Abstract
Logarithmic oscillations superimposed on the temporal relaxation patterns of complex systems are considered from the standpoint of their hierarchical origin. We propose that a closer examination of experimental data should reveal logarithmic oscillations in systems that are characterized by a hierarchical structure of their dynamical degrees of freedom. On that footing, a new methodology of data analysis is proposed that may prove important for the dynamics of protein folding and of conformational fluctuations in proteins in which the relevant time scales of the dynamical evolution underlying the relaxation kinetics can be deduced from these oscillations.
Keywords: dynamics of complex systems, protein folding, logarithmic temporal oscillations, hierarchically constrained dynamics, ultrametric spaces
Complex systems characterized by “structure with variations” (1) have gained increasing interest in physics, chemistry, and biology. The structural and dynamical features of complexity are ubiquitous in a variety of systems, such as proteins, biopolymers, glasses, organisms, or whole ecosystems. The underlying structural characteristics of complex systems involve a large variety of elementary units and/or strongly interacting elementary units (2). Complex systems exhibit a surprisingly rich temporal behavior being manifested in a nonpredictable time evolution (1, 2). This dynamics is usually connected with a multitude of time scales described through a relaxation time distribution (3). The relaxation behavior of a broad range of complex systems deviates from the simple exponential Debye relaxation f(t) = f0 exp{−t/τ). Instead, many data sets obtained from relaxation experiments can be described by the Kohlrausch-Williams-Watts stretched exponential function f(t) = f0exp{−(t/τ)β}, where 0 < β < 1, or they follow an asymptotic power-law [Nutting law (4)] f(t) = f0/(1 + t/τ)n with n > 0 (3, 5–7). Often, one also observes a transition from one behavior to the other that can be described by fractional relaxation equations (7, 8).
Usually, only effective time scales such as the constant τ in the power-law or stretched exponential law defined above can be deduced from the continuous relaxation time distribution, where possible individual contributions are smeared out (3, 4, 6–8). Additionally, novel information can be inferred from the presence of logarithmic oscillations in the time evolution of the underlying system, as this systematic behavior can be related to single exponential relaxations with correlated characteristic time scales. Thus, in their analysis of experimental patch-clamp data of fast chloride channels, Blatz and Magleby (9) found logarithmic oscillations, superimposed on a power-law trend. They demonstrated that a sum of single exponential functions describes well the distinct shoulders in their data, which is displayed on a double-logarithmic scale. This analysis shows the existence of distinct and discrete time scales governing the gating process. The data of Iben et al. (10) show oscillations around the asymptotic power-law trend, especially for the lower temperature range. Similar observations might be anticipated for various complex systems, such as proteins.
In this paper we address some mechanisms leading to logarithmic oscillations in the temporal relaxation patterns of some hierarchical model systems that have been shown to be relevant to the dynamics of proteins, or other complex systems. After the discussion of a Markoff chain model on the basis of which logarithmic oscillations emerge, we argue that measurements of this oscillatory time dependence will result in new information on the hierarchical dynamic structure in complex systems.
Hierarchically Constrained Dynamics
Nonexponential relaxation behavior can be related to internal barriers that are arranged in hierarchies. Such hierarchically constrained dynamics (11) depends on the time scale of the measurement, the hierarchies being typified by the constraints that the “spins” in a given tier n can only relax if a certain portion of spins in the above tier n − 1 have already relaxed. Depending on the specific laws imposed on this system, a power-law, or a stretched exponential relaxation pattern, can be derived. Alternatively, models with (hierarchical) energy barriers that are based on the properties of ultrametric spaces (12–16) are used for the description of relaxation phenomena in complex systems. In a treatment bridging constrained dynamics models and hierarchical barrier models, it was shown (17) that barrier models are as versatile as constraint models and that both are closely connected. This similarity, i.e., the same relaxation patterns resulting from the different physical models, was noted by Klafter and Shlesinger (6). It is common to all of these models that they exhibit a broad relaxation time spectrum (3) combining the cascades of discrete time scales that typify an individual barrier. A prototype example for such a hierarchical system is given by proteins.
The current conceptual basis for protein folding (18, 19) focuses on the description of the energy landscapes, their roughness, and the existence of funnels (18–30). A central issue in the structure-dynamics-function relations for protein folding pertains to the multiple time scales for the process. The kinetic inhomogeneity of the sequential domains is envisioned in terms of a “downhill folding” scenario (31–34) cascading through a hierarchy of protein substates. These substates, which have already been inferred from molecular dynamics simulations (35), can be described in terms of a Bethe tree (37), portrayed in Fig. 1. Similarly, inhomogeneous kinetics, originating from static disorder, was documented for other related processes in globular proteins, i.e., low-temperature CO or O2 recombination to myoglobin or hemoglobin (36). A sequential dynamics in the reordering of proteins after a “proteinquake,” following a hierarchy of protein substates that are arranged in a Bethe tree manner, as well as the existence of equilibrium fluctuations between protein states along the branches of this tree, was suggested by Frauenfelder and colleagues (38–42).
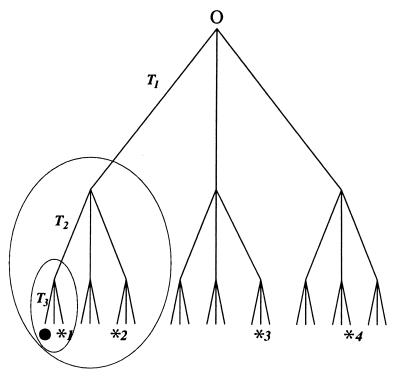
Figure 1.
Bethe tree and corresponding ultrametric space, with a branching factor 3. The tiers T1, T2, and T3 are shown. At each branching point, three possible paths on the down-cascading way are considered to be open. Only the terminal points on the bottom actually belong to the ultrametric space; the connecting lines represent (energy) barriers between the sites (see text). The dynamics on an ultrametric space infers that a relaxation of an excited state ● can be achieved by reaching one of the traps *, by a random walk on the branches. The ellipsoids denote the growing radius (ultrametric distance) embracing all sites the excitation can reach within the next step. Note that the sites *3 and *4 have the same ultrametric distance D from the original state ●.
A hierarchical space in the form of a Bethe tree, with a branching factor of three, is shown in Fig. 1. Note that only the end points on the bottom of the tree actually belong to the corresponding ultrametric space. The branches connecting these terminal points characterize (energy) barriers that have to be overcome to reach another point of the ultrametric space. Therefore, the system is hierarchical, and the barrier height d(x, y) between two sites x and y can be used as a metric on the ultrametric space, fulfilling the strong triangle inequality d(x, y) ≤ max{d(x, w), d(y, w)} for all sites w. Note the conditions d(•,*1) < d(•,*2) < d(•,*3) = d(•,*4).
In the case of the protein landscape picture developed by Wolynes and colleagues (27–34) for protein folding, the top point O in Fig. 1 represents the global state of the native folded protein, which acts as a funnel for a hierarchy of higher energy substates. For CO recombination in myoglobin (Mb) (38), the top point O refers to the bound MbCO configuration. This state is subdivided into several conformational substates that are further subdivided in a like manner. Equilibrium conformational fluctuations then take place on this Bethe tree structure (38). Similar tree structures involve the ultrametric topology encountered in the modeling of spin glasses (12, 14). There the different tiers of Fig. 1 resemble energy barriers arranged in a hierarchical way. A transition as displayed in Fig. 1 then means the following: Suppose the bullet represents a non-native state reached by the protein, for instance, through fluctuations, or by freezing it in. If *1, within the same tier, is another state, it can be reached by comparatively small energy fluctuations: i.e., by crossing a relatively low energy barrier. The activation has to be increasingly larger to achieve a transition for possible native states designated by *2, *3, etc.; i.e., it increases with growing ultrametric distance d.
A possible relaxation process on the ultrametric space is also sketched in Fig. 1. To relax, the excitation, symbolized by •, has to encounter a site that is occupied by one of the traps *. To find the traps designated *1, *2, etc., barriers of heights 1, 2, or 3, respectively, have to be overcome. This leads to the data obtained by Zumofen and colleagues for the kinetics in ultrametric spaces used for the modeling of reaction dynamics in complex systems (15, 16). If the bullet in Fig. 1 corresponds to a molecule of the species A, and the asterisks to either a molecule of the same species or to a substance B, reactions of the type A + A → 0 or A + B → 0, respectively, can be described.
Another system in which logarithmic oscillations were found is the Sierpiński gasket on which diffusion processes were studied by Klafter et al. (44). The structure-averaged autocorrelation function that is proportional to the density of the returning probability to the origin, P(0,t), shows oscillations that are the more distinct the higher the embedding dimension D is, thus mirroring the self-similar nature of the fractal Sierpiński gasket (44). This is connected with the random walker being confronted with geometric hierarchies, i.e., bottlenecks on a diversity of length scales, leading to temporal hierarchies of propagation. The typical time spent on a substructure of iteration N is related to the next iteration by τn+1/τn = D + 3 (44). A similar geometrical influence is found in branching data of the lung (45). Under some conditions, chaotic systems are characterized by the existence of islands in the corresponding phase space. These islands are hierarchically structured (46). Calculating the escape times from these archipelago-like, nested islands for the standard map, Zumofen et al. (47) relate the resulting logarithmic oscillations with the number of hierarchies of different island sizes. Recently, a hierarchical model of traders in a stock market has been shown to give rise to logarithmic oscillations, superimposed to the power-law trend, which are considered important in the stock market data before big crashes (48).
These examples for hierarchical topologies suggest that, also for proteins, logarithmic oscillations might be observed and thus information on relevant time scales deduced. In fact, one might suspect oscillations in the relaxation data from pressure-release experiments on myoglobin reported by Iben et al. (10) or in those for protein folding obtained by Gruebele and coworkers (43). In general, discrete hierarchies in systems give rise to oscillations in the decay patterns, expressed in the shoulders in a double-logarithmic plot. Each of these shoulders represents, in the course of time, the contribution of yet another relaxation channel in the decay cascades. Hierarchical dynamics can be caused by energetic barriers, by the underlying geometry, or by both.
Phenomenological Description of Logarithmic Oscillations
Let us now turn to a phenomenological description of logarithmic oscillations. Consider a system whose data roughly follow a power-law trend, the power-law being superimposed by oscillations, the period of which is logarithmic in time, i.e., the relaxation function follows the functional form
 |
1 |
where the “amplitude” Am(t) is, at least over some time range, periodic or quasiperiodic in the logarithm of time. If, for short times, Am(t) goes like Am(t) ∼ tμ, the relaxation function f(t) goes to a constant, as it should if the initial quantity described by f is normalized. Conversely, if, for long times, Am(t) decays exponentially, with a characteristic time τ*, the power-law window will be terminated for times longer than τ*. Unterminated power-law behavior with superimposed log-oscillations can be easily described by the amplitude
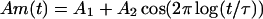 |
2 |
It is immediately clear that the oscillations are the more dominant the larger A2 is in comparison to A1. Note, however, that we require A1 > A2, and both are supposed to be positive real numbers.
To get a deeper insight into the coming into existence of such complicated relaxation behaviors, we consider the sum (7, 49, 50)
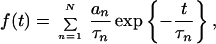 |
3 |
with N open relaxation channels. Through this representation, the time series is decomposed into N single Debye contributions of amplitudes an and characteristic times τn, which are a priori independent. Altogether, the system has 2N − 1 free parameters, as one parameter is determined either through the normalization condition or through the initial condition (50). In systems with hierarchical structures, typical for many complex systems, one finds that the individual constants are coupled through scaling relations: an = aαn−1 (0 < α < 1) and τn = τλn−1 (λ > 1); i.e., higher order relaxation modes decay slower, have a larger characteristic time constant, and contribute with a lower amplitude:
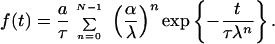 |
4 |
The scaling reduces the number of free parameters in the model to merely four. The analytical behavior of this hierarchically built sum, often referred to as Markoff chain (7, 50), can be studied through application of the Poisson summation formula (52)
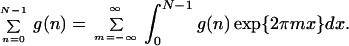 |
5 |
Introducing the latter relation into Eq. 4, and remembering the definition γ(a, x) ≡ ∫0x dy ya−1e−y of the incomplete γ-function, the following result can be inferred (refs. 7 and 50; M. O. Vlad and J. Ross, personal communication):
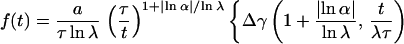 |
6 |
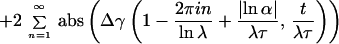 |
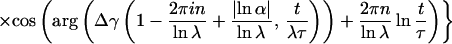 |
where Δγ(a, t/λτ) ≡ γ(a, t/λτ) − γ(a, t/λNτ). Thus, the power-law trend t−1−μ of index μ = |lnα|/lnλ is superimposed by the logarithmic oscillations given through the cosine terms. Consequently, the logarithmic oscillations emerge from Eq. 3 because of the scaling relations imposed on successive amplitudes an and characteristic times τn. The incomplete γ-functions let f(t) go to a constant value for t → 0+ and terminate the power-law region after the Nth shoulder, leading to the turnover to a fast exponential. On the footing of this Markoff chain model, the data of Blatz and Magleby (9) were investigated by Glöckle and Nonnenmacher (51).
Given the Markoff chain, Eq. 3, with the above scaling relations, the relaxation pattern exhibits a power-law with regularly spaced logarithmic oscillations. This situation is displayed in Fig. 2 A and B. Note that, due to Eq. 3 and the scaling relations for an and τn, the relaxation function f(t) is strictly monotonic, so that the corresponding relaxation time spectrum is positive. The populations themselves do not oscillate; the observed oscillations that are superimposed on the overall relaxation trend are attributable to the hierarchically ordered individual relaxations.
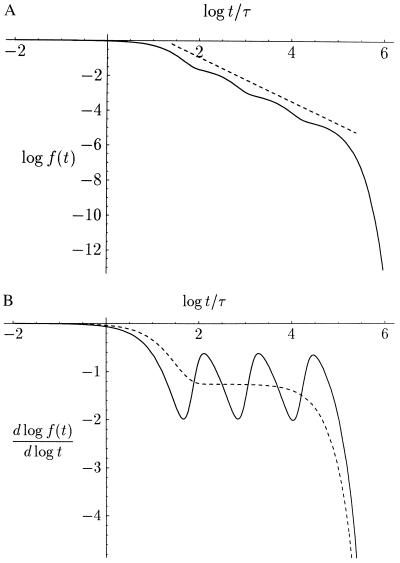
Figure 2.
(A) Decadic, double-logarithmic plot of the Markoff chain (Eq. 3) for n = 4, α = 0.5, and λ = 15. For short times t ≪ λτ, the power-law is terminated by the initial value; for long times t ≫ λNτ, a fast exponential decay takes over. The power-law trend has the slope 1 + μ = 1 + |lnα|/lnλ ≈ 1.26, according to τn+1/τn = λ. The dashed line corresponds to a power-law ∝ t−1.26. (B) Logarithmic derivative of the functions in A. The oscillations are clearly pronounced and wiggle around the dashed line representing the trend without the cosine function (second term in the braces of Eq. 6). The spacing of the humps is logλ ≈ 1.2 (τn+1/τn = λ). The plateau value is −1 −logα/logλ ≈ −1.26.
If the amplitudes still follow the scaling conditions an = aαn but the characteristic time scales grow like τn = τ0nκ, κ > 0, the overall relaxation trend follows a stretched exponential behavior (6). The situation is somewhat more intricate, as is revealed by Fig. 3 A and B. The nice symmetry of equidistant humps is broken. However, the logarithmic derivative still reveals the distinct shoulders.
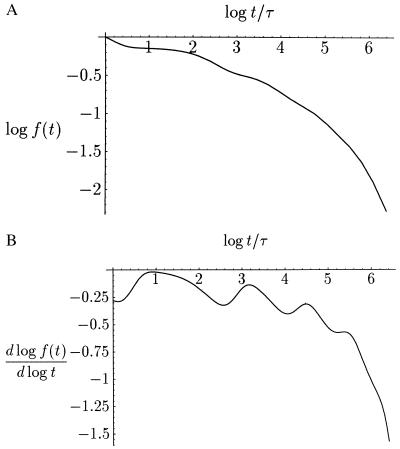
Figure 3.
(A) Stretched law with superimposed logarithmic oscillations in a decadic, double-logarithmic plot. We have plotted Σ062−n exp(−t/n8). (B) Logarithmic derivative of the plot in A. The irregular array of the humps is obvious, and the second and the sixth are smeared out with the preceding ones.
Scaling Conditions for Successive Time Scales
How does one detect logarithmic oscillations in the relaxation data? On first inspection, the measured data may not indicate any kind of oscillations, or it cannot be clearly distinguished from noise (43), even on a double-logarithmic plot. This is exemplified in the model calculations for a Markoff chain depicted in Fig. 2A. The log-oscillations can be considerably amplified by taking the logarithmic derivative of the function (Fig. 2B), with the oscillations being much more pronounced.
What information about the system can be obtained from this analysis? If the extended data analysis proposed above reveals a power-law or a stretched exponential trend in a given time window, and it is superimposed by log-oscillations, the system is (i) assembled in certain hierarchies: i.e., it possesses different domains in its dynamical response, and each domain is characterized by its own characteristic time; consequently, certain relations exist, connecting successive time scales: i.e., τn+1/τn ≈ const. for the power-law trend and τn+1/τn ≈ exp{const. × log([n + 1]/n)} for the stretched exponential trend. (ii) The individual amplitudes an and characteristic times τn are scaled in such a way that contributions with a larger characteristic time have a smaller amplitude. Consequently, only a comparatively small number of independent parameters exists for the characterization of this kind of relaxation dynamics of the complex system. Especially for the case of a power-law trend underlying the oscillations, the parameter λ can be deduced from the distance between successive humps. Either from the slope in Fig. 2A, or from the plateau value in Fig. 2B, one obtains the value for the power-law slope: i.e., 1+μ. Thus, one can infer α and can completely determine the relevant relaxation modes and their respective weights.
In conclusion, we have proposed a method of data analysis by help of which additional information on complex systems might be inferred from their dynamical response. This is important because the experimental techniques are more and more refined and open the window for even broader ranges of data. Thus, complex systems that are a compound of several tiers responsible for a multiple down-cascading relaxation are predicted to feature logarithmic oscillations. These oscillations, being of a large enough amplitude and periodicity that is comparable with the data window, can be extracted by plotting the logarithmic derivative of the data in respect to time. We believe that logarithmic oscillations are a ubiquitous feature for many systems in physics, chemistry, and biology and that their study, and hierarchical interpretation, make it possible to deduce relevant time scales for the relaxation dynamics of complex systems.
Acknowledgments
We are indebted to Prof. P. G. Wolynes for his perceptive comments on the manuscript. R.M. thanks Markus Porto and Israela Becker for technical assistance. Financial assistance from the German–Israeli Foundation (GIF) is gratefully acknowledged. R.M. is supported through an Amos de Shalit fellowship from the Minerva foundation.
References
- 1.Goldenfeld N, Kadanoff L P. Science. 1999;284:87–89. doi: 10.1126/science.284.5411.87. [DOI] [PubMed] [Google Scholar]
- 2.Guerra F, Peliti L, Vulpiani A. In: Measures of Complexity. Peliti L, Vulpiani A, editors. Berlin: Springer; 1988. [Google Scholar]
- 3.Metzler R, Klafter J, Jortner J, Volk M. Chem Phys Lett. 1998;293:477–484. [Google Scholar]
- 4.Reiner M. Rheologie in Elementarer Darstellung. Leipzig: VEB Fachbuchverlag; 1969. [Google Scholar]
- 5.Ngai K L, White C T. Phys Rev B. 1979;20:2475–2486. [Google Scholar]
- 6.Klafter J, Shlesinger M F. Proc Natl Acad Sci USA. 1986;83:848–852. doi: 10.1073/pnas.83.4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nonnenmacher T F, Metzler R. In: Applications of Fractional Calculus in Physics. Hilfer R, editor. Singapore: World Scientific; 1998. [Google Scholar]
- 8.Glöckle W G, Nonnenmacher T F. Rheol Acta. 1994;33:337–343. [Google Scholar]
- 9.Blatz A L, Magleby K L. J Physiol (London) 1986;378:141–174. doi: 10.1113/jphysiol.1986.sp016212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iben I E T, Braunstein D, Doster W, Frauenfelder H, Hong M K, Johnson J B, Luck S, Ormos P, Schulte A, Steinbach P J, et al. Phys Rev Lett. 1989;62:1916–1919. doi: 10.1103/PhysRevLett.62.1916. [DOI] [PubMed] [Google Scholar]
- 11.Palmer R G, Stein D L, Abrahams E, Anderson P W. Phys Rev Lett. 1984;53:958–961. [Google Scholar]
- 12.Mézard M, Parisi G, Sourlas N, Toulouse G, Virsaoro M. Phys Rev Lett. 1984;52:1156–1159. [Google Scholar]
- 13.Hubermann B A, Kerszberg M. J Phys A. 1985;18:L331–L336. [Google Scholar]
- 14.Ogielski A T, Stein D L. Phys Rev Lett. 1985;55:1634–1637. doi: 10.1103/PhysRevLett.55.1634. [DOI] [PubMed] [Google Scholar]
- 15.Zumofen G, Blumen A, Klafter J. J Chem Phys. 1986;84:6679–6686. [Google Scholar]
- 16.Blumen A, Zumofen G, Klafter J. Ber Bunsenges Phys Chem. 1986;90:1048–1053. [Google Scholar]
- 17.Kumar D, Shenoy S R. Solid State Commun. 1986;57:927–931. [Google Scholar]
- 18.Creighton T F, editor. Protein Folding. New York: Freeman; 1992. [Google Scholar]
- 19.Onuchic J N, Luthey-Schulten Z, Wolynes P G. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 20.Sali A, Shakhnovich E I, Karplus M. Nature (London) 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 21.Zwanzig R, Szabo A, Bagchi B. Proc Natl Acad Sci USA. 1992;89:20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwanzig R. Proc Natl Acad Sci USA. 1995;92:9801–9804. doi: 10.1073/pnas.92.21.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryngelson J D, Wolynes P G. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shakhnovich E I, Gutin A M. Europhys Lett. 1989;9:569–574. [Google Scholar]
- 25.Frauenfelder H, Sligar S G, Wolynes P G. Science. 1991;254:159–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 26.Berry R S, Elmaci N, Rose J P, Vekhter B. Proc Natl Acad Sci USA. 1997;94:9520–9524. doi: 10.1073/pnas.94.18.9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panchenko A, Luthey-Schulten Z, Wolynes P G. Proc Natl Acad Sci USA. 1996;93:2008–2013. doi: 10.1073/pnas.93.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolynes P G. In: Physics of Biological Systems. Flyvbjerg H, Hertz J, Jensen M H, Mouritsen O G, Sneppen K, editors. Berlin: Springer; 1998. (1998). [Google Scholar]
- 29.Wang J, Plotkin S S, Wolynes P G. J Phys (Paris) 1997;7:395–421. [Google Scholar]
- 30.Plotkin S S, Wang J, Wolynes P G. J Chem Phys. 1997;106:2932–2948. [Google Scholar]
- 31.Bryngelson J D, Wolynes P G. J Phys Chem. 1989;93:6902–6915. [Google Scholar]
- 32.Nymeyer H, García A E, Onuchic J N. Proc Natl Acad Sci USA. 1998;95:5921–5928. doi: 10.1073/pnas.95.11.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skorobogatiy M, Guo H, Zukerman M. J Chem Phys. 1998;109:2528–2535. [Google Scholar]
- 34.Onuchic J N, Wolynes P G, Luthey-Schulten Z, Socci N D. Proc Natl Acad Sci USA. 1995;92:3626–3630. doi: 10.1073/pnas.92.8.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker O M, Karplus M. J Chem Phys. 1997;106:1495–1517. [Google Scholar]
- 36.Austin R H, Beeson K W, Eisenstein L, Frauenfelder H, Gunsalus I C. Biochemistry. 1975;14:5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- 37.Hughes B D. Random Walks and Random Environments. Vol. 1. Oxford: Oxford Univ. Press; 1995. [Google Scholar]
- 38.Ansari A, Berendzen J, Bowne S F, Frauenfelder H, Iben I E T, Sauke T B, Shyamsunder E, Young R D. Proc Natl Acad Sci USA. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frauenfelder H, Petsko G A, Tsernoglou D. Nature (London) 1979;280:558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 40.Frauenfelder H, Wolynes P G. Science. 1985;229:337–345. doi: 10.1126/science.4012322. [DOI] [PubMed] [Google Scholar]
- 41.Frauenfelder H, Wolynes P G. Phys Today. 1994;47:58–64. [Google Scholar]
- 42.Frauenfelder H. In: Physics of Biological Systems. Flyvbjerg H, Hertz J, Jensen M H, Mouritsen O G, Sneppen K, editors. Berlin: Springer; 1998. [Google Scholar]
- 43.Ervin, J., Sabelko, J. & Gruebele, M. (1999) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 44.Klafter J, Zumofen G, Blumen A. J Phys A. 1991;24:4835–4842. [Google Scholar]
- 45.Shlesinger M F, West B J. Phys Rev Lett. 1991;67:2106–2108. doi: 10.1103/PhysRevLett.67.2106. [DOI] [PubMed] [Google Scholar]
- 46.Reichl E. The Transition to Chaos. Berlin: Springer; 1992. [Google Scholar]
- 47.Zumofen G, Klafter J, Shlesinger M F. In: Anomalous Diffusion, From Basics to Applications. Pekalski A, Sznajd-Weron K, editors. Berlin: Springer; 1999. [Google Scholar]
- 48.Sornette D, Johansen A. Physica. 1998;261A:581–598. [Google Scholar]
- 49.Sakmann B, Neher E. Single-Channel Recording. New York: Plenum; 1983. [Google Scholar]
- 50.Nonnenmacher T F, Nonnenmacher D J F. Phys Lett A. 1989;140:323–326. [Google Scholar]
- 51.Glöckle W G, Nonnenmacher T F. In: Fractals in Biology and Medicine. Nonnenmacher T F, Losa G A, Weibl E R, editors. Basel: Birkhäuser; 1993. [Google Scholar]
- 52.Morse P M, Feshbach H. Methods of Theoretical Physics. New York: McGraw–Hill; 1953. [Google Scholar]