Abstract
The pyridine nucleotides NAD and NADP play vital roles in metabolic conversions as signal transducers and in cellular defence systems. Both coenzymes participate as electron carriers in energy transduction and biosynthetic processes. Their oxidized forms, NAD+ and NADP+, have been identified as important elements of regulatory pathways. In particular, NAD+ serves as a substrate for ADP-ribosylation reactions and for the Sir2 family of NAD+-dependent protein deacetylases as well as a precursor of the calcium mobilizing molecule cADPr (cyclic ADP-ribose). The conversions of NADP+ into the 2′-phosphorylated form of cADPr or to its nicotinic acid derivative, NAADP, also result in the formation of potent intracellular calcium-signalling agents. Perhaps, the most critical function of NADP is in the maintenance of a pool of reducing equivalents which is essential to counteract oxidative damage and for other detoxifying reactions. It is well known that the NADPH/NADP+ ratio is usually kept high, in favour of the reduced form. Research within the past few years has revealed important insights into how the NADPH pool is generated and maintained in different subcellular compartments. Moreover, tremendous progress in the molecular characterization of NAD kinases has established these enzymes as vital factors for cell survival. In the present review, we summarize recent advances in the understanding of the biosynthesis and signalling functions of NAD(P) and highlight the new insights into the molecular mechanisms of NADPH generation and their roles in cell physiology.
Keywords: ADP-ribosylation, calcium signalling, NAD(P)(H), NAD kinase (NADK), oxidative stress
Abbreviations: cADPr, cyclic ADP-ribose; ALDH, aldehyde dehydrogenase; ES, embryonic stem; G6PD, glucose-6-phosphate dehydrogenase; IDP, NADP-specific isocitrate dehydrogenase; IDPc, cytosolic IDP; IDPm, mitochondrial IDP; ME, malic enzyme; NA, nicotinic acid; NAAD, NA–adenine dinucleotide; NAADP, NA–adenine dinucleotide phosphate; NADK, NAD kinase; NADPase, NADP phosphatase; Nam, nicotinamide; NAMN, NA mononucleotide; NRK, Nam riboside kinase; N(A)MNAT, Nam/NA mononucleotide adenylyltransferase; OAADPr, O-acetyl ADP-ribose; PARP, poly(ADP-ribose) polymerase; PARsylation, poly(ADP-ribosyl)ation; poly(P), polyphosphate; QA, quinolinic acid; QAPRT, QA phosphoribosyltransferase; RNI, reactive nitrogen intermediate; ROS, reactive oxygen species; Sir2, silent information regulator-2; TRPM2, transient receptor potential cation channel melastatin 2
INTRODUCTION
Cells throughout all organisms have established a universal set of small molecules that carry out versatile functions both in metabolic pathways and in regulatory processes. Among the best characterized of these molecules is ATP. It has been termed the universal energy currency of the cell, but, in addition, serves in signalling pathways as a substrate for protein phosphorylation as well as precursor of cAMP. It appears, however, that another molecule might have even more facets in its repertoire and has become subject of intense investigations, NAD. NAD and its phosphorylated form NADP were well known cofactors of cellular metabolism and were characterized as electron carriers in oxidoreductase reactions before other properties were unravelled and the involvement of the dinucleotides in signalling reactions moved into focus [1].
Although it is recognized to have important regulatory roles, NADP has so far received less attention. It is generally accepted that its reduced form, NADPH, is vital to the cellular oxidative defence systems and reductive syntheses. How delicately these processes are regulated by the availability of NADPH has become of increasing interest over the past few years. In particular, the mechanisms and subcellular compartmentation of NADP+ and NADPH generation appear to critically influence the physiological state of the cell and thereby cellular survival.
In the present review, we will briefly summarize the biosynthetic pathways and signalling mechanisms of NAD(P) and then elaborate on the importance and pathways of NADPH generation. Finally, we will focus on the key enzyme of NADP synthesis, NADK (NAD kinase), whose vital role has been highlighted in a number of recent studies.
BIOSYNTHESIS OF NAD(P)
The participation of NAD(P) in electron transfer reactions, that is, the reversible conversion between oxidized (NAD+, NADP+) and reduced (NADH, NADPH) forms does not result in a net consumption of the nucleotides:
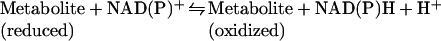 |
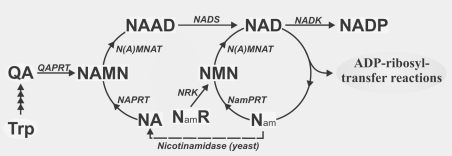
Consequently, except for cell divisions, the constant requirement for pyridine nucleotide re-synthesis does not arise from their function as redox carriers, but rather from their involvement in signalling reactions. The latter are accompanied by the cleavage of the glycosidic bond between the nicotinamide and ADP-ribose moieties. The liberated nicotinamide is presumed to be recycled by a salvage pathway (Figure 1). Two principal routes of NAD biosynthesis are known (Figure 1; reviewed in [2,3]). The de novo biosynthesis includes several steps to generate QA (quinolinic acid) from either L-tryptophan in animals and some bacteria, or L-aspartate in some bacteria and plants [4]. The subsequent enzymatic step catalysed by QAPRT (QA phosphoribosyltransferase) appears to be common, at least in eukaryotes. The salvage pathways utilize degradation products, namely NA (nicotinic acid) and Nam (nicotinamide), to regenerate NAD (Figure 1). Both pathways converge at the transfer of NMN or its acid form NAMN (NA mononucleotide) on to the adenylyl group of ATP under pyrophosphate release. The reaction is catalysed by N(A)MNAT (Nam/NA mononucleotide adenylyltransferase). In humans, three isoforms of N(A)MNAT were identified and recently shown to be expressed in the nucleus, in the Golgi apparatus and the mitochondria [5]. When NAMN is used in the transfer reaction, the product is NAAD (NA–adenine dinucleotide), which is subsequently amidated to NAD by NAD synthetase. Despite the presence of the same enzyme activities, mammalian cells exhibit a clear preference towards Nam compared with yeast, for which NA appears to be the preferred precursor. In fact, yeast express an enzyme, nicotinamidase, which deamidates Nam to NA before it can be recycled into NAD (Figure 1). With regard to the overall activity of NAD synthesis, the de novo synthesis from tryptophan (via QA) is of minor importance.
Figure 1. Schematic overview of NAD(P) biosynthetic pathways.
The major known pathways of NAD(P) synthesis are presented. Main endogenous precursors of NAD(P) synthesis are Nam, NA and L-tryptophan. Nicotinamidase is only found in yeast. NamR, Nam riboside; NA/NamPRT, NA/Nam phosphoribosyltransferase; NADS, NAD synthetase.
Niacin or vitamin B3, which is synonymous with both Nam and NA, was long thought to be the only external precursor for de novo synthesis of NAD. Recently, an alternative pathway has been established by the molecular identification of NRK (Nam riboside kinase) [6]. Although such an activity had been detected in mammalian tissues before [7], the recent study [6] demonstrated the importance of this enzyme for NAD biosynthesis in yeast and potentially in humans. Moreover, the presence of nicotinamide riboside in food sources was demonstrated, further supporting the importance of this novel pathway.
Finally, some of the cellular NAD is converted into NADP by NADK. As for all other enzymes of NAD(P) biosynthesis, this enzymatic activity has long been known, but the molecular identification and characterization has only been achieved within the past few years. It turns out that, at least by exerting a direct influence on NAD(P) turnover, many of these enzymes have a critical influence on major cellular events. Specifically, as will be highlighted in a subsequent section, the activities of NADKs determine the availability of NADPH and are therefore vital.
REGULATORY REACTIONS INVOLVING NAD+ AND NADP+: ADP-RIBOSE – A MODULE OF VARIOUS SIGNALLING REACTIONS
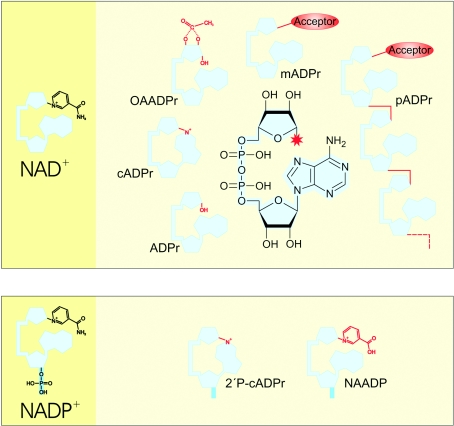
Both NAD+ and NADP+ are involved in signal transduction as precursors of messenger molecules. In addition, NAD+ serves also as substrate for covalent modifications of target molecules. All known derivatives share structural similarity owing to their ADP-ribose backbone (Figure 2). Thus, NAD+-mediated signalling reactions can be referred to as ADP-ribosyl transfers, because the initial cleavage of Nam (leaving the ADP-ribosyl moiety) is common to all of them. The eventual acceptors of the transfer reaction are rather diverse, including macromolecules such as proteins, or small molecules such as water (yielding ADP-ribose), acetate [yielding OAADPr (O-acetyl ADP-ribose)] or the adenine ring leading to intramolecular cyclization into cADPr (cyclic ADP-ribose; Figure 2).
Figure 2. Signalling derivatives of NAD(P).
The ADP-ribosyl moiety commonly shared by all derivatives is illustrated in structural detail and adumbrated in light blue, the red asterisk indicates the site of ADP-ribosyl attachment to the acceptor. The individual portion of each derivative is presented in red and the 2′-phosphate group of NADP+ in its derivatives is shown in dark blue. ADPr, ADP-ribose; mAPDr, mono-ADPr; pADPr, poly-ADPr; 2′P-cADPr, 2′-phosphate cADPr.
The first observations of NAD+-mediated protein modification by poly-ADP-ribose date back to the 1960s [8,9]. Mono-ADP-ribosyltransferase activities were identified originally in a group of bacterial toxins [10] and were later detected in eukaryotic cells [11]. The target molecules are usually proteins, but modification of DNA has also been described [12]. The physiological consequence of mono-ADP-ribosylation appears to be inhibition of the target proteins [13]. For example, modification by the mitochondrial transferase SIRT4 [14] inhibits glutamate dehydrogenase [15].
PARsylation [poly(ADPribosyl)ation] constitutes the attachment of ADP-ribose polymers to target proteins and therefore represents a rather elaborate protein modification. It is catalysed by PARPs [poly(ADP-ribose) polymerases], a protein family comprising 17 members in humans [16,17]. The founding and most catalytically active member, PARP-1, is a nuclear enzyme involved, among others, in DNA repair and apoptotic pathways [1,17,18]. PARsylation has also important functions in telomere dynamics [19], transcriptional regulation [20], cell division [21] and trafficking of endosomal vesicles [22].
ADP-ribose transfer has also an important function in tRNA splicing, which is mediated by NAD+. The 2′-phosphate of the splice intermediate is transferred to NAD+ thereby forming ADP-ribose 1″-2″ cyclic phosphate and Nam [23]. Whether this product has a physiological function remains unknown.
Several NAD(P)+ derivatives act as second messenger molecules. Following cleavage of the Nam, the ADP-ribose moiety undergoes a reaction with a small modifying group (e.g. water, NA or an intramolecular group, cf. Figure 2) resulting in a biologically active messenger. Remarkably, so far all known NAD(P)+ derivatives have been associated with calcium signalling. Even ADP-ribose itself triggers Ca2+ influx into the cytosol by activating TRPM2 (transient receptor potential cation channel melastatin 2), a Ca2+ channel in the plasma membrane [24].
ADP-ribosyl cyclases generate cADPr and 2′-phospho-cADPr from NAD+ and NADP+ respectively. Both molecules trigger cytosolic [Ca2+] elevation, presumably by activating the ryanodine receptor in the endoplasmic/sarcoplasmic reticulum [25]. In the presence of adenine, ADP-ribosyl cyclases can form other adducts of cADPr, which might also serve as Ca2+-mobilizing molecules [26]. The mammalian ADP-ribosyl cyclases (CD38 and CD157), homologues of the enzyme originally identified in Aplysia californica [27], are ecto-enzymes which exhibit their catalytic activity at the cell surface [28]. Extracellular generated cADPr can possibly be taken up by certain cell types [29], but there is also evidence for the existence of alternative intracellular enzymatic activities [30].
Today, the most potent intracellular Ca2+-mobilizing messenger known is NAADP (NA–adenine dinucleotide phosphate), a derivative of NADP+. It was shown to trigger calcium release from lysosomal Ca2+ stores, which are independent of the stores activated by cADPr or inositol 1,4,5-trisphosphate [31]. Astonishingly, the only enzymatic activity shown to be capable of generating NAADP is associated with ADP-ribosyl cyclases, which, in this case, exchange the Nam moiety of NADP+ with NA. Whether alternative pathways of NAADP generation exist, for example by phosphorylation of NAAD, have yet to be solved [32]. On the other hand, a 2′-specific Ca2+-dependent phosphatase has been characterized that degrades NAADP and thereby inactivates this messenger [33].
It is interesting to note that none of the conversions of NAD(P)+ and their consequences described above has been found in yeast, although even some prokaryotes express ADP-ribosylating or ADP-ribosyl cyclase activities. Nevertheless, yeast was the first organism in which yet another NAD+-mediated regulatory mechanism was detected that includes protein deacetylation and the generation of a potential messenger molecule, that is, the formation of OAADPr [34]. The corresponding enzyme activity was originally discovered for the Sir2 (silent information regulator-2) protein of Saccharomyces cerevisiae, which was identified as an NAD+-dependent histone deacetylase (reviewed in [35]). In the meantime, mammalian Sir2 homologues, sirtuins, have become of major interest owing, in part, to the remarkable effect of Sir2 on lifespan regulation by gene silencing [35]. The closest human relative to yeast Sir2, hSIRT1, exhibits the same enzymatic activity [35]. Besides histones, it also deacetylates other target proteins, including p53. Although closely related, not all mammalian sirtuins are in fact protein deacetylases, but some represent mono-ADP-ribosyltransferases [14,36].
The reaction mechanism of Sir2 involves the cleavage of the Nam moiety and subsequent transfer of the originally protein-bound acetyl group on to ADP-ribose. Owing to intramolecular isomerization, either the 2′- or the 3′-C atom of the terminal ribose is acetylated (Figure 2) [37]. Not only the deacetylation of the target protein, but also the generation of OAADPr appears to have signalling characteristics. OAADPr has Ca2+-mobilizing properties by activating the TRPM2 channel [38] and affects the structure of the Sir2–Sir3–Sir4 silencing complex [39].
Taken together, NAD(P)+ is extensively used in signal transfer reactions, both by modification of target molecules or generation of small signalling molecules. Thereby, the pyridine nucleotides influence virtually all vital cellular functions. It is important to note that all of the conversions described above require the oxidized form of NAD(P)+. The reduced form is not a substrate for these signalling reactions. However, as discussed below, the redox state of the dinucleotides is of great importance in the cellular environment. In fact, depending on the redox state of NAD, transcriptional activity can be reciprocally regulated by direct binding of either NADH or NAD+ to transcriptional co-repressor C-terminal binding protein [40].
REDOX STATE AND CELLULAR NAD(P) CONCENTRATIONS
The term redox state is commonly used to describe the balance of NAD+/NADH and NADP+/NADPH in the cell. It is reflected by several sets of metabolites (lactate and pyruvate, β-hydroxybutyrate and acetoacetate), that is, by the interconvertible oxidized and reduced forms of specific redox couples [41,42]. The cellular redox state influences many metabolic, signalling and transcriptional processes in the cell. It always has to be maintained such that sufficient reducing equivalents are available, for example, to counteract the potential damage by free radical intermediates, namely ROS (reactive oxygen species) and RNIs (reactive nitrogen intermediates). The acute shift of the intracellular redox balance towards oxidation, known as oxidative stress, is due to increased production or insufficient degradation of ROS and RNI and represents a major damaging factor in inflammatory processes, ischaemia/reperfusion injury, other pathological states and ageing [43–48].
Although the critical role of NAD(P) and its redox state have long been appreciated, there are still considerable uncertainties regarding their cellular dynamics, that is, overall concentration, free and protein-bound state, subcellular compartmentation and the redox state.
The assessment of these parameters is hampered by technical difficulties relating to ‘visualization’ in living cells and even to the accurate determination of overall concentrations. For example, owing to the instability of reduced and oxidized pyridine nucleotides under acidic or alkaline conditions respectively, commonly used extraction procedures are accompanied by a loss or breakdown of the nucleotides. Moreover, redox shifts can be brought about rather quickly and changes in concentration of the oxidized form (which is most commonly measured in acidic extracts) might not be due to usage or breakdown, but simply due to alteration of the redox state just prior to extraction. Therefore, when examining changes of pyridine nucleotide concentrations, both the oxidized and reduced nucleotides are to be taken into consideration.
In order to circumvent some of the methodological problems, the lactate/pyruvate ratio and the pyruvate/malate ratio were measured assuming a direct proportionality to NAD+/NADH and NADP+/NADPH ratios respectively (e.g. [41,42,49]). Based on the fluorescence properties of reduced pyridine nucleotides, advanced technologies now enable more exact estimates of the intracellular NAD(P) distribution. Two-photon-excitation microscopy and fluorescence lifetime imaging have been used to measure intracellular NAD(P)H concentrations to distinguish between protein-bound and free nucleotides in living cells [41,50]. Comfortingly, the concentration values obtained with advanced methods are in good accordance with the values obtained by ‘classical methods’ [51].
Estimates of the total cellular NAD vary, but are commonly in the submillimolar range. The intracellular concentrations of NADP+ and NADPH are low in comparison with the non-phosphorylated forms, and under normal metabolic conditions the NADP pool is predominantly in the reduced state. Thereby, a reserve for the regeneration of anti-oxidative defence systems is normally available. In contrast, the redox ratio of NAD+/NADH is rather high (∼3–10; [40]). In addition, it is most likely that the majority of the cellular NAD and NADP pools are not free, but are protein-bound [40,52]. Therefore, estimates of actual substrate availability, for example, for NAD(P)+-mediated signalling processes are to be viewed with caution, at least, when based upon pyridine nucleotide determinations in cellular extracts.
It is interesting to note that the NAD concentration does not change when enzymes of its biosynthesis, e.g. NMNAT or NamPRT, are overexpressed [53,54], suggesting that the pyridine nucleotides can be turned over at a considerable rate. On the other hand, conditions such as the overactivation of PARP-1 by extensive DNA damage can lead to a rapid decline of the cellular NAD+ content by up to 80% [55] and, if at all, cells re-establish the original concentration only within hours or days. A major contributing factor to this severity is the ensuing oxidative stress imposed by most DNA-damaging agents, which aggravates ATP depletion and thereby limits the capacity of NAD+ re-synthesis. The loss of NAD+ by PARsylation further compromises the capacity to restore energy transduction by glycolysis [56]. In these scenarios, NADPH also has a critical role, as it provides the reducing power to oxidative defence systems. Although this function has long been appreciated, only over the past few years have the processes of its generation and maintenance been analysed in detail, in particular with regard to the molecular mechanisms, subcellular organization and regulation. The exploration of NADP synthesis and the mechanisms of maintaining its reduced state has provided important insights into the molecular details of its protective role; methods have been established to shift the activities of key enzymes regulating the concentration or redox state of NADP. Several new studies indicate the feasibility to increase the cellular content or reduced state of NADP thereby improving the cellular resistance towards oxidative stress. Below, we will summarize the major cellular systems that keep NADP in its reduced state. Then, we will discuss the roles and properties of NADKs, the key enzymes of NADP synthesis that have become the focus of intense investigation over the past few years.
THE MAJOR SYSTEMS OF NADPH GENERATION
Until recently, the major source of NADPH generation was thought to be the pentose phosphate pathway located in the cytosol, in particular the first and rate-limiting step catalysed by G6PD (glucose-6-phosphate dehydrogenase). Perhaps to the disappointment of some textbook editors, other, at least as important, enzyme activities have been described more recently (although the original notion might still hold true for erythrocytes). Among them, the NADP+-specific forms of IDP (isocitrate dehydrogenase), ME (malic enzyme), ALDH (aldehyde dehydrogenase) as well as NADK have received major attention. Furthermore, a direct trans-hydrogenation between NAD and NADP takes place in mitochondria, although evidence for such a system in S. cerevisiae is lacking [57].
Critical NADP-dependent dehydrogenases
G6PD is constitutively expressed in all organisms and cell types. It had been regarded as a housekeeping enzyme, but research in several laboratories demonstrated that this enzyme is highly regulated [58–60]. G6PD deficiency, a rather common genetic abnormality in humans, is the cause of haemolytic anaemia [61,62], while cancer cells may exhibit significantly increased activity [63].
Yeast cells disrupted for the G6PD gene, ZWF1, grow normally [64] and mouse ES (embryonic stem) cells are viable after knockout of the gene [65]. Although G6PD is obviously not essential for a single cell, mouse embryos lacking G6PD fail to develop normally [66]. Moreover, ΔG6PD yeast cells show enhanced sensitivity to oxidizing agents owing to depletion of NADPH [64]. Similarly, mouse ES cells deleted for the G6PD gene are highly sensitive to agents perturbing the intracellular redox status [65,67]. These cells were able to maintain a high NADPH/NADP+ ratio. However, upon treatment with diamide, an oxidant of thiol groups, the NADPH and GSH contents decreased rapidly [67] and led to apoptotic cell death [68]. Transient up-regulation of G6PD activity and mRNA levels has been observed in various human cell lines following treatment with hydrogen peroxide or diamide [69], which is presumably triggered by the oxidation of NADPH to NADP+ [67]. Accordingly, overexpression of G6PD increased the resistance to hydrogen peroxide [70,71], whereas inhibition of G6PD potentiated cell death [71]. It is interesting to note that G6PD activity is also subject to physiological regulation. For example, stimulation of cell growth is accompanied by an increase in G6PD activity [58,60].
Not only the surprisingly mild phenotype of G6PD-deficient cells, but also the fact that NADPH is produced by a cytosolic enzyme that is unavailable to mitochondria, the major site of superoxide production, prompted investigations of alternative routes of NADPH generation. Thus, other NADP-specific dehydrogenases, including IDP, ME and ALDH, have been identified as significant contributors to the reduced state of NADP. At least as important are several NADKs which can phosphorylate NADH and thereby generate NADPH directly (Figure 3). Although different organisms rely on the same enzyme activities to regenerate NADPH, there are differences, particularly with regard to subcellular localization and subsets of isoforms. Therefore, we will discuss the major NADP+-reducing enzymes separately for yeast and mammalian cells.
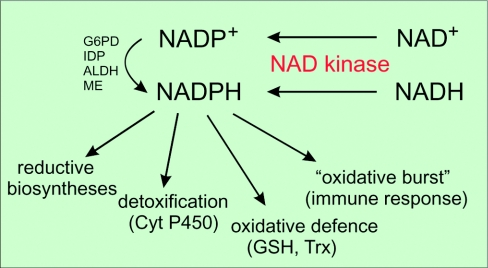
Figure 3. Generation and utilization of NADPH in eukaryotic cells.
Cyt P450, cytochrome P450; Trx, thioredoxin.
NADP-dependent dehydrogenases in yeast
Yeast
The isocitrate dehydrogenase family [isocitrate+NAD(P)++H+⇌α-oxoglutarate+CO2+NAD(P)H] consists of a mitochondrial NAD+-dependent isoform involved in the citric acid cycle and NADP+-dependent isoforms. Three IDP homologues, Idp1p, Idp2p and Idp3p, were identified in yeast and localized to mitochondria [72], cytosol [73] and peroxisomes [74] respectively. Both mitochondrial and cytosolic isoforms are not essential, even in the context of ZWF1 (encoding G6PD) deletion, since the triple mutant ΔIDP1ΔIDP2ΔZWF1 is viable [75]. Disruption of the peroxisomal IDP impaired growth on unsaturated fatty acids as the carbon source [74], indicating involvement of this enzyme in their metabolic utilization by providing NADPH in the peroxisomes.
Disruption of the gene encoding ME [malate+NAD(P)+⇌pyruvate+CO2+NAD(P)H], a mitochondrial protein, did not result in an observable growth phenotype [76], nor did it worsen the growth phenotypes of Idp and Zwf1 mutant strains [75]. The fact that the presumed major generators of NADPH, G6PD and IDP, were not essential under many growth conditions [75] prompted the search for alternative sources of NADPH. An isoform of the ALDH family [RCHO+NAD(P)++H2O⇌RCOOH+NAD(P)H+H+], Ald6p, a cytosolic enzyme, was identified as a suppressor of the ZWF1-deletion phenotype. Initially, disruption of both ZWF1 and ALD6 was thought to be lethal, whereas ALD6 deletion alone displayed no distinct phenotype [77]. However, the double mutant strain in this study was grown under conditions that also suppressed Idp2p expression. Indeed, it was later shown that co-disruption of only ALD6 and ZWF1 is not lethal [78]. Therefore, although contributing significantly to the reduction of NADP+, none of the NADP-specific dehydrogenases appears to be essential.
NADP-dependent dehydrogenases in mammals
In contrast with yeast, mammals possess two NADP-dependent isocitrate dehydrogenases, a mitochondrial (IDPm) and a cytosolic (IDPc) isoform [79], although the latter has also been ascribed to peroxisomes [80]. Both isoforms are major sources of NADPH supply in mammals. IDPm activity is transiently increased after treatment of cells with hydrogen peroxide [81] and both IDP activities are induced by γ-irradiation [82]. IDPc activity and protein content were also demonstrated to increase during differentiation of adipocytes indicating a role in supplying NADPH for fat and cholesterol biosyntheses [83]. Furthermore, overexpression of either IDP isoform improved the cells' ability to cope with oxidative stress, whereas decreased expression aggravated oxidative damage [81,84,85]. It is interesting to note that deletion of G6PD in mouse cells did not cause an up-regulation of IDPc activity [67]. However, this could be due to the fact that IDPc is, perhaps, generally more active in NADPH production than G6PD. For example, in rat liver, the contribution by IDPc exceeds that of G6PD 16–18-fold [42].
Two NADP+-dependent ME isoforms have been described in mammalian tissues and were localized to the cytosol and the mitochondria [86,87]. Both activities are not essential for cellular NADPH maintenance. However, cytosolic ME activity is increased in rat liver cirrhogenesis [88] and affected in acute hepatic injury [89], presumably by providing NADPH for detoxification reactions. Like IDPc, cytosolic ME is also not up-regulated in mouse cells deleted for G6PD [67]. The mitochondrial isoform plays an important role in the pyruvate-recycling pathway and in maintaining the intramitochondrial GSH in the brain [90]. In pancreatic islets, the mitochondrial pyruvate–malate shuttle could contribute far more to NADPH generation than the cytosolic pentose phosphate pathway [91].
The ALDH family consists of multiple forms in mammals being important for detoxification via NAD(P)-dependent conversion of endogenous and exogenous aldehyde substrates. ALDHs are up-regulated in response to oxidative stress, and several mutations in ALDH genes have been linked to diseases such as cancer and Alzheimer's disease [92]. The most abundant isoform 3A1 in mammalian cornea [93], a cytosolic protein, may play a role in cellular defence mechanisms that protect the corneal epithelium from oxidative damage [94]. However, whether mammalian ALDHs have a general role in the maintenance of reduced NADP has to be explored further.
The enzyme pyridine nucleotide transhydrogenase [H+out+NADH+NADP+⇌H+in+NAD++NADPH] is located in the membrane of many bacteria and in the inner membrane of animal mitochondria, where the catalytic site is exposed to the matrix side. It transports protons across the membrane in concert with hydride exchange between NAD and NADP, a reaction that couples the protonmotive force to the concentration of reducing equivalents [95]. Under physiological conditions the enzyme maintains mitochondrial NADP in the reduced state. Thereby, the transhydrogenase secures a high GSH/GSSG ratio in the mitochondria and modulates protein activities by regulating NADPH-dependent protein thiols [96]. Although knockout of the transhydrogenase gene in Escherichia coli, Caenorhabditis elegans and mouse was not lethal, these studies supported the important role of the transhydrogenase in defence against ROS (reviewed in [97]).
Taken together, none of the NADP+-reducing enzymes is essential by itself for maintaining the cellular redox balance. Besides G6PD, there are other key contributors to NADPH generation in the majority of cells. Still, so far it has remained unclear exactly how the NADPH pools in different organelles, cells and tissues are generated and maintained. It is plausible that cells do not rely on a single NADPH-regenerating system given the permanent threat of oxidative damage. Despite the important new insights obtained over the past few years, several fundamental questions have still to be answered. These relate above all to the regulation of both the identified dehydrogenases, for example by post-translational modifications, and the availability of substrates according to the physiological demands. In any case, the key step regulating the capacity of cellular NADPH generation lies with the synthesis and availability of NADP by NADKs (Figure 3).
NADK
The biosynthesis of NADP requires the phosphorylation of NAD catalysed by NADK (ATP:NAD 2′-phosphotransferase). This enzyme is essential as has been demonstrated both in prokaryotes such as E. coli, Mycobacterium tuberculosis, Bacillus subtilis and Salmonella enterica [98–101] and in the yeast S. cerevisiae [102,103]. Apparently, mice lacking NADK are also not viable (unpublished work cited in [103]). NADK activity has been known for decades. NADP formation was first observed in yeast homogenates [104,105] and the enzyme purified 10 years later [106]. Meanwhile, enzymes from animals [107–109], yeast [110,111] and plants [112] have been characterized. However, conclusive structural information has only recently become available following the identification of amino acid sequences of Micrococcus flavus and M. tuberculosis NADKs [113]. Based on this information, NADKs from a variety of organisms were soon identified and then studied in great detail (reviewed in [114]). At least for mammalian tissues a major obstacle for the isolation of the NADKs had been their relatively low abundance. NADKs are ubiquitous enzymes which apparently constitute a protein family distantly related to other kinases of metabolic intermediates [115]. In both yeast and plants, three NADK isoforms were identified with distinct subcellular locations. Strikingly, so far only a single isoform has been found in mammals.
Structural properties of NADKs
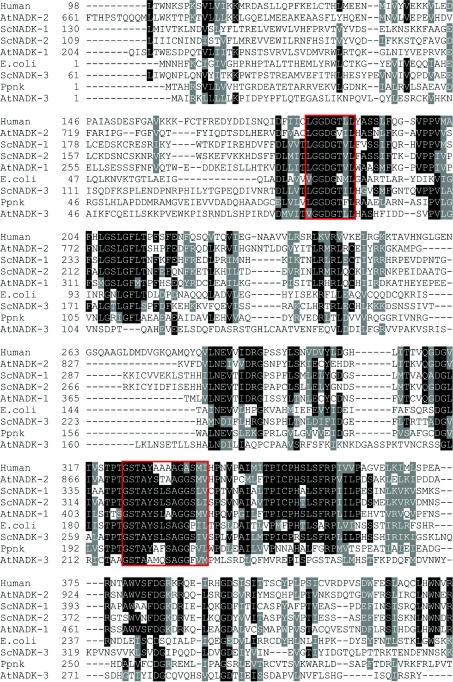
Almost all known NADKs are oligomeric proteins consisting of two to eight identical subunits of 30–60 kDa (Table 1). The comparison of amino acid sequences of several known prokaryotic and eukaryotic NADKs revealed a general structural organization consisting of a conserved catalytic domain (PFAM 01513) within the C-terminus and variable N-terminal parts. The catalytic domain includes two highly conserved motifs, an L/VGGDG motif and a glycine-rich motif, which are involved in substrate binding (Figure 4). The importance of these motifs for the catalytic activity of NADKs was confirmed by site-directed mutagenesis of several amino acid residues within the first and second nucleotide-binding motifs [115–117] and was supported further by the three-dimensional structures of prokaryotic NADKs [116,118–120]. The overall structure, as determined for M. tuberculosis NADK [116,118], is organized into an N-domain, a C-domain and a C-terminal tail. The N-terminal domain resembles a classical Rossmann fold, known to be involved in dinucleotide binding [121], consisting of a single parallel β-sheet flanked by α-helices. The C-terminal domain adopts a novel fold with structural similarity to the human Ki67 fork-head-associated domain [118]. The structure of Archaeoglobus fulgidus NADK [119] is similarly comprised of two major domains. The substrates NAD+ and ATP, and the product NADP+ are all bound to a cleft between the N- and C-domains and intersubunit contacts. Accordingly, NADKs are homo-oligomers whose subunit interactions are required for ligand binding. No other larger putative structural motifs have been discerned, except for a calmodulin-binding domain within the long N-terminal part of Arabidopsis thaliana NADK-2 [122].
Table 1. Physical and kinetic properties of prokaryotic and eukaryotic NADKs.
Activities using either NAD+ or NADH as substrate are presented. The values are given as μmol of product (NADP+ or NADPH respectively) formed per min per mg of protein. −, not detectable; poly(P) designates polyphosphate as the substrate instead of ATP.
| Organism | Subunit mass and number | Activity NAD+/NADH | Km NAD+ | Km NADH | Km ATP | Reference |
|---|---|---|---|---|---|---|
| Homo sapiens | ||||||
| hNADK | 49 kDa tetramer | 6.7/− | 0.54 | − | 3.3 | [127] |
| S. cerevisiae | ||||||
| ScNADK-1b (Utr1p) | 60 kDa hexamer | 16.2-51.5/∼2 | 0.5 | 3.9 | 0.6 | [102,126,129,152] |
| ScNADK-2b (Yef1p) | 56 kDa octamer | 3.3–9.5/∼1 | 1.9 | 2 | 0.17 | [102,126,129] |
| ScNADK-3b (Pos5p) | 46 kDa | 0.2/0.4, but see [151] | a | a | a | [151,155] |
| A. thaliana | ||||||
| AtNADK-1 | 58 kDa | <0.2/<0.1 | 0.52 | a | 0.73 | [122,125] |
| AtNADK-2 | 109 kDa | 0.2/− | 0.43 | − | 0.74 | [122] |
| AtNADK-3 | 35 kDa dimer | 41.2/23.2 | 2.39 | 0.042 | NAD+: 0.19 | [160] |
| NADH: 0.06 | ||||||
| E. coli | ||||||
| YfjB | 30 kDa hexamer | 13/− | 2 | − | 2.5 | [126,128] |
| M. tuberculosis | ||||||
| Ppnk | 33 kDa tetramer | 0.5–4.3/0.7 | 0.9–3.3 | a | 1.8–2.5 | [113,117,126] |
| (poly(P): 1.7/0.2) | (poly(P): 1.2–2.9) | (poly(P): 1.3–1.6) | ||||
| Micrococcus flavus | ||||||
| Mfnk | 34 kDa dimer | 18.3/8.7 | 0.53–0.83 | 0.45 | NAD+: 0.23 | [113,126] |
| (poly(P): 7.4/3.0) | (poly(P): 0.26–0.58) | (poly(P): 0.35) | NADH: 0.32 | |||
| (poly(P): NAD+: 0.33 | ||||||
| NADH: 0.58) |
aNot reported.
bProposed name.
Figure 4. Partial multiple sequence alignment of NADKs from several organisms.
Amino acid sequences of human NADK (protein ID NP_075394); A. thaliana NADK-1, NADK-2 and NADK-3 (NP_974347, NP_564145 and NP_177980); S. cerevisiae NADK-1/Utr1p, NADK-2/Yef1p and NADK-3/Pos5p (P21373, NP_010873 and NP_015136); E. coli YfjB (NP_417105); and M. tuberculosis Ppnk (BAB21478) were compared using Clustal W [170]. Identical and similar residues are highlighted in black and grey respectively. The two conserved NADK motifs are boxed in red.
Structural analyses of the NADK from A. fulgidus complexed with substrates indicated a critical role for the GGDG motif [119]. The GGDG motif has been shown to be a highly conserved motif in a superfamily including NADK, diacylglyceride kinase, sphingosine kinase and 6-phosphofructokinase [115]. Moreover, structural studies of prokaryotic NADKs at higher resolution suggest a particular mechanism of phosphate transfer based on substrate-assisted catalysis (G. Poncet-Montange and G. Labesse, Centre de Biochimie Structurale INSERM U554-CNRS UMR 5048-UM1, Montpellier, France, personal communication).
Substrate specificity and catalytic properties of NADKs
Table 1 summarizes physical and kinetic properties of NADKs from various organisms. NADKs from both prokaryotes and eukaryotes utilize nucleoside triphosphates, preferentially ATP, as phosphoryl donor for catalysis. Interestingly, some bacterial enzymes, for example from M. tuberculosis, Micrococcus flavus and B. subtilis, can utilize both inorganic poly(P) (polyphosphate) and nucleoside triphosphates as phosphoryl donors (reviewed in [114]). Poly(P) is considered to be an ‘ancient’ energy carrier preceding ATP [123]. Even glucose 6-phosphate-dependent bacterial NADKs have been described [124], but were not investigated further. How do poly(P)/ATP–NADKs distinguish between the phosphoryl donors poly(P) and ATP and which amino acid residues determine the difference? This important question has yet to be answered and may represent a key to the design of new antibacterial drugs. In particular the mycobacterial NADK could be a promising target for the treatment of tuberculosis since its NADK is essential and exhibits higher affinity towards poly(P) than ATP [117].
Relatively high Km values for ATP have been reported for E. coli, and human NADKs (Table 1, and also for Salmonella enterica), suggesting a regulation of their activity according to physiological changes of the cellular ATP concentration, which is in the millimolar range. Several recombinant enzymes have been shown to phosphorylate only NAD+, but not NADH, thus displaying high selectivity. Others exhibit a far less stringent substrate specificity and use both the oxidized and reduced nucleotide (see Table 1). Differences in substrate selectivity as reported for plant NADKs [122,125] may have arisen from the assay conditions. Moreover, native enzymes in their cellular context may interact with other factors that might influence substrate specificity. Recently, it was suggested that an arginine residue in the conserved domain of NADKs plays a major role in conveying a strict substrate preference towards NAD+ [126]. Replacement of this arginine by glycine or polar amino acid residues converted highly selective prokaryotic NADKs to relaxed ones. It should be noted, however, that, compared with the original activity, the converted E. coli NADK used the substrate NADH far less efficiently. Conversely, Micrococcus flavus relaxed NADK did not become more selective by a reciprocal single amino acid replacement, but exhibited a significant loss of activity regardless of the substrate used. Therefore, this amino acid position appears to be important for catalysis, but is not the only determinant of substrate specificity. The amino acid sequence alignment of NADK homologues (cf. Figure 4) revealed that primarily polar amino acid residues are found in eukaryotic homologues at the investigated position. This would imply relaxed substrate specificity in contrast with prokaryotic NADKs. However, although the human enzyme harbours glutamine, a polar amino acid, in this position, NADH phosphorylation was hardly detectable (N. Pollak and M. Ziegler, unpublished work).
The discovery of NAADP suggested the possibility of NADKs involvement in the synthesis of a second messenger. As can be inferred from Figure 1, NAAD is a physiological intermediate whose phosphorylation would result in the generation of NAADP. Recently, Bieganowski et al. [102] reported NAAD kinase activity for recombinant yeast NADKs, which constituted about 1% of the corresponding NADK activities. The physiological significance of this observation remains unclear, since no NAADP-responsive system in yeast has been described, while phosphorylation of NAAD by human NADK was not detected [127]. Still, other cellular factors could influence the affinity of human NADK towards NAAD.
Modulators of NADK activity
The catalytic activity of several NADKs is inhibited by the reduced pyridine nucleotides [99,117,128,129], by high concentrations of NADP+ [117,130] or by HgCl2 [128–130]. Interestingly, NADPH, NADH and NADP+ inhibit S. cerevisiae NADKs to different extents suggesting a possible regulation by these compounds [129]. NADK of Salmonella enterica is inhibited by NADPH during normal growth and is released from the inhibition in response to metabolic changes induced by UV irradiation or oxidative stress [99]. QA, a precursor of de novo NAD+ synthesis was shown to inhibit NADK of Salmonella enterica serotype Typhimurium [131] and to activate B. subtilis NADK [130], but had no effect on human NADK (N. Pollak and M. Ziegler, unpublished work).
Regulation of NADK activity by calcium and calmodulin
Although the molecular characterization of NADKs was only achieved recently, it had been known for almost three decades that at least the enzymes from plants, sea urchin eggs and human neutrophils are stimulated by calcium/calmodulin [132–134]. In fact, NADK was the first enzyme identified in plants to be regulated by the ubiquitous calcium-sensing protein calmodulin [135,136]. NADK activity was actually used as a tool to study calmodulins in plants, e.g. to determine their distribution in maize [137]. Plants possess both calmodulin-dependent and -independent NADK isoforms that differ in their subcellular localization [138–140]. In eggs from the sea urchins Lytechinus pictus and Strongylocentrotus purpuratus, calcium-mediated activation of NADK occurs early after fertilization and can be prevented by the calcium/calmodulin antagonists, EGTA/trifluoperazine [133]. Enhanced NADK activity results in an increase of the NADPH/NADP ratio, which is mediated by enzymes of the pentose phosphate pathway [141]. Accordingly, the need for elevated NADK activity during fertilization has been explained by the requirement for NADPH of ribonucleotide reductase [142] and the effect on eukaryotic initiation factor eIF-2B [143]. Moreover, increased NADPH levels lead to increased hydrogen peroxide production via NADPH oxidase [144,145], which is important for the post-fertilization hardening of the fertilization envelope [146]. In human neutrophils, the activation of NADK by calcium and calmodulin, and the ensuing increase of the NADP content [134] are probably related to cellular activation in response to bacterial challenges, which includes superoxide production by NADPH oxidase. A similar ‘oxidative burst’ was reported for thyroid cells [147] and for the calcium-mediated defence response in plants [148]. So far, the molecular mechanism of the calcium/calmodulin-dependent activation of NADKs has remained unknown. Compared with a partially purified calcium- and calmodulin-dependent enzyme from A. thaliana, the activity of two recombinant isoforms, AtNADK-1 and AtNADK-2, was not influenced by calcium/calmodulin, although AtNADK-2 exhibited some affinity for calmodulin [122]. Calmodulin had also no detectable effect on the activity of human recombinant NADK [127]. These observations suggest that the activation of NADK by calcium/calmodulin is indirect and requires additional factors which could mediate, for example, post-translational modifications. Moreover, the mechanism could differ depending on the cell type or species. Since higher plants express calmodulin-dependent and -independent NADKs as well as various calmodulin isoforms [149], different calmodulin-mediated pathways could exist to regulate individual NADK activities. In contrast, only a single NADK isoform is known in mammals along with an invariant calmodulin. There have been no reports regarding a potential influence of calcium and/or calmodulin on the recombinant yeast NADK isoforms. Interestingly, ScNADK-1/Utr1p has been functionally linked to Ald6p due to the involvement of both enzymes in salt stress, and since mutants lacking either protein show similar transcriptional responses [150]. Salt stress in yeast leads to an influx of calcium, which in turn might activate calmodulin-mediated modulation of ScNADK-1/Utr1p activity.
Properties and subcellular distribution of eukaryotic NADKs
The complexity of eukaryotic cells requires elaborate regulatory mechanisms to adjust to alterations in the environment. Owing to the essential functions of NADP, some of these mechanisms should be reflected in the modulation of NADK activities. Although so far very little is known about the transcriptional regulation of NADKs, recent studies have provided important insights into the alterations of NADK activities in response to changes in growth conditions. In addition, since NADP is membrane-impermeable, the pathways supplying different organelles with the pyridine nucleotide are of particular interest. Strikingly, while in yeast and plants three compartment-specific isoforms have been identified, only a single NADK isoform has so far been detected in mammals. Below, we will summarize the current knowledge about NADKs in yeast, plants and mammals. To avoid confusion and to emphasize the actual enzymatic activity, we propose to adopt the terminology established for the plant enzymes also for yeast NADKs based on sequence similarity and subcellular localization. Thus, below we will use the abbreviations ScNADK-1, ScNADK-2, and ScNADK-3 for the yeast NADKs Utr1p, Yef1p and Pos5p respectively.
NADKs in yeast
Of the three NADK isoforms in S. cerevisiae (see Table 1 and Figure 5), NADK-3 localizes to the mitochondrial matrix and prefers NADH as substrate [151]. NADK-1 [152] and NADK-2 [129] are presumably cytosolic proteins and exhibit relaxed substrate specificity with some preference towards NAD+. According to a global analysis of protein expression in yeast only ∼300 NADK-2 molecules per cell are present compared with ∼5000 molecules of NADK-1 and NADK-3 [153]. Disruption of both NADK-3 and NADK-1 is synthetic lethal, with or without the deletion of NADK-2 [102,103]. The double mutant can, however, be rescued by overexpression of any of the yeast NADK isoforms [102] or human NADK [103]. Therefore, yeast requires a certain level of NADK activity for survival, which is in close agreement with the reportedly essential functions of NADKs in prokaryotes [98–101].
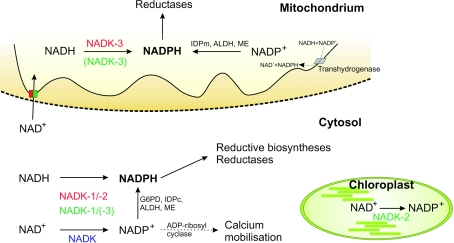
Figure 5. Subcellular distribution of NADP metabolism in eukaryotic cells.
NADK isoforms described in mammals, yeast and Arabidopsis are highlighted in blue, red and green respectively. Transhydrogenase and ADP-ribosyl cyclase have not been detected in yeast.
Yeast cells deleted for the mitochondrial NADK-3 are highly sensitive towards oxidative stress [154] and grow poorly on glycerol or in medium lacking arginine [151]. Deletion strains accumulate iron in the mitochondria and are defective in mitochondrial Fe–S cluster-containing enzymes [151] leading to up-regulation of genes involved in iron transport [103]. NADK-3 mutants also exhibit increased frameshift mutations in the mitochondrial DNA and increased petite colony formation [155]. In contrast, deletion of NADK-1 or NADK-2 does not result in severe growth defects or in hypersensitivity to elevated oxygen levels [151].
These observations establish NADK-3, the mitochondrial isoform, as the most critical generator of NADPH in yeast. In the mitochondrion, NADPH is needed for arginine synthesis (when absent from the growth medium), but its major physiological function relates to the protection against oxidative damage by feeding mitochondrial reducing systems. The role of NADK-1 is presumably to provide NADP for cytosolic NADP-dependent dehydrogenases and detoxifying systems. Given the low expression level and the absence of a phenotype of the deletion mutant, the role of NADK-2 remains rather unclear.
It has been demonstrated for yeast and plant cells that NAD+ can be exchanged between the cytosol and mitochondria (Figure 5; [156,157]), whereas the cytosolic and mitochondrial NADP pools appear to be maintained independently. The occurrence of mitochondrial and cytosolic NADK isoforms is therefore plausible, which is also indicated by the severe phenotype of mutants lacking NADK-1 and -3. In contrast with these observations, Shi et al. [129] reported viable yeast cells upon disruption of all three yeast NADK genes, which would imply the existence of a hitherto unidentified NADK isoform or even an alternative pathway of NADP generation.
NADKs in plants
NADK activity in plants has been found in the cytoplasm [140], mitochondria [138] and chloroplasts [139,158] (Figure 5). An NADK isoform in the chloroplasts is not unexpected because of the importance of NADP in photosynthesis. Three A. thaliana NADK isoenzymes have been identified, and the recombinant proteins were characterized with regard to functional properties (Table 1) and subcellular localization (Figure 5). AtNADK-1 lacks any potential targeting sequence and is most probably cytosolic. AtNADK-2 and AtNADK-3 contain a putative chloroplast transit peptide and a mitochondrial targeting sequence respectively [159,160]. A green fluorescent protein–NADK-2 construct co-localized with chlorophyll [159]. The subcellular location of NADK-3 is less clear. The protein has been suggested to be cytosolic, because immunoblot analyses did not reveal a corresponding reactive band in mitochondrial fractions [160]. Still, the NADK-3 sequence is similar to the yeast mitochondrial NADK-3 and also prefers NADH to NAD+ as its substrate.
Consistent with the important role of yeast NADKs in oxidative defence, irradiation or treatment with hydrogen peroxide of plant cell suspensions induces NADK-1 on both the mRNA and protein levels. Moreover, mutant lines exhibit increased sensitivity to oxidative stress [125]. Knockout of NADK-2 in A. thaliana delays growth and development, resulting in reduced leaf size and seed production. Furthermore, these mutant plants have lower levels of chlorophylls a and b, and are more sensitive to various stress conditions [159].
Two important aspects of NADP generation in plants have so far remained unresolved. First, the individual contributions of each of the three isoforms to the NADPH pools in different subcellular compartments have not been established, except, perhaps, for NADK-2. Secondly, the ‘model organism’ for the calcium/calmodulin-dependent stimulation of NADK activity has still not revealed the secret regarding the mechanism of this potentially critical regulation.
NADKs in mammals
Considering the eminent roles of NADKs in yeast and plants one would predict that in mammalian cells these enzymes are also essential. However, so far this assumption has not been experimentally addressed, except for the fact that mouse embryonic lethality upon NADK loss was reported as an unpublished observation by Shianna et al. [103]. Still, Akella and Harris [161] investigated NADK activity during development of rat conceptus and their results show dramatic spatio-temporal variations, reflecting the importance of NADK in maintaining the cellular redox state during organogenesis.
Perhaps the most astounding fact to mention here again is the absence of any evidence that would indicate the existence of more than one NADK isoform in mammals. A single human NADK cDNA has been cloned, overexpressed and the protein characterized [127]. An orthologous gene in rodents has been predicted from its sequence similarity. The deduced amino acid sequences exhibit high similarity to the NADKs of A. thaliana and S. cerevisiae (Figure 4). Similar to the situation for the human NADK, so far no additional isoform has been discerned.
Given the vital roles of at least two NADK isoforms in yeast, one would certainly expect more than one mammalian NADK isoform. That is, even a lethal knockout of the known gene in mice does not necessarily indicate that it is the only one. Nevertheless, at least according to the currently available information, an additional NADK isoform in mammals should have a primary structure substantially different from virtually all those that have been identified so far.
NADP DEGRADATION
NADP-degrading enzymes are supposed to act in concert with NADKs to maintain an adequate balance between NAD and NADP. Two enzyme activities are known that convert NADP+, namely ADP-ribosyl cyclase and NADPase (NADP phosphatase), which dephosphorylates NADP. As described above, ADP-ribosyl cyclases have functions related to calcium signalling. NADPases, however, are not well studied. Their activity was observed in rat liver Golgi apparatus and mitochondria [162,163], in dormant seeds of Avena sativa L. [164] and to be necessary for NADP utilization in Haemophilus influenzae [165]. NADPase activity of the phytoflagellate Euglena gracilis ZC mutant shows circadian oscillation similar to NADK activity, suggesting that both enzymes represent clock ‘gears’ [166,167]. Two NADPase isozymes were purified from the Arthrobacter sp. strain KM [168]. Interestingly, the hyperthermophilic archaeon Methanococcus jannaschii possesses a protein, MJ0917, with both NADK and NADPase activities [169]. This novel bifunctional protein could be important for the regulation of the NADP+ concentration and thereby maintain a defined NAD+/NADP+ ratio.
CONCLUSION
The pyridine nucleotides play vital roles in all organisms. While their roles in metabolic redox reactions have been well characterized, their function in signalling pathways has only been recognized recently. NAD+ appears to be vital for various regulatory systems throughout the cell. On the other hand, NADP+ derivatives are important constituents of cytosolic calcium signalling pathways. In addition, NADPH holds a key position in the oxidative defence systems. The synthesis of NADP and the enzymes that maintain its reduced state are of critical importance not only under conditions of oxidative stress, but also to counteract oxidative damage under normal physiological conditions. Therefore, further understanding of the pathways and molecular mechanisms of NADPH generation will also provide new tools to cope with the many pathological conditions that involve oxidative damage.
Acknowledgments
We gratefully acknowledge financial support from the Norwegian Research Council, the Deutsche Forschungsgemeinschaft (ZI 541/3 and 4) and the Norwegian Cancer Society (A05128/005).
References
- 1.Berger F., Ramirez-Hernandez M. H., Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem. Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Magni G., Amici A., Emanuelli M., Raffaelli N., Ruggieri S. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. [DOI] [PubMed] [Google Scholar]
- 3.Magni G., Amici A., Emanuelli M., Orsomando G., Raffaelli N., Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell. Mol. Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katoh A., Uenohara K., Akita M., Hashimoto T. Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 2006;141:851–857. doi: 10.1104/pp.106.081091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger F., Lau C., Dahlmann M., Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 6.Bieganowski P., Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss–Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 7.Sasiak K., Saunders P. P. Purification and properties of a human nicotinamide ribonucleoside kinase. Arch. Biochem. Biophys. 1996;333:414–418. doi: 10.1006/abbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 8.Chambon P., Weill J. D., Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 9.Honjo T., Nishizuka Y., Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- 10.Paone G., Wada A., Stevens L. A., Matin A., Hirayama T., Levine R. L., Moss J. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corda D., Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takamura-Enya T., Watanabe M., Totsuka Y., Kanazawa T., Matsushima-Hibiya Y., Koyama K., Sugimura T., Wakabayashi K. Mono(ADP-ribosyl)ation of 2′-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12414–12419. doi: 10.1073/pnas.221444598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Girolamo M., Dani N., Stilla A., Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 14.Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 15.Herrero-Yraola A., Bakhit S. M., Franke P., Weise C., Schweiger M., Jorcke D., Ziegler M. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 2001;20:2404–2412. doi: 10.1093/emboj/20.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otto H., Reche P. A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber V., Dantzer F., Ame J. C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 18.Burkle A. Poly(ADP-ribose): the most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 19.d'Adda di Fagagna F., Hande M. P., Tong W. M., Lansdorp P. M., Wang Z. Q., Jackson S. P. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat. Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 20.Kraus W. L., Lis J. T. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 21.Kanai M., Tong W. M., Sugihara E., Wang Z. Q., Fukasawa K., Miwa M. Involvement of poly(ADP-ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi N. W., Lodish H. F. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 23.Culver G. M., McCraith S. M., Zillmann M., Kierzek R., Michaud N., LaReau R. D., Turner D. H., Phizicky E. M. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1″-2″ cyclic phosphate. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- 24.Perraud A. L., Fleig A., Dunn C. A., Bagley L. A., Launay P., Schmitz C., Stokes A. J., Zhu Q., Bessman M. J., Penner R., et al. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- 25.Vu C. Q., Lu P. J., Chen C. S., Jacobson M. K. 2′-Phospho-cyclic ADP-ribose, a calcium-mobilizing agent derived from NADP. J. Biol. Chem. 1996;271:4747–4754. [PubMed] [Google Scholar]
- 26.Basile G., Taglialatela-Scafati O., Damonte G., Armirotti A., Bruzzone S., Guida L., Franco L., Usai C., Fattorusso E., De Flora A., Zocchi E. ADP-ribosyl cyclases generate two unusual adenine homodinucleotides with cytotoxic activity on mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14509–14514. doi: 10.1073/pnas.0503691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellmich M. R., Strumwasser F. Purification and characterization of a molluscan egg-specific NADase, a second-messenger enzyme. Cell Regul. 1991;2:193–202. doi: 10.1091/mbc.2.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H. C. Mechanisms of calcium signaling by cyclic ADP-ribose and NAADP. Physiol. Rev. 1997;77:1133–1164. doi: 10.1152/physrev.1997.77.4.1133. [DOI] [PubMed] [Google Scholar]
- 29.De Flora A., Zocchi E., Guida L., Franco L., Bruzzone S. Autocrine and paracrine calcium signaling by the CD38/NAD+/cyclic ADP-ribose system. Ann. N.Y. Acad. Sci. 2004;1028:176–191. doi: 10.1196/annals.1322.021. [DOI] [PubMed] [Google Scholar]
- 30.Ceni C., Muller-Steffner H., Lund F., Pochon N., Schweitzer A., De Waard M., Schuber F., Villaz M., Moutin M. J. Evidence for an intracellular ADP-ribosyl cyclase/NAD+-glycohydrolase in brain from CD38-deficient mice. J. Biol. Chem. 2003;278:40670–40678. doi: 10.1074/jbc.M301196200. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki M., Masgrau R., Morgan A. J., Churchill G. C., Patel S., Ashcroft S. J., Galione A. Organelle selection determines agonist-specific Ca2+ signals in pancreatic acinar and β cells. J. Biol. Chem. 2004;279:7234–7240. doi: 10.1074/jbc.M311088200. [DOI] [PubMed] [Google Scholar]
- 32.Chini E. N., De Toledo F. G. Nicotinic acid adenine dinucleotide phosphate: a new intracellular second messenger? Am. J. Physiol. Cell Physiol. 2002;282:C1191–C1198. doi: 10.1152/ajpcell.00475.2001. [DOI] [PubMed] [Google Scholar]
- 33.Berridge G., Cramer R., Galione A., Patel S. Metabolism of the novel Ca2+-mobilizing messenger nicotinic acid-adenine dinucleotide phosphate via a 2′-specific Ca2+-dependent phosphatase. Biochem. J. 2002;365:295–301. doi: 10.1042/BJ20020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner K. G., Landry J., Sternglanz R., Denu J. M. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 36.Frye R. A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 37.Jackson M. D., Denu J. M. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 38.Grubisha O., Rafty L. A., Takanishi C. L., Xu X., Tong L., Perraud A. L., Scharenberg A. M., Denu J. M. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J. Biol. Chem. 2006;281:14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- 39.Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q., Piston D. W., Goodman R. H. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 41.Williamson D. H., Lund P., Krebs H. A. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veech R. L., Eggleston L. V., Krebs H. A. The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 1969;115:609–619. doi: 10.1042/bj1150609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finkel T., Holbrook N. J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 44.Droge W. Oxidative stress and aging. Adv. Exp. Med. Biol. 2003;543:191–200. doi: 10.1007/978-1-4419-8997-0_14. [DOI] [PubMed] [Google Scholar]
- 45.Klaunig J. E., Kamendulis L. M. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 46.Valko M., Izakovic M., Mazur M., Rhodes C. J., Telser J. Role of oxygen radicals in DNA damage and cancer incidence. Mol. Cell. Biochem. 2004;266:37–56. doi: 10.1023/b:mcbi.0000049134.69131.89. [DOI] [PubMed] [Google Scholar]
- 47.Berg D., Youdim M. B., Riederer P. Redox imbalance. Cell Tissue Res. 2004;318:201–213. doi: 10.1007/s00441-004-0976-5. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X., Lee H. G., Casadesus G., Avila J., Drew K., Perry G., Smith M. A. Oxidative imbalance in Alzheimer's disease. Mol. Neurobiol. 2005;31:205–217. doi: 10.1385/MN:31:1-3:205. [DOI] [PubMed] [Google Scholar]
- 49.Obrosova I., Faller A., Burgan J., Ostrow E., Williamson J. R. Glycolytic pathway, redox state of NAD(P)-couples and energy metabolism in lens in galactose-fed rats: effect of an aldose reductase inhibitor. Curr. Eye Res. 1997;16:34–43. doi: 10.1076/ceyr.16.1.34.5113. [DOI] [PubMed] [Google Scholar]
- 50.Lakowicz J. R., Szmacinski H., Nowaczyk K., Johnson M. L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin S. J., Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 52.Canepa L., Ferraris A. M., Miglino M., Gaetani G. F. Bound and unbound pyridine dinucleotides in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Biochim. Biophys. Acta. 1991;1074:101–104. doi: 10.1016/0304-4165(91)90046-j. [DOI] [PubMed] [Google Scholar]
- 53.Mack T. G., Reiner M., Beirowski B., Mi W., Emanuelli M., Wagner D., Thomson D., Gillingwater T., Court F., Conforti L., et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 54.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., Lin S. S., Manchester J. K., Gordon J. I., Sinclair D. A. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 55.Sims J. L., Berger S. J., Berger N. A. Poly(ADP-ribose) polymerase inhibitors preserve nicotinamide adenine dinucleotide and adenosine 5′-triphosphate pools in DNA-damaged cells: mechanism of stimulation of unscheduled DNA synthesis. Biochemistry. 1983;22:5188–5194. doi: 10.1021/bi00291a019. [DOI] [PubMed] [Google Scholar]
- 56.Ying W., Garnier P., Swanson R. A. NAD+ repletion prevents PARP-1-induced glycolytic blockade and cell death in cultured mouse astrocytes. Biochem. Biophys. Res. Commun. 2003;308:809–813. doi: 10.1016/s0006-291x(03)01483-9. [DOI] [PubMed] [Google Scholar]
- 57.Rydstrom J., Hoek J. B., Ernster L. Nicotinamide nucleotide transhydrogenases. In: Boyer P. D., editor. The Enzymes. New York: Academic Press; 1976. pp. 51–88. [Google Scholar]
- 58.Kletzien R. F., Harris P. K., Foellmi L. A. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 1994;8:174–181. doi: 10.1096/fasebj.8.2.8119488. [DOI] [PubMed] [Google Scholar]
- 59.Stanton R. C., Seifter J. L., Boxer D. C., Zimmerman E., Cantley L. C. Rapid release of bound glucose-6-phosphate dehydrogenase by growth factors. Correlation with increased enzymatic activity. J. Biol. Chem. 1991;266:12442–12448. [PubMed] [Google Scholar]
- 60.Tian W. N., Pignatare J. N., Stanton R. C. Signal transduction proteins that associate with the platelet-derived growth factor (PDGF) receptor mediate the PDGF-induced release of glucose-6-phosphate dehydrogenase from permeabilized cells. J. Biol. Chem. 1994;269:14798–14805. [PubMed] [Google Scholar]
- 61.Beutler E., West C. Glucose-6-phosphate dehydrogenase variants in the chimpanzee. Biochem. Med. 1978;20:364–370. doi: 10.1016/0006-2944(78)90084-4. [DOI] [PubMed] [Google Scholar]
- 62.Luzzatto L. Glucose 6-phosphate dehydrogenase deficiency and hemolytic anemia. In: Nathan D. G., Oski D. F., editors. Hematology of Infancy and Childhood. Philadelphia: W. B. Saunders; 1993. pp. 674–691. [Google Scholar]
- 63.Weber G. Strongly conserved segment of gene expression in cancer cells. In: Reutter W., Popper H., Arias I. M., Heinrich P. C., Keppler D., Landmann L., editors. Modulation of Liver Cell Expression. Lancaster: MTP; 1987. pp. 303–314. [Google Scholar]
- 64.Nogae I., Johnston M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene. 1990;96:161–169. doi: 10.1016/0378-1119(90)90248-p. [DOI] [PubMed] [Google Scholar]
- 65.Pandolfi P. P., Sonati F., Rivi R., Mason P., Grosveld F., Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Longo L., Vanegas O. C., Patel M., Rosti V., Li H., Waka J., Merghoub T., Pandolfi P. P., Notaro R., Manova K., Luzzatto L. Maternally transmitted severe glucose 6-phosphate dehydrogenase deficiency is an embryonic lethal. EMBO J. 2002;21:4229–4239. doi: 10.1093/emboj/cdf426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Filosa S., Fico A., Paglialunga F., Balestrieri M., Crooke A., Verde P., Abrescia P., Bautista J. M., Martini G. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem. J. 2003;370:935–943. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fico A., Paglialunga F., Cigliano L., Abrescia P., Verde P., Martini G., Iaccarino I., Filosa S. Glucose-6-phosphate dehydrogenase plays a crucial role in protection from redox-stress-induced apoptosis. Cell Death Differ. 2004;11:823–831. doi: 10.1038/sj.cdd.4401420. [DOI] [PubMed] [Google Scholar]
- 69.Ursini M. V., Parrella A., Rosa G., Salzano S., Martini G. Enhanced expression of glucose-6-phosphate dehydrogenase in human cells sustaining oxidative stress. Biochem. J. 1997;323:801–806. doi: 10.1042/bj3230801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salvemini F., Franze A., Iervolino A., Filosa S., Salzano S., Ursini M. V. Enhanced glutathione levels and oxidoresistance mediated by increased glucose-6-phosphate dehydrogenase expression. J. Biol. Chem. 1999;274:2750–2757. doi: 10.1074/jbc.274.5.2750. [DOI] [PubMed] [Google Scholar]
- 71.Tian W. N., Braunstein L. D., Apse K., Pang J., Rose M., Tian X., Stanton R. C. Importance of glucose-6-phosphate dehydrogenase activity in cell death. Am. J. Physiol. 1999;276:C1121–C1131. doi: 10.1152/ajpcell.1999.276.5.C1121. [DOI] [PubMed] [Google Scholar]
- 72.Haselbeck R. J., McAlister-Henn L. Isolation, nucleotide sequence, and disruption of the Saccharomyces cerevisiae gene encoding mitochondrial NADP(H)-specific isocitrate dehydrogenase. J. Biol. Chem. 1991;266:2339–2345. [PubMed] [Google Scholar]
- 73.Loftus T. M., Hall L. V., Anderson S. L., McAlister-Henn L. Isolation, characterization, and disruption of the yeast gene encoding cytosolic NADP-specific isocitrate dehydrogenase. Biochemistry. 1994;33:9661–9667. doi: 10.1021/bi00198a035. [DOI] [PubMed] [Google Scholar]
- 74.Henke B., Girzalsky W., Berteaux-Lecellier V., Erdmann R. IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the β-oxidation of unsaturated fatty acids. J. Biol. Chem. 1998;273:3702–3711. doi: 10.1074/jbc.273.6.3702. [DOI] [PubMed] [Google Scholar]
- 75.Minard K. I., Jennings G. T., Loftus T. M., Xuan D., McAlister-Henn L. Sources of NADPH and expression of mammalian NADP+-specific isocitrate dehydrogenases in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:31486–31493. doi: 10.1074/jbc.273.47.31486. [DOI] [PubMed] [Google Scholar]
- 76.Boles E., de Jong-Gubbels P., Pronk J. T. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 1998;180:2875–2882. doi: 10.1128/jb.180.11.2875-2882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grabowska D., Chelstowska A. The ALD6 gene product is indispensable for providing NADPH in yeast cells lacking glucose-6-phosphate dehydrogenase activity. J. Biol. Chem. 2003;278:13984–13988. doi: 10.1074/jbc.M210076200. [DOI] [PubMed] [Google Scholar]
- 78.Minard K. I., McAlister-Henn L. Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 2005;280:39890–39896. doi: 10.1074/jbc.M509461200. [DOI] [PubMed] [Google Scholar]
- 79.Plaut G. W., Cook M., Aogaichi T. The subcellular location of isozymes of NADP-isocitrate dehydrogenase in tissues from pig, ox and rat. Biochim. Biophys. Acta. 1983;760:300–308. doi: 10.1016/0304-4165(83)90177-0. [DOI] [PubMed] [Google Scholar]
- 80.Yoshihara T., Hamamoto T., Munakata R., Tajiri R., Ohsumi M., Yokota S. Localization of cytosolic NADP-dependent isocitrate dehydrogenase in the peroxisomes of rat liver cells: biochemical and immunocytochemical studies. J. Histochem. Cytochem. 2001;49:1123–1131. doi: 10.1177/002215540104900906. [DOI] [PubMed] [Google Scholar]
- 81.Jo S. H., Son M. K., Koh H. J., Lee S. M., Song I. H., Kim Y. O., Lee Y. S., Jeong K. S., Kim W. B., Park J. W., et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 82.Lee S. H., Jo S. H., Lee S. M., Koh H. J., Song H., Park J. W., Lee W. H., Huh T. L. Role of NADP+-dependent isocitrate dehydrogenase (NADP+-ICDH) on cellular defence against oxidative injury by γ-rays. Int. J. Radiat. Biol. 2004;80:635–642. doi: 10.1080/09553000400007680. [DOI] [PubMed] [Google Scholar]
- 83.Koh H. J., Lee S. M., Son B. G., Lee S. H., Ryoo Z. Y., Chang K. T., Park J. W., Park D. C., Song B. J., Veech R. L., et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J. Biol. Chem. 2004;279:39968–39974. doi: 10.1074/jbc.M402260200. [DOI] [PubMed] [Google Scholar]
- 84.Kim H. J., Kang B. S., Park J. W. Cellular defense against heat shock-induced oxidative damage by mitochondrial NADP+ -dependent isocitrate dehydrogenase. Free Radical Res. 2005;39:441–448. doi: 10.1080/10715760500066265. [DOI] [PubMed] [Google Scholar]
- 85.Lee S. M., Koh H. J., Park D. C., Song B. J., Huh T. L., Park J. W. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radical Biol. Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 86.Bukato G., Kochan Z., Swierczynski J. Purification and properties of cytosolic and mitochondrial malic enzyme isolated from human brain. Int. J. Biochem. Cell Biol. 1995;27:47–54. doi: 10.1016/1357-2725(94)00057-3. [DOI] [PubMed] [Google Scholar]
- 87.Frenkel R. Bovine heart malic enzyme. I. Isolation and partial purification of a cytoplasmic and a mitochondrial enzyme. J. Biol. Chem. 1971;246:3069–3074. [PubMed] [Google Scholar]
- 88.Sanz N., Diez-Fernandez C., Valverde A. M., Lorenzo M., Benito M., Cascales M. Malic enzyme and glucose 6-phosphate dehydrogenase gene expression increases in rat liver cirrhogenesis. Br. J. Cancer. 1997;75:487–492. doi: 10.1038/bjc.1997.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diez-Fernandez C., Sanz N., Cascales M. Changes in glucose-6-phosphate dehydrogenase and malic enzyme gene expression in acute hepatic injury induced by thioacetamide. Biochem. Pharmacol. 1996;51:1159–1163. doi: 10.1016/0006-2952(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 90.McKenna M. C., Stevenson J. H., Huang X., Tildon J. T., Zielke C. L., Hopkins I. B. Mitochondrial malic enzyme activity is much higher in mitochondria from cortical synaptic terminals compared with mitochondria from primary cultures of cortical neurons or cerebellar granule cells. Neurochem. Int. 2000;36:451–459. doi: 10.1016/s0197-0186(99)00148-5. [DOI] [PubMed] [Google Scholar]
- 91.MacDonald M. J. Feasibility of a mitochondrial pyruvate malate shuttle in pancreatic islets. Further implication of cytosolic NADPH in insulin secretion. J. Biol. Chem. 1995;270:20051–20058. [PubMed] [Google Scholar]
- 92.Vasiliou V., Nebert D. W. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum. Genomics. 2005;2:138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pappa A., Sophos N. A., Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem. Biol. Interact. 2001;130–132:181–191. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 94.Pappa A., Chen C., Koutalos Y., Townsend A. J., Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radical Biol. Med. 2003;34:1178–1189. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 95.Fisher R. R., Kaplan N. O. Studies on the mitochondrial energy-linked pyridine nucleotide transhydrogenase. Biochemistry. 1973;12:1182–1188. doi: 10.1021/bi00730a026. [DOI] [PubMed] [Google Scholar]
- 96.Hoek J. B., Rydstrom J. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem. J. 1988;254:1–10. doi: 10.1042/bj2540001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rydstrom J. Mitochondrial NADPH, transhydrogenase and disease. Biochim. Biophys. Acta. 2006;1757:721–726. doi: 10.1016/j.bbabio.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 98.Gerdes S. Y., Scholle M. D., D'Souza M., Bernal A., Baev M. V., Farrell M., Kurnasov O. V., Daugherty M. D., Mseeh F., Polanuyer B. M., et al. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grose J. H., Joss L., Velick S. F., Roth J. R. Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7601–7606. doi: 10.1073/pnas.0602494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sassetti C. M., Boyd D. H., Rubin E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 102.Bieganowski P., Seidle H. F., Wojcik M., Brenner C. Synthetic lethal and biochemical analyses of NAD and NADH kinases in Saccharomyces cerevisiae establish separation of cellular functions. J. Biol. Chem. 2006;281:22439–22445. doi: 10.1074/jbc.M513919200. [DOI] [PubMed] [Google Scholar]
- 103.Shianna K. V., Marchuk D. A., Strand M. K. Genomic characterization of POS5, the Saccharomyces cerevisiae mitochondrial NADH kinase. Mitochondrion. 2006;6:94–101. doi: 10.1016/j.mito.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 104.Vestin R. Enzymic change from codehydrogenase I to codehydrogenase II. Naturwissenschaften. 1937;25:667–668. [Google Scholar]
- 105.von Euler H., Adler E. Enzymic intertransformation of codehydrogenase I and cohydrogenase II. Z. Physiol. Chem. 1938;252:41–48. [Google Scholar]
- 106.Kornberg A. Enzymatic synthesis of triphosphopyridine nucleotide. J. Biol. Chem. 1950;182:805–813. [Google Scholar]
- 107.Apps D. K. Kinetic Studies of pigeon liver NAD kinase. Eur. J. Biochem. 1968;5:444–450. doi: 10.1111/j.1432-1033.1968.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 108.Fernandes M. Properties of rat brain NAD-kinase. J. Neurochem. 1970;17:503–509. doi: 10.1111/j.1471-4159.1970.tb00528.x. [DOI] [PubMed] [Google Scholar]
- 109.Nemchinskaia V. L., Smirnova T. B. Isolation and properties of NAD kinase from rat liver. Biokhimiia. 1967;32:854–858. [PubMed] [Google Scholar]
- 110.Butler J. R., McGuinness E. T. Candida utilis NAD+ kinase: purification, properties and affinity gel studies. Int. J. Biochem. 1982;14:839–844. doi: 10.1016/0020-711x(82)90106-9. [DOI] [PubMed] [Google Scholar]
- 111.Tseng Y. M., Harris B. G., Jacobson M. K. Isolation and characterization of yeast nicotinamide adenine dinucleotide kinase. Biochim. Biophys. Acta. 1979;568:205–214. doi: 10.1016/0005-2744(79)90287-0. [DOI] [PubMed] [Google Scholar]
- 112.Yamamoto Y. NAD kinase in higher plants. Plant Physiol. 1966;41:523–528. doi: 10.1104/pp.41.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kawai S., Mori S., Mukai T., Suzuki S., Yamada T., Hashimoto W., Murata K. Inorganic polyphosphate/ATP-NAD kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 2000;276:57–63. doi: 10.1006/bbrc.2000.3433. [DOI] [PubMed] [Google Scholar]
- 114.Magni G., Orsomando G., Raffaelli N. Structural and functional properties of NAD kinase, a key enzyme in NADP biosynthesis. Mini Rev. Med. Chem. 2006;6:739–746. doi: 10.2174/138955706777698688. [DOI] [PubMed] [Google Scholar]
- 115.Labesse G., Douguet D., Assairi L., Gilles A. M. Diacylglyceride kinases, sphingosine kinases and NAD kinases: distant relatives of 6-phosphofructokinases. Trends Biochem. Sci. 2002;27:273–275. doi: 10.1016/s0968-0004(02)02093-5. [DOI] [PubMed] [Google Scholar]
- 116.Mori S., Yamasaki M., Maruyama Y., Momma K., Kawai S., Hashimoto W., Mikami B., Murata K. NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase–NAD complex. Biochem. Biophys. Res. Commun. 2005;327:500–508. doi: 10.1016/j.bbrc.2004.11.163. [DOI] [PubMed] [Google Scholar]
- 117.Raffaelli N., Finaurini L., Mazzola F., Pucci L., Sorci L., Amici A., Magni G. Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by site-directed mutagenesis. Biochemistry. 2004;43:7610–7617. doi: 10.1021/bi049650w. [DOI] [PubMed] [Google Scholar]
- 118.Garavaglia S., Raffaelli N., Finaurini L., Magni G., Rizzi M. A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis. J. Biol. Chem. 2004;279:40980–40986. doi: 10.1074/jbc.M406586200. [DOI] [PubMed] [Google Scholar]
- 119.Liu J., Lou Y., Yokota H., Adams P. D., Kim R., Kim S. H. Crystal structures of an NAD kinase from Archaeoglobus fulgidus in complex with ATP, NAD, or NADP. J. Mol. Biol. 2005;354:289–303. doi: 10.1016/j.jmb.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 120.Oganesyan V., Huang C., Adams P. D., Jancarik J., Yokota H. A., Kim R., Kim S. H. Structure of a NAD kinase from Thermotoga maritima at 2.3 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005;61:640–646. doi: 10.1107/S1744309105019780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 122.Turner W. L., Waller J. C., Vanderbeld B., Snedden W. A. Cloning and characterization of two NAD kinases from Arabidopsis. Identification of a calmodulin binding isoform. Plant Physiol. 2004;135:1243–1255. doi: 10.1104/pp.104.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kornberg A. Inorganic polyphosphate: a molecule of many functions. Prog. Mol. Subcell. Biol. 1999;23:1–18. doi: 10.1007/978-3-642-58444-2_1. [DOI] [PubMed] [Google Scholar]
- 124.Bark K., Kampfer P., Sponner A., Dott W. Polyphosphate-dependent enzymes in some coryneform bacteria isolated from sewage sludge. FEMS Microbiol. Lett. 1993;107:133–138. doi: 10.1111/j.1574-6968.1993.tb06019.x. [DOI] [PubMed] [Google Scholar]
- 125.Berrin J. G., Pierrugues O., Brutesco C., Alonso B., Montillet J. L., Roby D., Kazmaier M. Stress induces the expression of AtNADK-1, a gene encoding a NAD(H) kinase in Arabidopsis thaliana. Mol. Genet. Genomics. 2005;273:10–19. doi: 10.1007/s00438-005-1113-1. [DOI] [PubMed] [Google Scholar]
- 126.Mori S., Kawai S., Shi F., Mikami B., Murata K. Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution. J. Biol. Chem. 2005;280:24104–24112. doi: 10.1074/jbc.M502518200. [DOI] [PubMed] [Google Scholar]
- 127.Lerner F., Niere M., Ludwig A., Ziegler M. Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 2001;288:69–74. doi: 10.1006/bbrc.2001.5735. [DOI] [PubMed] [Google Scholar]
- 128.Kawai S., Mori S., Mukai T., Hashimoto W., Murata K. Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 2001;268:4359–4365. doi: 10.1046/j.1432-1327.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 129.Shi F., Kawai S., Mori S., Kono E., Murata K. Identification of ATP–NADH kinase isozymes and their contribution to supply of NADP(H) in Saccharomyces cerevisiae. FEBS J. 2005;272:3337–3349. doi: 10.1111/j.1742-4658.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 130.Garavaglia S., Galizzi A., Rizzi M. Allosteric regulation of Bacillus subtilis NAD kinase by quinolinic acid. J. Bacteriol. 2003;185:4844–4850. doi: 10.1128/JB.185.16.4844-4850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheng W., Roth J. R. Evidence for two NAD kinases in Salmonella typhimurium. J. Bacteriol. 1994;176:4260–4268. doi: 10.1128/jb.176.14.4260-4268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anderson J. M., Charbonneau H., Jones H. P., McCann R. O., Cormier M. J. Characterization of the plant nicotinamide adenine dinucleotide kinase activator protein and its identification as calmodulin. Biochemistry. 1980;19:3113–3120. doi: 10.1021/bi00554a043. [DOI] [PubMed] [Google Scholar]
- 133.Epel D., Patton C., Wallace R. W., Cheung W. Y. Calmodulin activates NAD kinase of sea urchin eggs: an early event of fertilization. Cell. 1981;23:543–549. doi: 10.1016/0092-8674(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 134.Williams M. B., Jones H. P. Calmodulin-dependent NAD kinase of human neutrophils. Arch. Biochem. Biophys. 1985;237:80–87. doi: 10.1016/0003-9861(85)90256-5. [DOI] [PubMed] [Google Scholar]
- 135.Anderson J. M., Cormier M. J. Calcium-dependent regulation of NAD kinase. Biochem. Biophys. Res. Commun. 1978;84:595–602. doi: 10.1016/0006-291x(78)90747-7. [DOI] [PubMed] [Google Scholar]
- 136.Muto S., Miyachi S. Properties of a protein activator of NAD kinase from plants. Plant Physiol. 1977;59:55–60. doi: 10.1104/pp.59.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stinemetz C. L., Kuzmanoff K. M., Evans M. L., Jarrett H. W. Correlation between calmodulin activity and gravitropic sensitivity in primary roots of maize. Plant Physiol. 1987;84:1337–1342. doi: 10.1104/pp.84.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dieter P., Marme D. A Ca2+, calmodulin-dependent NAD kinase from corn is located in the outer mitochondrial membrane. J. Biol. Chem. 1984;259:184–189. [PubMed] [Google Scholar]
- 139.Simon P., Bonzon M., Greppin H., Marme D. Subchloroplastic localization of NAD kinase activities: evidence for a Ca2+, calmodulin-dependent activity at the envelope and for a Ca2+, calmodulin-independent activity in the stroma of pea chloroplasts. FEBS Lett. 1984;167:332–338. [Google Scholar]
- 140.Simon P., Dieter P., Bonzon M., Greppin H., Marme D. Calmodulin-dependent and independent NAD kinase activities from cytoplasmic fractions of spinach (Spinacia oleracea L.) Plant Cell Rep. 1982;1:119–122. doi: 10.1007/BF00272368. [DOI] [PubMed] [Google Scholar]
- 141.Swezey R. R., Epel D. The in vivo rate of glucose-6-phosphate dehydrogenase activity in sea urchin eggs determined with a photolabile caged substrate. Dev. Biol. 1995;169:733–744. doi: 10.1006/dbio.1995.1183. [DOI] [PubMed] [Google Scholar]
- 142.Standart N. M., Bray S. J., George E. L., Hunt T., Ruderman J. V. The small subunit of ribonucleotide reductase is encoded by one of the most abundant translationally regulated maternal RNAs in clam and sea urchin eggs. J. Cell Biol. 1985;100:1968–1976. doi: 10.1083/jcb.100.6.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Akkaraju G. R., Hansen L. J., Jagus R. Increase in eukaryotic initiation factor 2B activity following fertilization reflects changes in redox potential. J. Biol. Chem. 1991;266:24451–24459. [PubMed] [Google Scholar]
- 144.Foerder C. A., Klebanoff S. J., Shapiro B. M. Hydrogen peroxide production, chemiluminescence, and the respiratory burst of fertilization: interrelated events in early sea urchin development. Proc. Natl. Acad. Sci. U.S.A. 1978;75:3183–3187. doi: 10.1073/pnas.75.7.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Heinecke J. W., Shapiro B. M. Respiratory burst oxidase of fertilization. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1259–1263. doi: 10.1073/pnas.86.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Foerder C. A., Shapiro B. M. Release of ovoperoxidase from sea urchin eggs hardens the fertilization membrane with tyrosine crosslinks. Proc. Natl. Acad. Sci. U.S.A. 1977;74:4214–4218. doi: 10.1073/pnas.74.10.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Perrild H., Loveridge N., Reader S. C., Robertson W. R. Acute stimulation of thyroidal NAD+ kinase, NADPH reoxidation, and peroxidase activities by physiological concentrations of thyroid stimulating hormone acting in vitro: a quantitative cytochemical study. Endocrinology. 1988;123:2499–2505. doi: 10.1210/endo-123-5-2499. [DOI] [PubMed] [Google Scholar]
- 148.Harding S. A., Oh S. H., Roberts D. M. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 1997;16:1137–1144. doi: 10.1093/emboj/16.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zielinski R. E. Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:697–725. doi: 10.1146/annurev.arplant.49.1.697. [DOI] [PubMed] [Google Scholar]
- 150.Butcher R. A., Schreiber S. L. Identification of Ald6p as the target of a class of small-molecule suppressors of FK506 and their use in network dissection. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7868–7873. doi: 10.1073/pnas.0402317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Outten C. E., Culotta V. C. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2003;22:2015–2024. doi: 10.1093/emboj/cdg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kawai S., Suzuki S., Mori S., Murata K. Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol. Lett. 2001;200:181–184. doi: 10.1111/j.1574-6968.2001.tb10712.x. [DOI] [PubMed] [Google Scholar]
- 153.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 154.Krems B., Charizanis C., Entian K. D. Mutants of Saccharomyces cerevisiae sensitive to oxidative and osmotic stress. Curr. Genet. 1995;27:427–434. doi: 10.1007/BF00311211. [DOI] [PubMed] [Google Scholar]
- 155.Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryotic Cell. 2003;2:809–820. doi: 10.1128/EC.2.4.809-820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Todisco S., Agrimi G., Castegna A., Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- 157.Douce R., Neuburger M. The uniqueness of plant mitochondria. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989;40:371–414. [Google Scholar]
- 158.Muto S., Miyachi S. Light-induced conversion of nicotinamide adenine dinucleotide to nicotinamide adenine dinucleotide phosphate in higher plant leaves. Plant Physiol. 1981;68:324–328. doi: 10.1104/pp.68.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chai M. F., Chen Q. J., An R., Chen Y. M., Chen J., Wang X. C. NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol. Biol. 2005;59:553–564. doi: 10.1007/s11103-005-6802-y. [DOI] [PubMed] [Google Scholar]
- 160.Turner W. L., Waller J. C., Snedden W. A. Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress) Biochem. J. 2005;385:217–223. doi: 10.1042/BJ20040292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Akella S. S., Harris C. Developmental ontogeny of NAD+ kinase in the rat conceptus. Toxicol. Appl. Pharmacol. 2001;170:124–129. doi: 10.1006/taap.2000.9093. [DOI] [PubMed] [Google Scholar]
- 162.Navas P., Minnifield N., Sun I., Morre D. J. NADP phosphatase as a marker in free-flow electrophoretic separations for cisternae of the Golgi apparatus midregion. Biochim. Biophys. Acta. 1986;881:1–9. doi: 10.1016/0304-4165(86)90089-9. [DOI] [PubMed] [Google Scholar]
- 163.Richter C. NADP+ phosphatase: a novel mitochondrial enzyme. Biochem. Biophys. Res. Commun. 1987;146:253–257. doi: 10.1016/0006-291x(87)90718-2. [DOI] [PubMed] [Google Scholar]
- 164.Gallais S., de Crescenzo M. A., Laval-Martin D. L. Evidence of active NADP(+) phosphatase in dormant seeds of Avena sativa L. J. Exp. Bot. 2000;51:1389–1394. [PubMed] [Google Scholar]
- 165.Reidl J., Schlor S., Kraiss A., Schmidt-Brauns J., Kemmer G., Soleva E. NADP and NAD utilization in Haemophilus influenzae. Mol. Microbiol. 2000;35:1573–1581. doi: 10.1046/j.1365-2958.2000.01829.x. [DOI] [PubMed] [Google Scholar]
- 166.Goto K., Laval-Martin D. L., Edmunds L. N., Jr Biochemical modeling of an autonomously oscillatory circadian clock in Euglena. Science. 1985;228:1284–1288. doi: 10.1126/science.2988128. [DOI] [PubMed] [Google Scholar]
- 167.Laval-Martin D. L., Carre I. A., Barbera S. J., Edmunds L. N., Jr Rhythmic changes in the activities of NAD kinase and NADP phosphatase in the achlorophyllous ZC mutant of Euglena gracilis Klebs (strain Z) Arch. Biochem. Biophys. 1990;276:433–441. doi: 10.1016/0003-9861(90)90742-h. [DOI] [PubMed] [Google Scholar]
- 168.Kawai S., Mori S., Mukai T., Murata K. Cytosolic NADP phosphatases I and II from Arthrobacter sp. strain KM: implication in regulation of NAD+/NADP+ balance. J. Basic Microbiol. 2004;44:185–196. doi: 10.1002/jobm.200310362. [DOI] [PubMed] [Google Scholar]
- 169.Kawai S., Fukuda C., Mukai T., Murata K. MJ0917 in archaeon Methanococcus jannaschii is a novel NADP phosphatase/NAD kinase. J. Biol. Chem. 2005;280:39200–39207. doi: 10.1074/jbc.M506426200. [DOI] [PubMed] [Google Scholar]
- 170.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]