison within the highly clonal S. enterica serovar Typhi. The sequence revealed that 84 S. enterica serovar Typhi strain CT18 genes were missing, or altered, in the Ty2 genome, whereas 29 genes were Ty2 specific (8).
A microarray was constructed with sequences from the annotated open reading frames (ORFs) in STM supplemented with annotated chromosomal ORFs from STY that are divergent from Typhimurium (>10% DNA sequence divergence) (19). This array consisted of genes or gene fragments representing 471 STY-specific genes cleared of paralogous sequences, which were added to an STM array that represents 4,442 STM genes and gene fragments (including 104 pSLT plasmid genes). Overall, STM genome coverage is 96.6%; coverage of the STY genome is 94.5% (4,348 genes), excluding plasmids (19).
Comparative genomic analysis utilizing microarray technology has been used to provide insights into the evolution and virulence of a number of pathogenic bacteria including Campylobacter jejuni, Helicobacter pylori, Escherichia coli, Mycobacterium tuberculosis, S. enterica, Vibrio cholerae, and Streptococcus pyogenes (2, 7, 9-11, 23, 27). In the present study we utilize the S. enterica serovar Typhimurium and Typhi microarray to assay gene content differences among nine different Typhi isolates. We identify additional CT18 genomic regions of divergence within the Typhi, and characterize regions whose absence in every Typhi strain except CT18 indicates true CT18 specificity.
MATERIALS AND METHODS
The nonredundant microarray was probed with labeled genomic DNA from nine serovar Typhi strains of interest (Table 1). Briefly, genomic DNAs of S. enterica serovar Typhi strains listed in Table 1 were prepared from fresh overnight cultures by using the GenElute Bacterial Genomic DNA kit (Sigma, St. Louis, Mo.). Cells were grown in Luria broth at 37°C. The harvested nucleic acid was labeled by using Cy3- and Cy5-dCTP (Amersham, Piscataway, N.J.) and Klenow enzyme as previously described (21). Probes were purified by using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) as suggested by the manufacturer, eluted in 1 mM Tris-HCl (pH 8.0), dried, and resuspended in 10 μl of sterile water. Immediately before use, the labeled probes of Typhi CT18 (control sample) and one of the eight query Typhi strains (experimental sample) were mixed with equal volumes of 2× hybridization buffer containing 50% formamide, 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.2% sodium dodecyl sulfate and then boiled for 5 min. Probes were hybridized to the nonredundant Salmonella microarray overnight at 42°C by using a hybridization chamber (Corning, Inc., Corning, N.Y.) submerged in water. Protocols suggested by the manufacturer for hybridizations in formamide buffer (http://www.corning.com/Lifesciences/technical_information/techDocs/gaps_ii_manual_protocol_5_02_cls_gaps_005.pdf) were applied for prehybridization, hybridization, and posthybridization wash processes. Scans were performed on a ScanArray 5000 laser scanner (Packard BioChip Technologies, Billerica, Mass.) by using ScanArray 2.1 software. Signal intensities were quantified by using the QuantArray 3.0 software package (Packard BioChip Technologies, Billerica, Mass.). Spots were analyzed by adaptive quantitation, and the local background was subsequently subtracted from the recorded spot intensities. Ratios of the contribution of each spot to total signal in each channel were calculated (data normalization). Negative values (i.e., local background intensities higher than spot signal) were considered no data. Since the array was spotted in triplicate, one hybridization resulted in three datum points per gene. The median of the three ratios per gene was recorded. The lowest 5% of gene specific signals were determined for the control sample (Typhi CT18) and excluded from the graphical representation in Fig. 1A.
TABLE 1.
S. enterica serovar Typhi strains used in this study
| Strain | SGSC no.a | Date (yr) | Origin | Source or reference |
|---|---|---|---|---|
| CT18 | 4072 | 1993 | Vietnam | 18 |
| 26T25 | 3148 | 1994 | Canada | R. Khakria |
| ST24A | 2771 | 1986 | Malaysia | T. Pang |
| Ty2 | 2408 | NAb | NA | 13 |
| 3125 | 3185 | 1983 | Chile | T. Pang |
| In15 | 2783 | 1994 | Indonesia | T. Pang |
| CDC1707 | 2661 | NA | Liberia | 22 |
| SARB63 | 2520 | 1988 | Senegal | 5 |
| SARB64 | 2521 | 1988 | Senegal | 5 |
SGSC, Salmonella Genetic Stock Centre (http://www.ucalgary.ca/∼kesander/index.html).
NA, not available.
FIG.1.
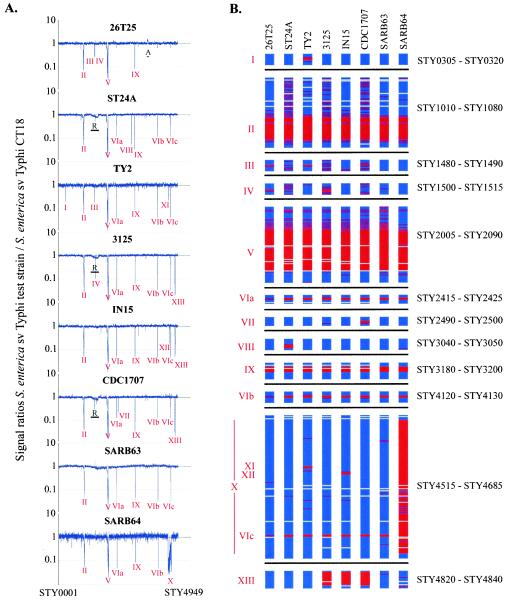
Detection of deleted regions on S. enterica serovar Typhi chromosomes. (A) Comparative genomic hybridization of eight S. enterica serovar Typhi strains versus serovar Typhi CT18. The eight plots display medians of signal ratios of the query S. enterica serovar Typhi strain over Typhi CT18. Genes are plotted in order of their position on the Typhi CT18 genome. The 13 regions containing genes deleted from other Typhi strains are indicated. The region with abberant replicative behavior in strains ST24A, 3125, and CDC1707 is marked with an “R.” The amplified segment in strain 26T25 is marked with an “A.” (B) Presence or absence of S. enterica serovar Typhi CT18 protein coding sequence homologues in other Typhi strains. Gene status is color coded: blue, present; purple, uncertain; red, absent; and gray, low signal. Only the 13 regions that harbor genes that are missing in other Typhi strains, along with some adjacent genes, are shown.
The presence and absence of genes were evaluated based on comparison of normalized hybridization signal ratios of the query Typhi strain versus Typhi CT18 for the respective gene spot on the array. The median of the ratios of the genes neighbored on the STY genome by elements with ratios higher than 0.67 on both sides (presence baseline P) and the standard deviation (SDP) of these ratios were calculated for each query strain. Similarly, the medians and standard deviations of genes neighbored on STY by elements with ratios lower than 0.5 were determined (absence baseline A and SDA). Ratios which were higher than the “presence” threshold set at 2 SDP below the baseline P were scored as “present,” whereas genes with ratios lower than the “absence” threshold set at 2 SDA above the baseline A were scored as “absent.” Genes that were outside these thresholds were scored as uncertain.
RESULTS AND DISCUSSION
A microarray containing 94.5% of the ORFs from the Typhi CT18 genome (4,348 genes) was used to determine the shared genome content of eight other Typhi strains. The strains were picked for their temporal and geographic diversity and included the two known MLEE types, as represented by the two Typhi strains in the SARB collection (5). More than 4,000 of the CT18 genes were shared by all of the Typhi strains examined. However, 13 STY regions had variable present or absent patterns in the Typhi strains examined (Table 2 and Fig. 1). Nine of these regions were Typhi-specific regions, i.e., found in the Typhi CT18 sequence but not in the Typhimurium LT2 genome. Six regions were absent from a single Typhi strain (Table 2).
TABLE 2.
S. enterica serovar Typhi CT18 genomic regions absent from various other Typhi isolates
| Region | Corresponding designation in Deng et al. (8) | No. of genes | STY gene no. (specificity) | Protein function(s) | No. of strains |
|---|---|---|---|---|---|
| I | g | 2 | 0311-0312 (STY specific) | Hypothetical proteins | 1 |
| II | f | 23 | 1048-1071 (STY specific) | Prophage | 8 |
| III | e | 2 | 1484-1485 | NarV | 3 |
| IV | 2 | 1508-1509 | Transporter | 2 | |
| V | 42 | 2038-2077 (STY specific) | Prophage | 8 | |
| VIa | a | 2 | 2419-2420 (STY specific) | IS1 InsAB | 7 |
| VIb | m | 2 | 4124-4125 (STY specific) | IS1 InsAB | 7 |
| VIc | o | 2 | 4657-4658 (STY specific) | IS1 InsAB | 7 |
| VII | 2 | 2494-2495 | Sensor kinase, regulator of capsule synthesis | 1 | |
| VIII | 3 | 3046-3048 | Hypothetical proteins | 1 | |
| IX | i | 5 | 3188-3193 (STY specific) | Hypothetical proteins, Int | 8 |
| X | 149 | 4521-4680 (STY specific) | Vi antigen, type IV pilus, SopE, phage proteins | 1 | |
| XI | n | 2 | 4580-4582 (STY specific) | Hypothetical proteins | 1 |
| XII | 2 | 4587-4588 (STY specific) | Hypothetical proteins | 1 | |
| XIII | 14 | 4821-4834 (STY specific) | P4-like phage | 3 |
The microarray experiment suggested that region I, which is Typhi specific and comprises two genes (STY0311 to STY0312), is absent from Typhi strain Ty2 (Fig. 1). This result was subsequently confirmed by the decoded Ty2 genome sequence (8). Region I encodes a hypothetical protein and a secreted protein. Since it is known that bacterial secreted proteins tend to be hypervariable, the pattern of hybridization noted may indicate divergence of this region from strain CT18 (4, 14).
Region II, comprising 24 genes (STY1048 to STY1071), encodes Typhi CT18-specific bacteriophage gene homologues and is absent or divergent from all other Typhi strains examined. The decoded Ty2 sequence confirmed this finding and showed that STY1072 and STY1073, which had intermediate scores in the microarray experiment, were also missing (8). The region from STY1071 to STY1073 is present in two copies on the CT18 genome (the respective duplicate genes are STY2015 to STY2013), and the C-terminal 588 nucleotides of STY1070 are duplicated in STY2016. The copy number difference for these genes in Ty2 compared to CT18 was correctly detected by the microarray, although the distinction between true absence and altered copy number is not always possible for multicopy genes. Miscalls due to cross-hybridization are a general limitation for phage regions since these genes can be very similar to elements present on other phages located elsewhere on the genome. Caution must always be applied in the interpretation of microarray hybridization results for genes that are present in more than one copy on the reference genome. Upstream of region II, ORFs STY1011 to STY1044 show significant homology to the lambdoid phage Gifsy-2 from S. enterica serovar Typhimurium LT2; therefore, it is possible that, in the Typhi strains examined, region II has been replaced by divergent phage sequence. Recombination between lambdoid-like phage and the generation of unique phage genomes has been well documented (3, 6).
Region III consists of STY1484, which was found to be divergent or absent in CDC1707, and gene narV (STY1485), which was detected as absent from strains Ty2 and 26T25 (Fig. 1). The Ty2 genome sequence revealed that, in fact, narV and narW (STY1486) are fused by a deletion in this strain, retaining only 98 nucleotides of narV, but 531 nucleotides of narW (8). Comparative hybridization of genomic DNA to the microarray did not detect the deletion in narW, but the remaining sequence of narV was small enough to observe the deletion event.
STY1508 (region IV) is a pseudogene and is absent from strains 26T25 and 3125. In strain 3125, the adjacent gene STY1509 was also predicted to be absent or else diverged. Another CT18 pseudogene, STY2012, is absent from the Ty2 genome sequence. However, our current microarray version does not contain a representative spot for this pseudogene, and we were therefore unable to report on the status of STY2012 in the other Typhi strains.
Region V (STY2038 to STY2077) is another Typhi CT18-specific prophage region that is absent or divergent in all Typhi strains examined. All genes of this region except STY2038 and STY2039 were absent from the Ty2 genome sequence (8). Similarities between CT18 and Ty2 for these two genes were 96 and 95% in any given 100-bp sequence window, respectively, a value that is just below the 97% sequence identity required to ascertain identical hybridization behavior (19). In addition, the lower hybridization ratios obtained in the microarray experiment for STY2038 and STY2039 in Ty2 were also generated by their sequence identity to STY1048 to STY1049, genes which belong to a phage region (region II) that is missing in Ty2. The same scenario resulted in the “undercall” of STY2035 in Ty2, i.e., the gene was reported as missing or divergent, when in fact only its cross-hybridizing counterpart (STY1052) was missing. Overall, Parkhill et al. (18) identified seven prophage elements within Typhi strain CT18. The absence, or divergence, of two of these prophage elements (regions II and V) in all other Typhi strains examined suggests possible continuing phage mobility.
Region VI is present on the Typhi CT18 genome in three copies and encodes the two genes (insAB) of an IS1 element. IS1 is also present in five copies on the Typhi CT18 plasmid pHCM1. This element is absent, or present in substantially lower copy numbers, in all isolates examined except for Typhi strain 26T25, which apparently retains several copies of insAB. The microarray has only one representative spot for each of the two IS1 genes; therefore, the ratios reported for the three copies (regions VIa [STY2419 to STY2420], VIb [STY4124 to STY4125], and VIc [STY4657 to STY4658], respectively) are identical. The Ty2 genome sequence confirmed the absence of IS1 insAB in this Typhi strain (8).
Region VII (STY2494 and STY2495) encodes a kinase and a regulator of the capsule synthesis B component (rcsB) and is absent from Typhi strain CDC1707.
STY3046 to STY3048, which comprises region VIII, encodes a putative decarboxylase, as well as two conserved hypothetical proteins, and is absent from strain ST24A.
Region IX (STY3188 to STY3193), a Typhi CT18-specific region, encodes several hypothetical proteins, as well as an integrase, and is absent from all Typhi isolates examined, suggesting this region to be a recent addition to the CT18 genome. Its absence in Ty2 was confirmed by the genome sequence (8).
Region X is a remarkable deletion in strain SARB64. It comprises more than 133 kb and encompasses 149 genes, from STY4521 to STY4680, almost none of which have homologs in Typhimurium LT2. Among the genes of this region are the tvi and vex operons, which are responsible for the synthesis and export, respectively, of the Typhi-specific Vi capsular antigen, and the pil locus involved in type IV pilus formation. Furthermore, the invasion-associated secreted protein SopE is encoded on this island, as are numerous hypothetical proteins and phage elements. Six of the 149 ORFs are pseudogenes: STY4537, STY4541, STY4541a, STY4598, STY4608, and STY4633. The microarray reported all but one of the genes of region X as absent or uncertain. The spot that represented the exception, STY4606, has high homology (89% over the entire sequence) to two other genes on the STY CT18 genome, STY2887 and STY3697. Therefore, cross-hybridization prevented a correct call for this gene. This region appears to be inserted at the pheU tRNA gene (12), and the presence of a homologous pheU tRNA pseudogene on the other end of the region could be the basis of a deletion event by homologous recombination (K. Sanderson, unpublished data). SARB64 is a relatively rare MLEE type, originally from West Africa (5). It is possible that the large deletion is correlated with this MLEE type.
Regions XI (STY4580 to STY4582) and XII (STY4587 and STY4588) are Typhi-specific regions that are absent in Typhimurium LT2. They both encode hypothetical proteins and are absent from strain Ty2 and In15, respectively. Absence of region XI in Ty2 was also noted by Deng et al. (8).
Finally, the Typhi-specific region XIII (STY4821 to STY4834), which encodes P4-like genes, is absent, or divergent, in three strains (3125, CDC1707, and In15), suggesting a P4-like phage to have entered, and possibly left, Typhi genomes after their evolutionary separation from serovar Typhimurium.
In strain 26T25 there is an amplification of 31 genes (STY3709 to STY3742) (region A in Fig. 1A). The amplified region encodes gene clusters required for thiamine biosynthesis and 50S ribosomal subunit synthesis and is located between ribosomal gene clusters. rRNA clusters are known to recombine with one another, particularly in host-specific S. enterica, as in serovar Typhi (15), and duplications of inter-rrn fragments have also been observed in several S. enterica serovar Typhimurium strains (S. Porwollik, unpublished data).
STY1360 to STY1639, encompassing a total of 257 genes shows an aberrant hybridization pattern in three strains—3125, CDC1707, and ST24A—compared to the rest of the Typhi genome (Fig. 1A, region R). This region shows a progressive reduction in apparent DNA content from STY1360 to STY1639 in all three strains; however, the downward trend is not significant enough to be considered a deletion of this region but may instead reflect aberrant replication patterns in some cells of these isolates. Interestingly, the region is adjacent to the Typhi CT18 terminus of the replication site (ter), STY1360, and the locus corresponding to the Typhimurium LT2 ter site (STY1655). The ter site was previously shown to undergo extensive inversions involving IS200 elements in serovar Typhi strains (1, 17). The abberant region encodes several bacteriophage structural and assembly proteins, as well as putative virulence genes and transporter systems.
Two genes, lppA and lppB (STY1745 and STY1746), which encode outer membrane lipoprotein, show an intermediate hybridization pattern in Typhi strain In15. Since protein exposed to the external environment tend to be more variable, the intermediate hybridization pattern may reflect divergence of these proteins in response to selective pressure.
In addition to these differences, there were three deviations in gene content of the eight Typhi strains compared to Typhi CT18 that were detected among genes present in Typhimurium LT2 but were absent in Typhi CT18. The deletion of yneH (STM1525) in Typhi CT18 is strain specific, since all other Typhi strains examined still retained this gene in their genome. Second, one of the Typhi strains, strain CDC1707, apparently contained homologs of STM4491/STM4492 and STM4495/STM4496. These genes are part of an 11-gene Typhimurium LT2 region that is frequently deleted in other Salmonella strains (20). This area is inserted into the Typhimurium LT2 genome at a tRNA site (leuX) and starts with a gene (STM4489) that encodes an integrase, suggesting foreign origin. Third, the gene cluster of STM1568 to STM1572 encoding nitrate inducible formate dehydrogenase subunits (fdnIH), a molybdopterin-dependent oxidoreductase (fdnG), a permease (yddG), and a predicted porin (nmpC or ompD) have homologues in both SARB63 and SARB64 but in none of the other Typhi isolates. Previous observations had suggested the absence of ompD from all clinical Typhi isolates (24).
Finally, it is important to point out that only genes that are present on the array can be assayed. For example, the 29 genes present in Ty2 that are not present in CT18 (8) have not been assayed, and their status in the other strains remains unknown. Another general caveat for the assessment of the genetic repertoire of related bacteria by using microarrays is the fact that the functionality of the detected genes cannot be characterized. However, the enormous potential of microarrays to quickly and cost-effectively survey genome contents makes this technique a valuable tool for comparative genomics studies.
In conclusion, by utilizing a nonredundant microarray for the S. enterica serovars Typhimurium and Typhi, we examined several recent isolates of the highly clonal serovar Typhi, whose only known host is humans. We uncovered significant differences in gene content among the isolates examined. Prophage genes make up the major differences in gene content among Typhi strains. Contrary to previous analysis, the present study demonstrated the underappreciated diversity among serovar Typhi isolates. The polymorphic regions could be useful targets for epidemiological classification of Typhi strains by microarrays, PCR, or immunological methods.
Acknowledgments
We thank Ken Sanderson for critical reading of the manuscript and helpful discussions.
This work was supported, in part, by NIH grant AI34829 (M.M.) and the generosity of Sidney Kimmel and by an Operation for Economic Cooperation and Development fellowship (E.F.B.).
REFERENCES
- 1.Alokam, S., S. L. Liu, K. Said, and K. E. Sanderson. 2002. Inversions over the terminus region in Salmonella and Escherichia coli: IS200s as the sites of homologous recombination inverting the chromosome of Salmonella enterica serovar Typhi. J. Bacteriol. 184:6190-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139(Pt. 6):1125-1132. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 7.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed]
- 12.Hansen-Wester, I., and M. Hensel. 2002. Genome-based identification of chromosomal regions specific for Salmonella spp. Infect. Immun. 70:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hone, D. M., A. M. Harris, S. Chatfield, G. Dougan, and M. M. Levine. 1991. Construction of genetically defined double aro mutants of Salmonella Typhi. Vaccine 9:810-816. [DOI] [PubMed] [Google Scholar]
- 14.Li, J., H. Ochman, E. A. Groisman, E. F. Boyd, F. Solomon, K. Nelson, and R. K. Selander. 1995. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc. Natl. Acad. Sci. USA 92:7252-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, S. L., and K. E. Sanderson. 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS. Microbiol. Lett. 164:275-281. [DOI] [PubMed] [Google Scholar]
- 16.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 17.Ng, I., S. L. Liu, and K. E. Sanderson. 1999. Role of genomic rearrangements in producing new ribotypes of Salmonella typhi. J. Bacteriol. 181:3536-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 19.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 31:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porwollik, S., R. M. Wong, S. H. Sims, R. M. Schaaper, D. M. DeMarini, and M. McClelland. 2001. The ΔuvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat. Res. 483:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. Farmer III. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santiviago, C. A., C. S. Toro, S. A. Bucarey, and G. C. Mora. 2001. A chromosomal region surrounding the ompD porin gene marks a genetic difference between Salmonella typhi and the majority of Salmonella serovars. Microbiology 147:1897-1907. [DOI] [PubMed]
- 25.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. D. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selander, R. K., J. Li, and K. Nelson. 1996. Evolutionary Genetics of Salmonella enterica, p. 2691-2707. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 27.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]