Abstract
The diagnostic performances of the clinical case definition of influenza virus infection based on the combination of fever and cough and of two rapid influenza diagnostic tests, the Directigen Flu A+B test (Directigen; BD Diagnostic Systems, Sparks, Md.) and the QuickVue influenza test (QuickVue; Quidel, San Diego, Calif.), were compared to those of viral culture and an in-house reverse transcription (RT)-PCR during the 2000-2001 flu season. Two hundred consecutive nasopharyngeal aspirates were analyzed from 192 patients, including 122 adults and 70 children. Viral culture identified influenza virus A in 16 samples and influenza virus B in 55 samples, whereas RT-PCR identified influenza virus A in 21 samples and influenza virus B in 64 samples. When RT-PCR was used as the reference standard, the likelihood ratios for a positive test were 40.0 for Directigen, 8.6 for QuickVue, and 1.4 for the combination of fever and cough, whereas the likelihood ratios for a negative test were 0.22, 0.16, and 0.48, respectively. Our study suggests that (i) the poor specificity (35 to 58%) and the poor positive predictive value (41 to 60%) of the clinical case definition of influenza preclude its use for prediction of influenza virus infections during epidemics, especially when infection control decision making in the hospital setting is considered; (ii) Directigen has a higher diagnostic yield than QuickVue but is associated with a larger number of invalid results; (iii) the sensitivities of the rapid diagnostic tests are significantly lower with samples from adults than with samples from children, with the rates of false-negative results reaching up to 29%; and (iv) RT-PCR detects more cases of influenza than viral culture, and this greater accuracy makes it a more useful reference standard.
Influenza syndrome is defined by a rapid-onset systemic illness, with patients presenting with fever, chills, cough, myalgias, headache, and sore throat. During the peak of the influenza season, it is estimated that up to 70% of flu-like illnesses are caused by influenza viruses. However, patients infected with other respiratory viruses that circulate frequently in the community at the same time that influenza virus is circulating could present with similar symptoms.
Differentiation of influenza virus from the other respiratory viruses is of prime importance because the illness caused by influenza virus is associated with higher rates of morbidity and mortality, is potentially preventable by vaccination, and can now be managed with specific antivirals (3). It has been reported that, with the knowledge that influenza virus is circulating in a community, physicians can correctly diagnose influenza virus infections in 77 to 87% of their patients on the basis of the presence of fever and cough (3, 26, 46).
The recent advent of new treatments for influenza has stimulated the development of rapid diagnostic methods because these treatments have shown clinical benefit only when they are administered within 36 to 48 h of the appearance of symptoms (1, 41). The “gold standard” for the diagnosis of influenza is tissue culture isolation, which takes from 2 to 14 days. Detection of virus-infected cells in nasopharyngeal secretions by direct or indirect immunofluorescent staining is widely used but is quite technique and technician dependent and its completion still requires 2 h (23). Rapid diagnosis of influenza permits the institution of antiviral treatment, helps to control nosocomial transmission of the infection, and contributes to reductions in the cost and the length of the hospital stay. Rapid diagnosis has previously been shown to be cost-effective in a pediatric hospital (29, 43) and useful for controlling influenza epidemics in geriatric institutions (8, 24).
At least five different rapid enzyme-linked immunosorbent assay kits are available for the diagnosis of influenza: Directigen Flu A and Directigen Flu A+B (the latter of which is referred to herein as Directigen; Becton Dickinson Diagnostic Systems, Sparks, Md.), QuickVue influenza test (referred to herein as QuickVue; Quidel, San Diego, Calif.), Flu OIA (Bio Star, Inc., Boulder, Colo.,) and Zstat (ZymeTx, Inc., Oklahoma City, Okla.). Most of them have been compared with culture (8, 12, 20, 23, 24, 29, 33, 34, 36, 39, 44, 45) or culture and reverse transcription (RT)-PCR (2, 18, 21, 22, 25), but in some studies RT-PCR has occasionally been used to clarify the results for samples with discrepant results (6, 11, 28). This study aimed to evaluate the accuracy of the clinical case definition of influenza for prediction of influenza virus infections during epidemics and to compare the diagnostic performances of two new enzyme-linked immunosorbent assays, Directigen and QuickVue, to those of viral culture and an in-house RT-PCR.
MATERIALS AND METHODS
Clinical specimens.
The first 200 nasopharyngeal aspirates submitted to our hospital microbiology laboratory for testing for influenza virus during the 2000-2001 flu epidemic were prospectively included in the study. Due to their costs, diagnostic tests for influenza were recommended only for hospitalized patients and outpatients at high risk for cardiac or pulmonary complications following influenza. Age, sex, and whether the patient was hospitalized were recorded for each patient with clinically suspected influenza. The patients' charts were retrospectively reviewed for the presence of fever (≥38°C) and cough. A nasopharyngeal aspirate of 2 ml was obtained by a respiratory therapist by using a small flexible tube attached to a sterile mucus trap. If no secretions were obtained, 2 ml of sterile saline was introduced and reaspirated. The samples were sent to the laboratory, kept at 4°C, and processed within 12 h of receipt. Mucoid specimens were diluted 1:4 in saline; the samples were vortexed and then split into four aliquots for testing by viral culture, Directigen, QuickVue, and RT-PCR.
Viral culture.
An aliquot of 200 μl was inoculated into two wells each of Madin-Darby canine kidney (MDCK) cells and primary rhesus monkey kidney cells (BioWhittaker) seeded in 24-well plates. The plates were examined every other day for the presence of a cytopathic effect. Hemagglutination assay was performed with the monkey cells at 7 days of incubation, and hemadsorption was performed with MDCK cells on days 7 and 14 of incubation. Respiratory viruses isolated in culture were identified by indirect immunofluorescence (VRK Bartels).
Directigen.
Directigen is a membrane-based enzyme immunoassay which differentiates between influenza viruses A and B. The test was performed with fresh clinical specimens according to the instructions of the manufacturer. In brief, 200 μl of each nasopharyngeal aspirate was gently mixed with 8 drops (approximately 120 μl) of extraction buffer in the tube provided with the assay kit. Four drops (approximately 60 μl) of the specimen extract was then added to each well of the test device. Subsequently, specific conjugate, washing buffer, and substrate solutions were added within a 10-min period. The results were read at 5 min, the stop solution was added, and the test result was read again. The control dot needed to be visible (unless it was obscured by an intense purple triangle) for a valid test, and if the dot was absent, the result was regarded as indeterminate. A purple triangle was required for a positive result.
QuickVue. QuickVue is a lateral-flow immunoassay which detects both influenza virus A and influenza virus B but does not differentiate between the two viruses. A technician who was blinded to the Directigen results performed QuickVue within 7 days of receipt of the sample (the manufacturer recommends testing within 1 h of sampling) using 200-μl aliquots of samples kept at 4°C. QuickVue first involves the extraction of influenza virus A and B antigens by detergents. The patient specimen is placed in an extraction reagent tube, during which time the virus particles in the specimen are disrupted, exposing internal viral nucleoproteins. After extraction, a test strip is placed in the extraction reagent tube, where nucleoproteins in the specimen react with the lyophilized buffer and mouse monoclonal anti-influenza virus A and anti-influenza virus B antibodies contained in the test strip. If the extracted specimen contains influenza virus antigens, a pink or red test line along with a blue control line appears on the test strip, indicating a positive result. If influenza virus A or B antigens are not present or are present at very low levels, only a blue control line appears. The test is read after 10 min.
RT-PCR.
Primers targeting the matrix protein gene were chosen after the most conserved regions were identified in GenBank and by consideration of previously published primers (7, 9, 10, 13, 15, 18, 37, 40, 42, 47) (Table 1). The forward and reverse primers used for influenza virus A detection were described by Cooper and Subbarao (10) and Zhang and Evans (47), respectively. The forward primer used for influenza virus B detection was previously published by Zhang and Evans (47), and the reverse primer was designed by our team.
TABLE 1.
Primers used in RT-PCR for detection of the influenza virus A and B matrix genes
| Influenza virus type and primers | Sequence | 5′ position (bp) | Product size (bp) | GenBank accession no. |
|---|---|---|---|---|
| Influenza virus A | ||||
| AM149 | 5′-CTCATGGAATGGCTAAAGACA-3′ | 149 | 352 | AF258522 |
| AM501R | 5′-TGCTGGGAGTCAGCAATCTG-3′ | 501 | VO1099 | |
| Influenza virus B | ||||
| BM26 | 5′-TGTCGCTGTTTGGAGACACA-3′ | 26 | 444 | AF100392 |
| BM470R | 5′-TGTGATGCTTGTTTCTCGCA-3′ | 470 | AF100392 |
Samples of 100 μl previously frozen at −70°C were treated with sputolysin (1:1) and pelleted. RNA was extracted from the pellet by using a monophasic solution of phenol and guanidinium isothiocyanate (Trizol LS reagent; Gibco BRL) and resuspended in 30 μl of TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). cDNA synthesis was accomplished with the QIAGEN OneStep RT-PCR kit, which uses both reverse transcriptase and Taq DNA polymerase. Ten microliters of the RNA samples or 1 μl of a positive control and 9 μl of water were used in a final reaction volume of 25 μl. All four primers (0.6 μM each) were included in the mix. cDNA was amplified by 50 cycles of PCR with 30-s steps at 94, 55, and 72°C. Positive controls (influenza virus A/Texas/36/91 H1N1 and influenza virus B Beijing/185/93) and a negative control (water for resuspension of the samples) were included in each series. Following electrophoresis on 2% agarose gels, bands were detected by ethidium bromide staining. The two amplicons of influenza viruses A and B were easily distinguished by their molecular weights of 352 and 444 bp, respectively. This RT-PCR protocol was validated by testing with three patient samples confirmed by Health Canada to be similar to A/Texas/36/91 (H1N1), A/Wuhan/359/95 (H3N2), and B/Beijing/184/93, respectively, and two Health Canada reference samples, A/New Caledonia/20/99 (H1N1) and A/Panama/2007/99 (H3N2). The protocol was subsequently confirmed with a quality control panel from Health Canada, including 8 positive samples (including the same reference strains described above as well as B/Victoria/504/00) and 2 negative samples, as well as with 24 patient samples that had been tested by culture and Directigen. Initial studies involved the use of a nested PCR strategy with 40 cycles and then 25 cycles of PCR. Subsequently, we determined that all samples found to be positive by nested PCR were positive by use of 50 PCR cycles and one primer pair specific for influenza virus A and another primer pair specific for influenza virus B. No spurious bands except for a low-molecular-weight primer-dimer band, which was present in all samples, were visible for negative samples, even after 50 cycles of PCR (Fig. 1).
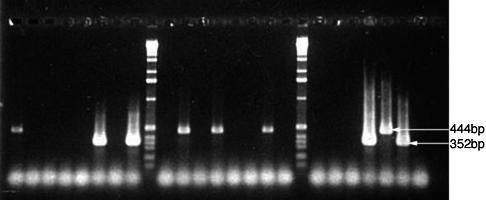
FIG. 1.
Detection of influenza virus RNA by RT-PCR. RNA was extracted from clinical samples or viral cultures by using Trizol before 50 cycles of amplification by RT-PCR. Amplicons were separated on 2% agarose gels and then stained with ethidium bromide. The 444-bp influenza virus B-specific and the 352-bp influenza virus A-specific amplicons produced from the controls are identified by arrows in the 24th and 25th lanes, respectively. A negative control was included in the 26th lane. Molecular weight markers were run in the 9th and 18th lanes. The results for clinical samples containing influenza virus B (1st, 11th, 13th, and 16th lanes) or influenza virus A (6th, 8th, and 23rd lanes) are also visible.
Statistical analyses.
We compared the performances of the clinical case definition (fever ≥38°C and cough), Directigen, and QuickVue for the diagnosis of influenza by using either viral culture or RT-PCR as the reference standard. We calculated the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), likelihood ratio (LR) for a positive test (LRP), and LR for a negative test (LRN) from two-by-two contingency tables for each test and case definition. All invalid results for rapid tests and contaminated cultures were excluded from the analyses.
LRP is given by sensitivity/(1 − specificity); conversely, LRN is given by (1− sensitivity)/specificity (38). By algebraically combining sensitivity and specificity, LRs describe more than the independent values themselves (38); they indicate by how much a given diagnostic test result will raise or lower the pretest probability of the target disorder (19). An LR of 1 means that the posttest probability is exactly the same as the pretest probability. LRPs of >1 increase the probability that the target disorder is present, and the higher the LR is, the greater this increase is. Conversely, LRNs of <1 decrease the probability of the target disorder, and the smaller the LR is, the greater the decrease in probability and the smaller its final value are. LRPs of >10 or LRNs of <0.1 generate large and often conclusive changes from pretest probability to posttest probability, LRPs of 5 to 10 and LRNs of 0.1 to 0.2 generate moderate shifts in pretest probability to posttest probability, LRPs of 2 to 5 and LRNs of 0.2 to 0.5 generate small (but sometimes important) changes in probability, and LRPs of 1 to 2 and LRNs of 0.5 to 1 alter probability to a small (and rarely important) degree (19).
Since the LR is increasingly considered the best index with which to evaluate a test or predictive method, the performances of the different tests were evaluated by comparing their LRs qualitatively and quantitatively (95% confidence intervals [CIs]). Of note, the comparison of the 95% CIs did not adjust for the fact that the tests were performed with samples from the same patients and therefore might have missed a statistical difference if one existed (type II error).
RESULTS
Clinical data.
The first case of influenza confirmed by viral culture in our laboratory occurred on 31 December 2000. The nasopharyngeal aspirates included in our study were obtained between 14 January and 13 February 2001. The population studied comprised 70 children and 122 adults aged from 1 day to 98 years (median age, 39 years). A total of 141 patients were hospitalized. Clinical data regarding the presence of fever and cough were available for 169 patients, of whom 128 were hospitalized. Most of the 31 patients for whom clinical data were not available had negative results for influenza virus infection by culture (77%) or RT-PCR (71%).
Comparison of rapid test and RT-PCR results to viral culture results.
In total, 71 (36%) of the 200 samples were positive for influenza virus by cell culture: 16 (including 9 children) for influenza virus A and 55 (including 27 children) for influenza virus B. Two specimens were positive for respiratory syncytial virus and one specimen each was positive for adenovirus, parainfluenza virus type 2, parainfluenza virus type 3, and herpes simplex virus type 1. One culture was contaminated, and 122 samples were negative. One sample showed a cytopathic effect on MDCK cells, but the presence of influenza virus could not be confirmed by immunofluorescence. Since the sample was positive for influenza virus B by Directigen and RT-PCR and positive by QuickVue, the culture was considered positive for influenza virus B.
Directigen detected influenza virus A in 13 samples confirmed to be positive by viral culture and influenza virus B in 44 samples confirmed to be positive by viral culture but did not detect virus in 9 specimens positive by culture (2 were positive for influenza virus A and 7 were positive for influenza virus B). It gave indeterminate results for 16 samples (8%), of which 1 was positive for influenza virus A, 4 were positive for influenza virus B, 1 was positive for parainfluenza virus type 2, and 1 was positive for parainfluenza virus type 3 by viral culture. Among the samples negative by viral culture, 7 samples were positive by Directigen (4 were positive for influenza virus A and 3 were positive for influenza virus B). Finally, all specimens positive for other respiratory viruses by viral culture were negative by Directigen.
QuickVue detected influenza virus A in 14 samples confirmed to be positive by viral culture and influenza virus B in 50 samples confirmed to be positive by viral culture but did not detect virus in 6 specimens positive by culture (1 was positive for influenza virus A and 5 were positive for influenza virus B). It gave only one indeterminate result for a sample positive for influenza virus A by viral culture. Among the samples negative by viral culture, 17 specimens were positive by QuickVue. Finally, 1 sample positive for respiratory syncytial virus by viral culture also tested positive for influenza virus by QuickVue.
All samples that were positive for influenza virus by culture except for one that was positive for influenza virus B also gave positive results by RT-PCR. The culture-positive, PCR-negative sample was also negative by both rapid tests. Of the additional 14 positive samples by RT-PCR (9 influenza virus B-positive samples, including 3 from children, and 5 influenza virus A-positive samples, including 3 from children) that gave negative results by viral culture, 5 (from 5 of the 6 children) were also positive by Directigen and 7 (including the 6 samples from the children) were also positive by QuickVue. All samples positive for viruses other than influenza virus gave negative results by RT-PCR. In order to control for false-positive and false-negative results, a second RT-PCR was performed with a new RNA extract for all RT-PCR-positive and culture-negative samples and RT-PCR-negative and culture-positive samples. The result of the second RT-PCR was always identical to the result of the initial RT-PCR.
Comparison of rapid test results to RT-PCR results.
Eighty-five (43%) of the 200 samples processed for RT-PCR were positive for influenza virus: 21 for influenza virus A and 64 for influenza virus B. Directigen detected virus in 63 specimens confirmed to be positive by RT-PCR (16 were positive for influenza virus A and 47 were positive for influenza virus B) but did not detect virus in 16 samples with positive RT-PCR results (3 of these were positive for influenza virus A and 13 were positive for influenza virus B). Among the 16 specimens with indeterminate results by Directigen, 6 were positive by RT-PCR (2 were positive for influenza virus A and 4 were positive for influenza virus B). Two samples positive by Directigen (one was positive for influenza virus A and one was positive for influenza virus B) gave negative results by RT-PCR.
QuickVue detected influenza virus A in 18 samples and influenza virus B in 54 samples found to be positive by RT-PCR but did not detect virus in 12 samples positive by RT-PCR (2 were positive for influenza virus A and 10 were positive for influenza virus B). One sample positive for influenza virus A by RT-PCR gave an invalid result by QuickVue. Eleven samples (6%) were positive by QuickVue but negative by RT-PCR.
Comparison of performances of clinical definition and rapid tests to those of viral culture and RT-PCR.
Table 2 compares the performances of the clinical definition of influenza (fever and cough), Directigen, and QuickVue to those of viral culture and RT-PCR with samples from all patients, hospitalized patients, adults, and children ≤5 years old.
TABLE 2.
Comparison of performances of clinical definition of influenza (fever and cough), Directigen, and QuickVue to those of viral culture and RT-PCR
| Patient group | Comparison to viral culture
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fever and cougha
|
Directigenb
|
||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LRP (95% CI) | LRN (95% CI) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LRP (95% CI) | LRN (95% CI) | ||||||||||
| All patients | 86 | 42 | 48 | 83 | 1.5 (0.8-1.8) | 0.33 (0.17-0.63) | 86 | 94 | 89 | 92 | 14.3 (6.9-29.6) | 0.15 (0.08-0.28) | |||||||||
| Hospitalized patients | 81 | 42 | 42 | 81 | 1.4 (0.9-1.8) | 0.45 (0.24-0.88) | 88 | 93 | 85 | 94 | 12.6 (5.8-27.4) | 0.13 (0.06-0.30) | |||||||||
| Adults | 94 | 37 | 41 | 93 | 1.5 (1.2-1.8) | 0.16 (0.04-0.63) | 83 | 97 | 94 | 92 | 27.7 (7.0-109.2) | 0.18 (0.09-0.37) | |||||||||
| Children ≤5 yr of age | 63 | 54 | 48 | 68 | 1.4 (0.8-2.3) | 0.69 (0.35-1.36) | 95 | 88 | 82 | 97 | 7.9 (3.1-19.9) | 0.06 (0.01-0.41) | |||||||||
| Comparison to viral culture
|
Comparison to RT-PCR
|
||||||||||||||||||||
| QuickVuec
|
Fever and coughd
|
||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LRP (95% CI) | LRN (95% CI) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LRP (95% CI) | LRN (95% CI) | ||||||||||
| 91 | 86 | 78 | 95 | 6.5 (5.3-8.0) | 0.10 (0.05-0.22) | 80 | 42 | 53 | 72 | 1.4 (1.1-1.7) | 0.48 (0.29-0.80) | ||||||||||
| 91 | 85 | 74 | 95 | 6.1 (3.7-10.0) | 0.11 (0.04-0.26) | 76 | 41 | 45 | 73 | 1.3 (1.0-1.6) | 0.58 (0.33-1.02) | ||||||||||
| 86 | 92 | 82 | 94 | 10.8 (4.7-19.6) | 0.15 (0.07-0.34) | 83 | 35 | 45 | 76 | 1.3 (1.0-1.6) | 0.26 (0.12-0.55) | ||||||||||
| 95 | 71 | 64 | 96 | 3.3 (1.9-5.6) | 0.07 (0.01-0.48) | 65 | 58 | 60 | 64 | 1.6 (0.9-2.7) | 0.60 (0.31-1.15) | ||||||||||
n = 168, 127, 102, and 47 patients for the four patient groups, respectively.
n = 183, 128, 110, and 53 patients for the four patient groups, respectively.
n = 198, 139, 121, and 52 patients for the four patient groups, respectively.
n = 169, 128, 103, and 47 patients for the four patient groups, respectively.
n = 184, 129, 111, and 53 patients for the four patient groups, respectively.
n = 199, 141, 122, and 54 patients for the four patient groups, respectively.
For all patients, the combination of fever ≥38°C and cough for the diagnosis of influenza had a moderate to high sensitivity (80 to 86%) when either RT-PCR or viral culture was used as the gold standard. The sensitivity was much lower with samples from children ≤5 years old (63 to 65%) and individuals ≥65 years old (58 to 77%; data not shown). However, the case definition was associated with a low specificity (35 to 58%) and a low PPV (41 to 60%) with samples from all categories of patients, regardless of the reference standard used. The LRPs and LRNs varied between 1.3 and 1.6 and between 0.16 and 0.69, respectively, indicating that the presence or absence of fever and cough changed the pretest probability of influenza minimally. In general, the LRPs for the case definition were significantly lower than the LRPs for Directigen (7.9 to 41.5) and QuickVue (3.3 to 10.9). Similarly, the LRNs for the case definition were generally higher than the LRNs for either Directigen or QuickVue, although the differences were not statistically significant.
The sensitivities of Directigen were high with samples from children ≤5 years old (95%) but moderate with samples from adults (71 to 83%) and hospitalized patients (83 to 88%). The test was highly specific with samples from all categories of patients, especially when RT-PCR was used as the reference standard (97 to 98%). Directigen had high PPVs with samples from all categories of patients but moderate NPVs, particularly with samples from adults. The LRPs were very high, most notably when RT-PCR was used as the gold standard (31.7 to 41.5), indicating that a positive test result had a major impact on the probability of influenza. The LRNs varied between 0.05 and 0.27, yielding large to moderate shifts in the probability of influenza when the test was negative; a negative test had the highest impact for children.
The sensitivity of QuickVue was similar to that of Directigen, with up to 24% of false-negative results occurring among adults. However, the test was slightly less specific than Directigen, especially among samples from children. The LRPs were relatively low with samples from children (3.3 to 5.1), indicating that a positive test yields small changes in the pretest probability of influenza. The LRP for QuickVue with samples from children appeared to be much lower than that of Directigen, although the difference was not statistically significant. The LRPs for samples from adults and hospitalized patients varied between 6.1 and 10.9, meaning that a positive test result yields moderate shifts in the pretest probability to the posttest probability of influenza. These values were much lower than those obtained by Directigen, but the difference did not reach statistical significance. The LRNs for QuickVue were comparable to the LRNs for Directigen with samples from all categories of patients, producing moderate to large changes in the posttest probability of influenza when the test was negative.
Both rapid tests were significantly more sensitive with samples from children than samples from adults, independently of the reference standard used (for Directigen and viral culture, 95% for samples from children versus 83% for samples from adults [P < 0.001]; for Directigen and RT-PCR, 95% for samples from children versus 71% for samples from adults [P < 0.001]; for QuickVue and viral culture, 95% for samples from children versus 86% for samples from adults [P < 0.001]; for QuickVue and RT-PCR, 96% for samples from children versus 76% for samples from adults [P < 0.001] [all P values were determined by the chi-square test]). Finally, the performances of Directigen and QuickVue were comparable when RT-PCR-positive and culture-negative samples were analyzed: both rapid tests identified virus in about 50% of positive samples (Directigen, 5 of 14 positive samples; QuickVue, 7 of 14 positive samples), while they detected virus in about 80% of the RT-PCR-positive and culture-positive specimens.
DISCUSSION
Rapid identification of influenza cases is essential, especially among hospitalized patients, not only for the initiation of antiviral treatment and avoidance of the unnecessary use of antibiotic therapy but also for the prevention of nosocomial transmission of influenza virus to patients at high risk of influenza-related complications. During the flu season, the combination of fever and cough was reported to be the best clinical definition of influenza, with sensitivities of 64 to 78%, specificities of 55 to 67%, and PPVs of 77 to 87% (3, 26, 46). Some investigators have suggested that physicians could use this association to rapidly enact specific antiviral treatment without using rapid testing for influenza virus antigen detection, which could be limited to patients with atypical presentations and cases occurring at the beginning of the flu season (3). However, most of those studies were done in the setting of randomized clinical trials evaluating the efficacies of neuraminidase inhibitors and might have overestimated the PPV of the clinical definition of influenza due to the selection of relatively young, healthy people with defined symptoms of influenza-like illness. Therefore, the pertinence of these results is questionable when they are applied to the target population served by practicing physicians in the community (patients with high-risk conditions who consult their physician for influenza-related complications) (16). Other, earlier studies reported that no clinical symptoms or signs are specific for influenza virus infections (5, 14, 27). Our study confirms the earlier findings and suggests that the poor specificity and PPV of the clinical case definition for influenza preclude its use for predicting influenza virus infections, especially when infection control decision making in the hospital setting is considered.
The 2000-2001 flu season allowed us to test the devices with both influenza virus A and influenza virus B, with an excess of influenza virus B. Only nasopharyngeal aspirates were considered because they usually provide the greatest sensitivity (11, 20, 34, 35, 45), although this may not be true for children (17). When the results of both rapid tests were compared with those of RT-PCR, both rapid tests appeared to be more sensitive for the detection of influenza virus A than for the detection of influenza virus B (for Directigen, 84 versus 78%; for QuickVue, 90 versus 84% [data not shown]). In our study, the performance of Directigen was equivalent or superior to what has previously been described for Directigen Flu A (12, 20, 23, 24, 28, 29, 33) and consistent with the performance of Directigen in two previous studies with nasopharyngeal aspirates from a pediatric population (6, 44). Our results are also consistent with those from previous studies of QuickVue, which reported sensitivities and specificities varying between 75 and 95% and between 76 and 93%, respectively (21, 31, 34, 45). Our use of QuickVue within 7 days of storage at 4°C rather than within 1 h, as suggested by the manufacturer, might have affected the performance of the test but reflects the limitations of a hospital laboratory to perform tests “stat.”
Globally, the LRPs for Directigen were two to three times higher than those for QuickVue when the results of viral culture were used and three to eight times higher than those for QuickVue when the results of RT-PCR were used. The LRNs for Directigen and QuickVue were comparable. We were unable to demonstrate a statistical difference between the LRPs for the two tests due to the low power of the study and the absence of statistical adjustment for the fact that both tests were performed with samples from the same patients. Nevertheless, we believe that the difference between the LRPs for the two tests is clinically significant and favors the use of Directigen as a rapid test for the diagnosis of influenza.
There were significantly more invalid results by Directigen than by QuickVue (16 and 1 samples, respectively). Most of these samples were very mucoid, even after they were diluted 1:4 in saline. Of note, the results for two samples which were culture positive for parainfluenza virus were invalid when they were tested by Directigen. More tests would be necessary to evaluate if this was only a coincidental finding or if the parainfluenza virus antigens inhibit the internal control reaction. No cross-reaction with other viruses was observed by Chan et al. (6) in their extensive evaluation. Finally, the interpretation of QuickVue results was often complicated by a difficulty in distinguishing negative samples from faintly positive samples, a problem also noted by Quach et al. (31). We have considered all faintly positive samples as true positive, as recommended by the manufacturer.
We, as others (2, 18, 21, 22), noted that RT-PCR identified more samples as positive than the other methods did. Viral culture may miss between 3 and 46% of influenza virus-positive samples, most often for patients whose clinical course of disease is more advanced (2, 4, 7, 9, 13, 18, 30, 32, 37, 42). Thus, some investigators now consider that RT-PCR should be the gold standard for the diagnosis of influenza (46). In the present study, 36% of clinical samples were influenza virus positive by culture; the diagnostic yield increased to 43% by RT-PCR. Specificity was also shown to be excellent with samples from children; among 43 samples from children, we found only 1 that had a positive RT-PCR result but that was not also positive by culture or by both rapid antigen tests. This RT-PCR-positive sample was positive by one of the two rapid tests and indeterminate by the other (and, thus, probably also had a true-positive result).
Although RT-PCR can detect nonviable organisms in minute quantities that might be present in respiratory secretions after symptoms resolve, our observations with samples from children do not show that this occurs frequently. Among the samples from adults, eight were RT-PCR positive but negative by culture and the rapid tests. When a second aliquot was extracted and analyzed by RT-PCR, these samples were again positive for the same influenza virus type. As it is not obvious why RT-PCR could be falsely positive with samples from adults but not samples from children, we presumed that these samples were falsely negative by the other tests due to the lower level of excretion of virus by adults compared to the level of excretion by children (23). Our study was limited by the absence of an internal control for RT-PCR to better identify possible inhibition or sample loss. However, among 71 samples cultured, only 1 culture-positive sample was not also RT-PCR positive, indicating inhibition or sample loss in, at most, only 1.4% of culture-positive samples.
Age was a critical factor in the diagnostic yields of the rapid tests. The sensitivities of both devices were dramatically higher with samples from children than with samples from adults, which probably reflects higher levels of viral excretion by children (23). The high false-negative rate of the rapid antigenic tests with samples from adults has important repercussions on the isolation and management of hospitalized adults with influenza, which argue in favor of the use of RT-PCR instead of viral culture as a rapid test for confirmation of influenza. In our hospital, we compared the efficacy of a surveillance program for influenza using Directigen and viral culture during the first year and Directigen and RT-PCR during the second year. The replacement of viral culture by RT-PCR as the reference standard allowed rapid identification of samples from patients with false-negative results by Directigen, which resulted in a 1.5-fold reduction in the rates of nosocomial influenza infection among adults by decreasing the median delay of isolation of adults from 60 to 24 h and by allowing prompt administration of antiviral prophylaxis to the room contacts of these patients (unpublished data).
In conclusion, our study suggests that the clinical case definition of influenza based on the presence of fever and cough is inaccurate for prediction of influenza virus infections, especially when infection control decision making in a hospital setting is considered. Directigen has a higher diagnostic yield than QuickVue because of its higher specificity and LRP and its ability to differentiate between influenza virus A and influenza virus B. The higher number of indeterminate results by Directigen than by QuickVue is, however, a drawback. The sensitivities of the rapid diagnostic tests are significantly lower with samples from adults than with those from children, with false-negative rates reaching up to 29%. The high false-negative rates of the rapid diagnostic tests with samples from adults have important implications for the management of hospitalized patients and on the infection control measures used for hospitalized patients. Finally, RT-PCR detects more influenza cases than viral culture, and this greater accuracy makes it a more useful reference standard.
Acknowledgments
This work was supported by Roche Canada. Some of the diagnostic kits were graciously offered by Becton Dickinson Biosciences and Quidel Corporation.
We thank Michel Lettre and team for outstanding technical work. We also thank Gina Bravo for help with the statistical analyses.
REFERENCES
- 1.Anonymous. 1998. Randomised trial of efficacy and safety of inhaled zanamivir in treatment of influenza A and B virus infections. Lancet 352:1877-1881. [PubMed] [Google Scholar]
- 2.Boivin, G., I. Hardy, and A. Kress. 2001. Evaluation of a rapid optical immunoassay for influenza viruses (FLU OIA test) in comparison with cell culture and reverse transcription-PCR. J. Clin. Microbiol. 39:730-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin, G., I. Hardy, G. Tellier, and J. Maziade. 2000. Predicting influenza infections during epidemics with use of a clinical case definition. Clin. Infect. Dis. 31:1166-1169. [DOI] [PubMed] [Google Scholar]
- 4.Carman, W. F., L. A. Wallace, J. Walker, S. McIntyre, A. Noone, P. Christie, J. Millar, and J. D. Douglas. 2000. Rapid virological surveillance of community influenza infection in general practice. BMJ 321:736-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrat, F., A. Tachet, C. Rouzioux, B. Housset, and A. J. Valleron. 1999. Evaluation of clinical case definitions of influenza: detailed investigation of patients during the 1995-1996 epidemic in France. Clin. Infect. Dis. 28:283-290. [DOI] [PubMed] [Google Scholar]
- 6.Chan, K. H., N. Maldeis, W. Pope, A. Yup, A. Ozinskas, J. Gill, W. H. Seto, K. F. Shortridge, and J. S. Peiris. 2002. Evaluation of the Directigen FluA+B test for rapid diagnosis of influenza virus type A and B infections. J. Clin. Microbiol. 40:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherian, T., L. Bobo, M. C. Steinhoff, R. A. Karron, and R. H. Yolken. 1994. Use of PCR-enzyme immunoassay for identification of influenza A virus matrix RNA in clinical samples negative for cultivable virus. J. Clin. Microbiol. 32:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church, D. L., H. D. Davies, C. Mitton, H. Semeniuk, M. Logue, C. Maxwell, and C. Donaldson. 2002. Clinical and economic evaluation of rapid influenza A virus testing in nursing homes in Calgary, Canada. Clin. Infect. Dis. 34:790-795. [DOI] [PubMed] [Google Scholar]
- 9.Claas, E. C., A. J. van Milaan, M. J. Sprenger, M. Ruiten-Stuiver, G. I. Arron, P. H. Rothbarth, and N. Masurel. 1993. Prospective application of reverse transcriptase polymerase chain reaction for diagnosing influenza infections in respiratory samples from a children's hospital. J. Clin. Microbiol. 31:2218-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, L. A., and K. Subbarao. 2000. A simple restriction fragment length polymorphism-based strategy that can distinguish the internal genes of human H1N1, H3N2, and H5N1 influenza A viruses. J. Clin. Microbiol. 38:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covalciuc, K. A., K. H. Webb, and C. A. Carlson. 1999. Comparison of four clinical specimen types for detection of influenza A and B viruses by optical immunoassay (FLU OIA test) and cell culture methods. J. Clin. Microbiol. 37:3971-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez, E. A., L. H. Taber, and R. B. Couch. 1993. Comparison of rapid diagnostic techniques for respiratory syncytial and influenza A virus respiratory infections in young children. J. Clin. Microbiol. 31:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, J. S., D. M. Fleming, and M. C. Zambon. 1997. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 35:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, M. T. 1977. The ′flu-like' illness. Practitioner 219:699-711. [PubMed] [Google Scholar]
- 15.Fouchier, R. A., T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hak, E., K. G. Moons, T. J. Verheij, and A. W. Hoes. 2001. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 161:1351-1352. [DOI] [PubMed] [Google Scholar]
- 17.Heikkinen, T., A. A. Salmi, and O. Ruuskanen. 2001. Comparative study of nasopharyngeal aspirate and nasal swab specimens for detection of influenza. BMJ 322:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann, B., C. Larsson, and B. W. Zweygberg. 2001. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription-PCR: comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA). J. Clin. Microbiol. 39:134-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaeschke, R., G. H. Guyatt, D. L. Sackett, et al. 1994. Users' guides to the medical literature. III. How to use an article about a diagnostic test. B. What are the results and will they help me in caring for my patients? JAMA 271:703-707. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser, L., M. S. Briones, and F. G. Hayden. 1999. Performance of virus isolation and Directigen Flu A to detect influenza A virus in experimental human infection. J. Clin. Virol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami, C., H. Shimizu, S. Watanabe, M. Saikusa, T. Munemura, K. Mitamura, N. Sugaya, and M. Imai. 2001. Evaluation of immunochromatography method for rapid detection of influenza A and B viruses. Kansenshogaku Zasshi. 75:792-799. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 22.Kehl, S. C., K. J. Henrickson, W. Hua, and J. Fan. 2001. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 39:1696-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landry, M. L., S. Cohen, and D. Ferguson. 2000. Impact of sample type on rapid detection of influenza virus A by cytospin-enhanced immunofluorescence and membrane enzyme-linked immunosorbent assay. J. Clin. Microbiol. 38:429-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonardi, G. P., H. Leib, G. S. Birkhead, C. Smith, P. Costello, and W. Conron. 1994. Comparison of rapid detection methods for influenza A virus and their value in health-care management of institutionalized geriatric patients. J. Clin. Microbiol. 32:70-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liolios, L., A. Jenney, D. Spelman, T. Kotsimbos, M. Catton, and S. Wesselingh. 2001. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J. Clin. Microbiol. 39:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monto, A. S., S. Gravenstein, M. Elliott, M. Colopy, and J. Schweinle. 2000. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 160:3243-3247. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson, K. G., J. Kent, V. Hammersley, and E. Cancio. 1997. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ 315:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noyola, D. E., B. Clark, F. T. O'Donnell, R. L. Atmar, J. Greer, and G. J. Demmler. 2000. Comparison of a new neuraminidase detection assay with an enzyme immunoassay, immunofluorescence, and culture for rapid detection of influenza A and B viruses in nasal wash specimens. J. Clin. Microbiol. 38:1161-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noyola, D. E., and G. J. Demmler. 2000. Effect of rapid diagnosis on management of influenza A infections. Pediatr. Infect. Dis. J. 19:303-307. [DOI] [PubMed] [Google Scholar]
- 30.Pregliasco, F., C. Mensi, L. Camorali, and G. Anselmi. 1998. Comparison of RT-PCR with other diagnostic assays for rapid detection of influenza viruses. J. Med. Virol. 56:168-173. [DOI] [PubMed] [Google Scholar]
- 31.Quach, C., D. Newby, G. Daoust, E. Rubin, and J. McDonald. 2002. QuickVue Influenza Test for rapid detection of influenza A and B viruses in a pediatric population. Clin. Diagn. Lab. Immunol. 9:925-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rebelo-de-Andrade, H., and M. C. Zambon. 2000. Different diagnostic methods for detection of influenza epidemics. Epidemiol. Infect. 124:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reina, J., M. Munar, and I. Blanco. 1996. Evaluation of a direct immunofluorescence assay, dot-blot enzyme immunoassay, and shell vial culture in the diagnosis of lower respiratory tract infections caused by influenza A virus. Diagn. Microbiol. Infect. Dis. 25:143-145. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, W. J., R. H. Schwartz, and M. M. Thorne. 2002. Evaluation of diagnostic tests for influenza in a pediatric practice. Pediatr. Infect. Dis. J. 21:193-196. [DOI] [PubMed] [Google Scholar]
- 35.Schmid, M. L., G. Kudesia, S. Wake, and R. C. Read. 1998. Prospective comparative study of culture specimens and methods in diagnosing influenza in adults. BMJ 316:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultze, D., Y. Thomas, and W. Wunderli. 2001. Evaluation of an optical immunoassay for the rapid detection of influenza A and B viral antigens. Eur. J. Clin. Microbiol. Infect. Dis. 20:280-283. [DOI] [PubMed] [Google Scholar]
- 37.Schweiger, B., I. Zadow, R. Heckler, H. Timm, and G. Pauli. 2000. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J. Clin. Microbiol. 38:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simel, D. L., G. P. Samsa, and D. B. Matchar. 1991. Likelihood ratios with confidence: sample size estimation for diagnostic test studies. J. Clin. Epidemiol. 44:763-770. [DOI] [PubMed] [Google Scholar]
- 39.St. George, K., N. M. Patel, R. A. Hartwig, D. R. Scholl, J. A. Jollick, Jr., L. M. Kauffmann, M. R. Evans, and C. R. Rinaldo, Jr. 2002. Rapid and sensitive detection of respiratory virus infections for directed antiviral treatment using R-Mix cultures. J. Clin. Virol. 24:107-115. [DOI] [PubMed] [Google Scholar]
- 40.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treanor, J. J., F. G. Hayden, P. S. Vrooman, R. Barbarash, R. Bettis, D. Riff, S. Singh, N. Kinnersley, P. Ward, R. G. Mills, et al. 2000. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. JAMA 283:1016-1024. [DOI] [PubMed] [Google Scholar]
- 42.van Elden, L. J., M. Nijhuis, P. Schipper, R. Schuurman, and A. M. van Loon. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo, P. C., S. S. Chiu, W. H. Seto, and M. Peiris. 1997. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J. Clin. Microbiol. 35:1579-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamazaki, M., K. Kimura, K. Mitamura, S. Watanabe, O. Komiyama, K. Yamamoto, M. Ichikawa, Y. Hashimoto, N. Hagiwara, T. Maezawa, M. Imai, and N. Sugaya. 2000. Clinical evaluation of rapid diagnostic kit detecting separately influenza A and B viruses. Kansenshogaku Zasshi 74:1032-1037. (In Japanese.) [DOI] [PubMed]
- 45.Yamazaki, M., K. Mitamura, K. Kimura, O. Komiyama, M. Nirasawa, K. Yamamoto, M. Ichikawa, K. Someya, T. Nakano, Y. Hashimoto, N. Hagiwara, T. Maezawa, S. Watanabe, H. Shimizu, and N. Sugaya. 2001. Clinical evaluation of an immunochromatography test for rapid diagnosis of influenza. Kansenshogaku Zasshi 75:1047-1053. (In Japanese.) [DOI] [PubMed]
- 46.Zambon, M., J. Hays, A. Webster, R. Newman, and O. Keene. 2001. Diagnosis of influenza in the community: relationship of clinical diagnosis to confirmed virological, serologic, or molecular detection of influenza. Arch. Intern. Med. 161:2116-2122. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, W. D., and D. H. Evans. 1991. Detection and identification of human influenza viruses by the polymerase chain reaction. J. Virol. Methods 33:165-189. [DOI] [PubMed] [Google Scholar]