Abstract
The human neurodegenerative and cancer predisposition condition ataxia-telangiectasia is characterized at the cellular level by radiosensitivity, chromosomal instability, and impaired induction of ionizing radiation-induced cell cycle checkpoint controls. Recent work has revealed that the gene defective in ataxia-telangiectasia, termed ATM, encodes an ≈350-kDa polypeptide, ATM, that is a member of the phosphatidylinositol 3-kinase family. We show that ATM binds DNA and exploit this to purify ATM to near homogeneity. Atomic force microscopy reveals that ATM exists in two populations, with sizes consistent with monomeric and tetrameric states. Atomic force microscopy analyses also show that ATM binds preferentially to DNA ends. This property is similar to that displayed by the DNA-dependent protein kinase catalytic subunit, a phosphatidylinositol 3-kinase family member that functions in DNA damage detection in conjunction with the DNA end-binding protein Ku. Furthermore, purified ATM contains a kinase activity that phosphorylates serine-15 of p53 in a DNA-stimulated manner. These results provide a biochemical assay system for ATM, support genetic data indicating distinct roles for DNA-dependent protein kinase and ATM, and suggest how ATM may signal the presence of DNA damage to p53 and other downstream effectors.
Ataxia-telangiectasia (A-T) is a human autosomal recessive disorder present at an incidence of ≈1 in 100,000 in the population. A-T is characterized by several debilitating symptoms, including progressive cerebellar degeneration, oculocutaneous telangiectasia, growth retardation, immune deficiencies, and characteristics of premature aging (1–3). At the cellular level, A-T is characterized by chromosomal instability, radio-resistant DNA synthesis, and hypersensitivity to ionizing radiation (IR) and radiomimetic drugs. In addition, A-T cells are defective in the radiation-induced G1-S, S, and G2-M cell cycle checkpoints that are thought to arrest the cell cycle in response to DNA damage to allow repair of the genome before DNA replication or mitosis (3). This may in part reflect the fact that A-T cells exhibit deficient or severely delayed induction of p53 in response to IR (4–6). The protein defective in A-T therefore acts upstream of p53 in an IR-induced DNA damage-signaling pathway.
The gene mutated in A-T, termed ATM, has been mapped and its cDNA cloned (7, 8). Sequence analyses reveal that the ATM gene encodes an ≈350-kDa polypeptide that is a member of the phosphatidylinositol (PI) 3-kinase family of proteins by virtue of a kinase catalytic domain in its carboxyl-terminal region. Classical PI 3-kinases, such as PI 3-kinase itself, are involved in signal transduction and phosphorylate inositol lipids that act as intracellular second messengers (9). However, ATM bears most sequence similarity with a subset of the PI 3-kinase family that comprises proteins that, like ATM, are involved in cell cycle control and/or in the detection and signaling of DNA damage (10, 11). Included in this subgroup is the DNA-dependent protein kinase (DNA-PK) catalytic subunit (DNA-PKcs) (12), defects in which lead to IR hypersensitivity, deficient double strand break rejoining, and an inability to perform site-specific V(D)J recombination. Other members of the ATM sub-group of the PI 3-kinase family include Saccharomyces cerevisiae Tel1p and Mec1p, together with the Mec1p homologues of Schizosaccharomyces pombe (Rad3), Drosophila melanogaster (mei-41), and humans (ATR) (10, 11). As with ATM, defects in these proteins lead to genomic instability, hypersensitivity to DNA damaging agents, and defects in DNA damage-induced cell cycle checkpoint controls (1, 13).
Although genetic data indicate an involvement of ATM-like proteins in DNA damage recognition and its repair, the mechanisms by which they function are not well understood. DNA-PKcs, however, has been subjected to detailed biochemical investigations. This polypeptide associates with the heterodimeric DNA end-binding protein Ku to form a protein serine-threonine kinase whose activity is stimulated by DNA double-strand (ds) breaks or DNA ds-to-single-strand (ss) transitions (14, 15). Recently, it was shown that DNA-PKcs also can interact with DNA ends in the absence of Ku at low salt concentrations (16, 17). In light of its activation by DNA ends, it is believed that DNA-PK functions as a DNA damage sensor and potentiates DNA repair by triggering DNA damage signaling pathways and/or by modulating the activity of the DNA repair machinery (18–20).
In contrast to DNA-PK, little is known about the mechanism(s) by which ATM functions. However, recent studies have revealed that ATM is expressed ubiquitously and is localized predominantly to the cell nucleus (21–24). These properties, together with the sequence similarity between ATM and DNA-PKcs, suggest that ATM might function directly in the detection of DNA damage. To investigate this and other potential roles for ATM, we have purified it from human cell nuclear extracts and have examined its biochemical properties. In addition, we have used the atomic force microscope (AFM) to observe purified ATM in the presence or absence of DNA. These studies reveal direct binding of ATM to DNA and show that ATM binds preferentially to DNA ends. Furthermore, and consistent with recent reports (25–27), we demonstrate that purified ATM phosphorylates p53 on serine-15 and additionally reveal that this activity is stimulated by DNA.
MATERIALS AND METHODS
DNA Interaction Studies.
Oligonucleotides. One DNA strand containing a 5′ biotin group (indicated by a “B” below) was annealed with its complementary oligonucleotide and was bound to streptavidin-coated iron-oxide particles (Dynabeads, Dynal, Oslo). HeLa nuclear extract (50 μg) or ATM-enriched extract (50 μg; Q-Sepharose pool; see below) was incubated on ice for 30 min with the DNA-iron oxide particles. After washing with 5 × 0.5 ml of D* buffer (25 mM Hepes⋅KOH, pH 7.6/20% glycerol/2 mM MgCl2/0.2 mM EDTA/1 mM DTT/0.5 mM PMSF/1 mM Na metabisulfite) containing 50 mM KCl, protein was eluted with 500 mM KCl D* buffer or in gradual stepwise manner with KCl concentrations of 100, 250, and 500 mM in buffer D*. Fractions were analyzed for ATM by Western blotting using rabbit polyclonal antiserum raised against amino acid residues 1,980–2,337 of ATM (22). The oligonucleotides used: ds 15-mer, 5′ B-CCTGCCCTTGCCTGA-3′ and its complement; ds 25-mer, 5′ B-CCTGCCCTTGCCTGACGCTATTAGT-3′ and its complement; ds 50-mer, 5′ B-TTGTAAAACGACGGCCAGTGAATTCATCATCAATAATATACCTTATTTTG-3′ and its complement; and ss 50-mer, as ds 50-mer but without its complement.
ATM Purification.
All steps were performed at 4°C. HeLa nuclear extract (20 ml; Computer Cell Culture Centre, Mons, Belgium) was applied to a Q-Sepharose column (35 ml, 1.5 × 20 cm) equilibrated in D* buffer containing 50 mM KCl. After washing with 2-column vol of 50 mM KCl D*, protein was eluted with a continuous salt gradient of 50–500 mM KCl in D* buffer. ATM eluted between 160 and 200 mM KCl. Fractions containing ATM and devoid of DNA-PK (as judged by Western blot analysis) were pooled and, after diluting to 100 mM KCl in D* buffer, were loaded onto a heparin agarose column (1.5 × 6 cm) preequilibrated in 100 mM KCl D* buffer. The column was washed with 2-column vol of 100 mM KCl in buffer D* before eluting with a continuous gradient of 50–500 mM KCl in buffer D*. ATM was followed by Western blot analysis and eluted between 200 and 220 mM KCl. Peak fractions were pooled and dialyzed against 50 mM buffer D*. Peak ATM fractions then were incubated with gentle mixing for 1 h with 200 μg of biotinylated 50-bp ds DNA conjugated to streptavidin iron-oxide particles. Unbound protein was rebound to fresh DNA-iron oxide particles. Particles were collected via a magnet and were washed five times with 0.5 ml of 50 mM KCl D* buffer before eluting with 2 × 75 μl of 250 mM KCl buffer D* and then with 2 × 75 μl of 500 mM KCl buffer D*. Purified ATM was stored at −70°C.
Immunological Methods.
Western immunoblot analysis was performed as described (22). Anti-Sp1 and anti-human p53 (Do-1) antibodies were from Serotec and Santa Cruz Biotechnology, respectively. Antisera against the 70-kDa subunit of replication protein and RNA polymerase II were generous gifts from R. Fishel (Thomas Jefferson Univ., Philadelphia) and D. Reinberg (Howard Hughes Medical Institute, Piscataway, NJ), respectively. Phosphospecific antibodies were raised against an 11-residue phosphopeptide (corresponding to residues 10–20 of human p53 and containing a phosphoserine at residue 15) coupled to KLH (Monrovian Antibody, Brno, Czech Republic). Serum was shown to recognize full-length wild-type p53 phosphorylated in vitro with DNA-PK but not unphosphorylated protein or DNA-PK-phosphorylated p53 that contains a point mutation converting serine-15 to alanine (28). Immunoprecipitations were performed by incubating ATM preparations with 2.5 μl of either PBS or monoclonal antibodies raised against either ATM (a kind gift from Y. Shiloh, Sackler School of Medizine, Tel Aviv Univ., Ramat Aviv, Israel) (25) or poly (ADP-ribose) polymerase (29) in 150 mM KCl D* buffer containing 0.1% Nonidet P-40 for 1 h at 4°C. Immunoprecipitations then were added to the equivalent of 50 μl of goat anti-mouse IgG iron oxide beads (Dynal) and were incubated at 4°C for 1 h with gentle rocking. Beads were removed by using a magnet and 5 μl of each immunoprecipitation assayed for p53 kinase activity as described below.
Atomic Force Microscopy.
Reactions were performed in 50 mM Hepes (pH 7.5), 200 mM KCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mM EDTA using a 5 μl reaction volume. After a 5-min incubation at 30°C, reactions were chemically fixed by the introduction of Hepes-buffered EM-grade glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA) to a final concentration of 0.1% and were incubated for at least 5 min at room temperature before mounting for AFM. Samples were mounted as described (30). Micrographs for statistical analysis were gathered at a scan size of 2 μm. Supercoiled DNA was pBluescript (Stratagene) used at a final concentration of 1.5 mg/ml (≈0.9 nM). This molar concentration of DNA was used for all DNA substrates examined. Two linear DNA substrates were examined and were found to have similar properties with regard to ATM binding. A 660-bp linear DNA fragment generated by PCR as described (30) was used at a final concentration of 0.3 mg/ml. A 2.1-kilobase fragment of the pGBT9 vector (CLONTECH) was generated by cleaving with EcoRV and ScaI. This fragment was used at a final concentration of ≈1 mg/ml. Linear DNAs were gel purified by using the Qiaex II gel extraction kit (Qiagen, Chatsworth, CA). Irradiated pBluescript SK(+) was a kind gift from M. Plumb (Horwell, Oxon, U.K.). Purified ATM was diluted into reaction mixtures to a final concentration of ≈0.2 μg/ml (≈0.6 nM). Volume measurements were performed by using the nanoscope iii 4.22r2 software package (Digital Instruments, Santa Barbara, CA). Statistical analysis was performed by using spss/mac (SPSS, Chicago) on a Macintosh 8600/300 (Apple Computer).
Kinase Assays.
Kinase assays were assembled in 1× kinase buffer (50 mM Hepes, pH 7.5/50 mM KCl/4 mM MnCl2/6 mM MgCl2/10% glycerol/1 mM DTT/1 mM NaF/1 mM NaVO4/2 μM microcystin-LR) containing either immunoprecipitates or purified ATM and 50–100 ng of purified recombinant p53 (32), either in the absence or presence of increasing amounts of sheared calf thymus DNA or BamHI-digested pG13CAT plasmid DNA. Reactions were preincubated on ice for 3 min before addition of 10 μCi of γ[32P]-ATP and ATP to a final concentration of 100 μM. For phosphoserine-15 analysis, ATP was added to a final concentration of 100 μM. For wortmannin inhibition studies, wortmannin was added to a final concentration as indicated and was preincubated at room temperature for 30 min before addition of ATP. Reactions were incubated at 30°C for 10 min, and proteins were separated by SDS/PAGE. Gels either were transferred for Western blotting or were submitted to autoradiography to detect phosphorylated p53.
RESULTS
ATM Binds to DNA.
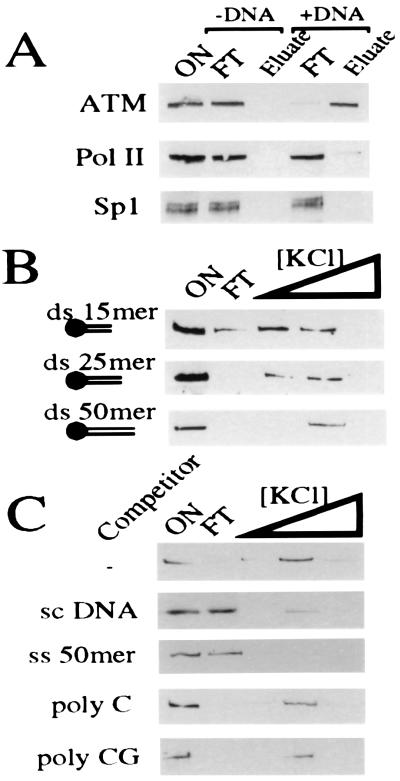
Given its nuclear location and role in genomic surveillance, we were intrigued by the possibility that ATM, or an ATM complex, could interact with DNA. To investigate this, a biotinylated ds 50-mer oligonucleotide of random sequence was coupled to streptavidin iron-oxide particles, and these were used to recover DNA binding proteins from HeLa cell nuclear extracts. This approach revealed that ATM indeed interacts with particles bearing this random piece of ds DNA (Fig. 1A). This binding is attributable to the presence of DNA because streptavidin iron-oxide particles alone (Fig. 1A) or iron oxide particles saturated with biotin (data not shown) are unable to bind ATM. In contrast, neither the sequence-specific DNA-binding protein Sp1 nor the nonspecific DNA-interacting protein complex containing RNA polymerase II is retained effectively by the random DNA fragment used in these studies (Fig. 1A). Under conditions in which >90% of ATM binds to the DNA-coupled particles, <2% of total nuclear protein is retained. Hence, the retention of ATM by DNA in these assays is highly specific. Additional studies revealed that ATM is also retained by particles containing other unrelated oligonucleotides (data not shown), suggesting that the interaction is not sequence-specific.
Figure 1.
ATM binds to DNA. (A) HeLa nuclear extract (ON) was bound to either streptavidin iron oxide beads (−DNA) or streptavidin iron oxide beads bearing a 50-mer ds DNA oligonucleotide (+DNA). After extensive washing, ATM was eluted in 500 mM KCl. Eluted proteins were subjected to 7% SDS/PAGE and ATM visualized by Western blotting using ATM.B antiserum. Equivalent fractions were probed for Sp1 and RNA polymerase II (Pol II). (B) ATM binding to DNA oligonucleotides depends on length. ATM-enriched extract (ON) was bound to streptavidin iron oxide beads attached to dsDNA of various sizes (15, 25, or 50 bp). After extensive washing, ATM was eluted sequentially with 100, 250, and 500 mM KCl. Eluates were analyzed as in A. (C) ATM is competed effectively by an excess of ssDNA and supercoiled DNA but not by poly-C or poly-CG. Washing, elution, and ATM detection was as above.
To investigate the DNA binding properties of ATM further, we tested a series of DNA molecules of different sizes for interaction with ATM. In these studies, binding and initial washes were conducted in 50 mM KCl, and then bound material was eluted by sequential washes at 100, 250, and 500 mM KCl. Fig. 1B demonstrates that the interaction between DNA and ATM depends on the size of the DNA molecule. With a ds 15-mer, some ATM is still present in the unbound fraction, and most bound material elutes in the lower salt wash. However, as the duplex size is increased, it becomes progressively more effective at binding ATM, such that when using a ds 50-mer oligonucleotide, ATM binding is almost quantitative and all bound ATM elutes in the higher salt wash (Fig. 1B). Furthermore, ATM binding to dsDNA in such experiments is competed by ssDNA and by circular supercoiled plasmid DNA but is not competed effectively by RNA (Fig. 1C). Taken together, these data show that ATM either directly or indirectly interacts with DNA molecules in an apparently nonsequence specific fashion.
Purification of ATM.
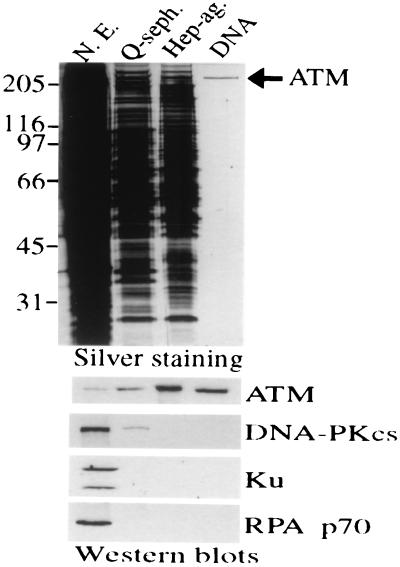
HeLa cell nuclear extract was used as starting material for purification, and ATM was monitored through the purification strategy by Western blot analysis using anti-ATM antibodies. In light of the DNA-binding properties of ATM, we used a final DNA affinity step in the purification scheme. Silver staining demonstrates that this leads to an essentially homogenous preparation of an ≈350-kDa polypeptide, and Western blotting reveals that this is recognized strongly by ATM antiserum ATM.B (ref. 22; Fig. 2) and two other antibodies raised to distinct regions of the ATM polypeptide (data not shown). Reprobing Western blots with antisera raised against Ku, DNA-PKcs, and replication protein A demonstrated that these abundant nonspecific DNA binding proteins are all removed efficiently during ATM purification (Fig. 2). Furthermore, Western blotting with antibodies against the ATM-related protein ATR revealed that this is also absent from purified ATM preparations (data not shown). Quantitative Western blotting and silver-staining reveal that the final yield of ATM is ≈25% and indicate that ATM is of relatively low abundance, comprising ≈0.005% of total nuclear protein by weight.
Figure 2.
Purification of ATM to essential homogeneity. Equivalent volumes (5 μl) of HeLa cell nuclear extract (50 μg protein) or pooled fractions after sequential Q-Sepharose, Heparin-agarose, or DNA affinity chromatography were subjected to 8% SDS/PAGE and proteins visualized by silver staining (Upper). Fractions also were subjected to Western blot analysis (Lower) using antibodies raised against ATM. Filters then were stripped and reprobed by using antisera raised against DNA-PKcs, Ku70 plus Ku80 (Ku), or the 70-kDa subunit of replication protein A, as indicated.
Analysis of ATM by Atomic Force Microscopy.
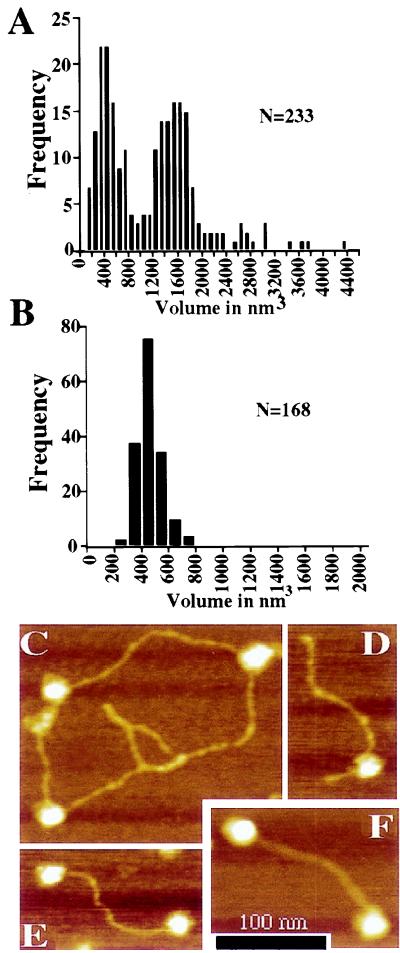
The AFM is well suited for biological imaging because of the instrument’s ability to visualize proteins and their complexes without metal coating or staining (reviewed in ref. 33). It provides images as three-dimensional topographs of the sample surface, and the three-dimensional nature of such images provides a means of measuring molecular volumes. When we used this technique to analyze our purified ATM preparations, two distinct peaks of particle size were evident (Fig. 3A). Although several factors contribute to the absolute volume observed (e.g., probe geometry and electrostatic interactions between probe and sample), the important observation here is the relative volumes of the two ATM species and their distribution. These volumes were measured from particle images gathered with the same AFM probe on the same day in consecutive scans, meaning that it is unlikely that the volume distribution is the result of two different probe geometries. Furthermore, the same distribution is observed if the data set is divided in half according to the sequence in which the data were collected, indicating that the results are not attributable to alterations in the probe shape over time. Perhaps most importantly, volume data from separate experiments yielded essentially indistinguishable results (data not shown). These results therefore suggest that ATM can exist in higher order assemblies.
Figure 3.
Analysis of ATM by atomic force microscopy. (A) The volume of ATM particles adsorbed to freshly cleaved micas was calculated from atomic force micrographs. Two distinct populations were observed in the sample at 200 mM KCl. Peaks in the volume distribution are apparent at ≈400 and ≈1,600 nm3. (B) The volume of free DNA-PKcs was measured as a control for volumetric analysis of the ATM particle. DNA-PKcs displays a single peak in the volume distribution with an average particle volume of 479 nm3. (C–F) Height-encoded atomic force micrographs of ATM in association with DNA. (C) ATM particles bound to supercoiled pBluescript plasmid. (D) A 660-bp linear DNA with blunt ends is bound by an ATM particle at an internal site. (E and F) The majority of linear DNAs bound by ATM are associated with the protein through an interaction involving a DNA end. (Bar = 100 nm.)
When the volume of DNA-PKcs was examined, it was found to be markedly different from that of ATM. Thus, as summarized in Fig. 3B, DNA-PKcs exists as a single species with an average volume of 479 nm3. This is roughly consistent with the predicted volume of the particle based on a protein density of 1.36 g/ml (≈560 nm3 predicted volume assuming a molecular weight of 465 kDa; the smaller observed volume is likely to be the result of the collapse of the DNA-PKcs particle after adsorption to the mica surface and the effects of drying during sample preparation (see ref. 31). The predicted volume of ATM based on the same assumptions would be ≈430 nm3, which is in close agreement with the smaller species observed in our volumetric analysis. Therefore, based on the relative volume of the particles observed, it is likely that purified ATM exists both as a monomer and in larger particles that represent ATM tetramers. However, other experimental approaches need to be performed to verify the precise nature of the multimeric species.
Next, we used the AFM to investigate in more detail the interaction between ATM with linear and supercoiled DNA substrates. Thus, reactions containing purified ATM and either of the two types of DNA were incubated at 30°C for 5 min in 200 mM KCl followed by chemical fixation with 0.1% glutaraldehyde, and then the resulting complexes were examined by AFM analysis (Table 1 and Fig. 3 C–F). At the molar ratios and salt conditions used, 37% of supercoiled DNA was bound to at least one ATM particle (n = 144). Notably, however, DNA binding was enhanced significantly on linear DNA. Analysis of micrographs collected from reactions containing a 660-bp linear DNA revealed that ≈57% (n = 251) of DNA molecules were bound under the same reaction conditions used for supercoiled DNA. This difference in DNA binding observed between linear and supercoiled DNAs was found to be significant by using a two-tailed t test (P < 0.001). Most strikingly, of the linear DNA molecules bound to ATM, 63% displayed end-bound protein. These data suggest that the increased ATM binding to linear versus supercoiled DNA is attributable to the preferential association of the protein with DNA ends. Representative molecules from reactions containing supercoiled or linear DNA are shown in Fig. 3 C–F, and quantitative data derived from atomic force micrographs are summarized in Table 1. We also examined ATM binding to plasmid DNA that had been exposed to 150 Gy of IR and consisted primarily of supercoiled and open circular forms (40% open circular, 58% supercoiled, 2% linear). ATM did not display increased association with the irradiated DNA (Table 1), consistent with the interpretation that ATM interacts with some preference for DNA ends rather than with ss nicks or other forms of damage induced by IR treatment.
Table 1.
Comparison of ATM Protein Binding to Different DNA Molecules by AFM
| DNA | Supercoiled plasmid DNA
|
Linear DNA
|
Irradiated plasmid DNA
|
|||
|---|---|---|---|---|---|---|
| Observed | Percent | Observed | Percent | Observed | Percent | |
| Free | 91 | 63.2 | 109 | 43.4 | 175 | 67.0 |
| Internally bound | 53 | 36.8 | 52 | 20.7 | 86 | 33.0 |
| End bound | — | — | 90 | 35.9 | — | — |
| N | 144 | 251 | 261 | |||
ATM protein was bound in the presence of supercoiled plasmid DNA, a 660-bp linear DNA PCR fragment, or plasmid DNA irradiated with 150 Gy of ionizing radiation. DNA molecules in 2 μM AFM scans were scored as bound or unbound.
A DNA-Stimulated p53 Serine-15 Kinase Activity Copurifies with ATM.
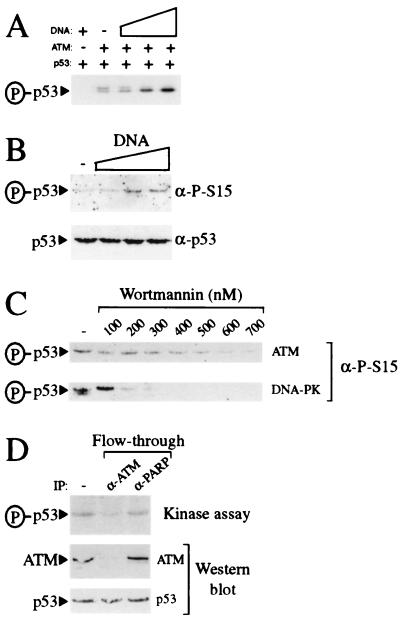
Recently, it has been reported that ATM phosphorylates p53 on serine-15 (25–27). To establish whether our ATM preparations contain such a kinase activity and to see whether it is influenced by DNA, purified ATM was incubated with p53 and γ[32P]-ATP in the absence or presence of increasing concentrations of sheared genomic DNA. As illustrated in Fig. 4A, weak but significant p53 kinase activity is detected in reactions to which DNA has not been added. Moreover, kinase activity is stimulated significantly on addition of sheared genomic DNA. Importantly, no kinase activity is apparent when p53 is incubated with DNA in the absence of ATM (Fig. 4A), illustrating that the kinase activity detected is mediated by the ATM preparation. Consistent with previous reports (25–27), our purified ATM preparation was found to mediate phosphorylation of p53 on serine-15, as visualized by using antibodies that recognize p53 only when it has been phosphorylated on serine-15 (Fig. 4B). Furthermore, this serine-15 kinase activity is stimulated significantly by the addition of sheared genomic DNA (Fig. 4B).
Figure 4.
Purified ATM contains DNA-stimulatable p53 kinase activity. (A) ATM preparations phosphorylate p53 in a DNA-stimulated manner. Kinase reactions containing γ[32P]-ATP were performed either in the absence of ATM and presence of 50 ng BamHI-digested pG13CAT plasmid DNA or in the presence of ATM preparations in the absence (−) or presence of 0.5, 5, or 50 ng of BamHI-digested pG13CAT plasmid DNA. All reactions contained recombinant p53 as a substrate. Phosphorylated p53 was detected by autoradiography. (B) ATM preparations phosphorylate p53 at serine-15 in a DNA-stimulatable manner. Kinase reactions containing ATM using p53 as a substrate were performed either in the absence (−) or presence of 1, 5, or 10 ng of sheared calf thymus genomic DNA. Serine-15 phosphorylation was detected by Western blotting with phosphospecific antibodies raised against p53 phosphorylated at serine-15. (C) ATM-associated kinase activity is inhibited by wortmannin. Kinase reactions using either DNA-PK or ATM preparations and recombinant p53 as a substrate were performed in the presence of 50 ng of BamHI-digested pG13CAT plasmid DNA. Reactions were preincubated either in the absence (−) or presence of increasing concentrations of wortmannin for 30 min before the addition of ATP. Serine-15 phosphorylation was detected as in B. (D) Immunodepletion of kinase activity from ATM preparations using ATM antibody. ATM preparations were incubated in either the absence (−) or presence of ATM or poly(ADP-ribose) polymerase monoclonal antibodies as indicated. After immunodepletion, ATM preparations were used in kinase assays in the presence of γ[32P]-ATP, BamHI-digested pG13CAT plasmid DNA, and recombinant p53. Phosphorylation of p53 was monitored by autoradiography. Amounts of ATM and p53 in kinase reactions were monitored by Western blotting with ATM and p53 antibody, respectively.
Recent work has indicated that ATM is inhibited by submicromolar concentrations of the PI 3-kinase inhibitor wortmannin (25, 34). Consistent with this, the DNA-stimulated p53 serine-15 kinase activity of our ATM preparations is inhibited by wortmannin, with almost complete loss of activity being achieved at ≈600 nM wortmannin (Fig. 4C Upper). Significantly, these inhibition characteristics are distinct from those of DNA-PK, which, under the assay conditions used in this study, is inhibited at between 100 and 200 nM wortmannin (Fig. 4C Lower), and from those of ATR, which is inhibited by wortmannin with an IC50 of 1.8 μM (34). These results are consistent with the fact that we are unable to detect DNA-PKcs or ATR in our most purified ATM samples, and provide additional support for the p53 serine-15 kinase activity in our ATM preparations being mediated by ATM. Furthermore, through conducting parallel titration studies with highly active purified DNA-PK, we have established that, if our ATM preparations do contain DNA-PK, it must be present at levels significantly below those necessary to generate detectable p53 kinase activity (data not shown). To further verify that the kinase activity in our preparations depends on ATM, we conducted immunoprecipitation experiments by using the previously characterized anti-ATM monoclonal antibody ATM132 (25). As illustrated in Fig. 4D, kinase activity is depleted from ATM preparations by using ATM antibody whereas control poly(ADP-ribose) polymerase antibody is relatively inefficient at depleting kinase activity. Taken together, these data reveal that, as with immunoprecipitated material (25–27), p53 serine-15 kinase activity in our preparations is mediated by ATM.
DISCUSSION
Given that ATM is related in sequence to DNA-PKcs and is involved in the detection and signaling of DNA damage, it has been proposed that ATM or an ATM complex might interact directly or indirectly with DNA. Here, we have demonstrated that ATM is retained on immobilized particles bearing DNA molecules, and studies using unrelated oligonucleotides suggest that this interaction is not sequence dependent. Furthermore, as determined by AFM analyses, ATM binds with preference to dsDNA ends. By exploiting these and other biochemical properties of ATM, we have developed a strategy to purify it from HeLa nuclear extracts to near homogeneity. The high purity of our final ATM preparations and the fact that ATM in such preparations can re-bind to DNA (data not shown) suggests strongly that ATM interacts with DNA directly. Although this appears different from the situation with DNA-PKcs, which requires Ku to associate with DNA stably (15, 17, 36), UV protein-DNA cross-linking has revealed that, in the context of the DNA-PKcs/Ku holoenzyme, DNA-PKcs does make close contacts with DNA (15). Furthermore, recent data indicate that, at low salt concentrations, DNA-PKcs can interact with DNA ends in the absence of Ku (16, 17).
Although displaying similarities to DNA-PK, ATM does exhibit important differences. For example, ATM appears to bind with a greater selectivity to DNA ends than to DNA ss breaks. This is not the case for the DNA-PK targeting subunit Ku, which has a similar affinity for nicked DNA as it has for a dsDNA end (37). Also, the binding of ATM to DNA appears more salt stable than that of DNA-PKcs. Thus, comparing our AFM data to those in a previous study (30), DNA-PKcs binds DNA poorly even at 100 mM KCl whereas we have shown that ATM binds DNA effectively at 200 mM KCl. Nevertheless, as is the case for DNA-PKcs, the binding of ATM to DNA is not strong or stable enough to allow ATM-DNA complexes to be visualized by the electrophoretic mobility shift assay. Based on their similarities at the protein sequence level and in their biological functions, it is tempting to speculate that ATM and DNA-PKcs (independent of Ku) bind to DNA through similar mechanisms, and it will be of great interest to identify and compare their DNA binding domains and modes of DNA interaction in more detail.
In line with recent reports (25–27), we have shown that a p53 serine-15 kinase activity consistently co-purifies with ATM. Furthermore, kinase activity associated with our purified ATM is inhibited by wortmannin at similar concentrations to those described for immunoprecipitated ATM and at concentrations of this agent that are distinct from those required to inhibit DNA-PK or ATR (see Fig. 4C; refs. 25, 28, 34, and 35). These data strongly suggest that the serine-15 kinase activity present in our ATM preparations is mediated by ATM, although we cannot formally exclude the possibility that additional polypeptides are present in our preparations that phosphorylate p53 on other sites. Notably, we have found that the kinase activity in our ATM preparations is stimulated by DNA. This is consistent with the work of Gately et al. (38), who have reported that kinase activity in ATM immunoprecipitates is stimulated by DNA. In contrast, Banin et al. (25) have reported that the p53 kinase activity in ATM immunoprecipitates is unaffected by DNA, possibly reflecting differences in conditions used for protein retrieval, the assay itself, or the substrate used.
Our data are consistent with models in which ATM protein kinase activity is triggered in vivo upon its association with DNA, resulting in signaling to downstream targets, including p53, that impinge on the cell cycle, transcription, and/or apoptotic machineries. In light of the DNA-PK paradigm, however, it seems likely that ATM will require additional polypeptides to perform its kinase activity efficiently. Given that the available genetic and biochemical data suggest that DNA-PK, ATM, and ATR act largely independently of one another, a key question is whether they compete or act in concert. An attractive model is that the DNA-PK, ATM, and ATR systems are all involved in recognizing DNA damage but either recognize different forms of DNA damage or recognize overlapping forms of DNA damage under different temporal or spatial constraints. The availability of biochemical assays for ATM, DNA-PK, and ATR should prove instrumental in addressing these important issues and determining how ATM activity is triggered in response to genotoxic insult.
Acknowledgments
We thank members of the SPJ lab for their advice and support. This work was funded by Grants SP2143/0103 and SP2143/0501 from the Cancer Research Campaign and by grants from the Kay Kendall Leukaemia Fund and the A-T Children’s Project. This work was also supported by National Institutes of Health Grant CA50519 to D.J.C. and by grants from the U.S. Department of Energy to R.B.C. and D.J.C. Queries on AFM analysis should be directed to R.B.C.
ABBREVIATIONS
- A-T
ataxia-telangiectasia
- IR
ionizing radiation
- PI
phosphatidylinositol
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs
DNA-PK catalytic subunit
- ds
double strand
- ss
single strand
- AFM
atomic force microscope
References
- 1.Hoekstra M F. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- 2.Lavin M F, Shiloh Y. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 3.Rotman G, Shiloh Y. Hum Mol Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 4.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J J. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 5.Khanna K K, Lavin M F. Oncogene. 1993;12:3307–3312. [PubMed] [Google Scholar]
- 6.Siliciano J D, Canman C E, Taya Y, Sakaguchi J, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 8.Savitsky K, Sfez S, Tagle D A, Ziv Y, Sartiel A, Collins F S, Shiloh Y, Rotman G. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 9.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 10.Keith C T, Schreiber S L. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 11.Zakian V A. Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 12.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 13.Jackson S P. Cancer Surv. 1996;28:261–279. [PubMed] [Google Scholar]
- 14.Dvir A, Stein L Y, Calore B L, Dynan W S. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 15.Gottlieb T M, Jackson S P. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 16.Yaneva M, Kowalewski T, Lieber M R. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammarsten O, Chu G. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieber M R, Grawunder U, Wu X, Yaneva M. Curr Opin Genet Dev. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- 19.Critchlow S, Jackson S P. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 20.Smith G C M, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Lee E Y-H P. J Biol Chem. 1996;271:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- 22.Lakin N D, Weber P, Stankovic T, Rottinghaus S T, Taylor A M, Jackson S P. Oncogene. 1996;13:2707–2716. [PubMed] [Google Scholar]
- 23.Brown K D, Ziv Y, Sadanandan S N, Chessa L, Collins F S, Shiloh Y, Tagle D A. Proc Natl Acad Sci USA. 1997;94:1840–1845. doi: 10.1073/pnas.94.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watters D, Khanna K K, Beamish H, Birrell G, Spring K, Kedar P, Gatei M, Stenzel D, Hobson K, Kozlov S, et al. Oncogene. 1997;14:1911–1921. doi: 10.1038/sj.onc.1201037. [DOI] [PubMed] [Google Scholar]
- 25.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 26.Canman C E, Lim D-S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 27.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, LeesMiller S P, Lavin M F. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 28.Lakin N D, Hann B C, Jackson S P. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 29.Smith G C M, d’Adda di Fagagna, Lakin N D, Jackson S P. Mol Cell Biol. 1999;19:6076–6084. doi: 10.1128/mcb.19.9.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cary R B, Peterson S R, Wang J, Bear D G, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu C Y, Cary R B, Chen D J, Peterson S R, Stewart P L. J Mol Biol. 1998;284:1075–1081. doi: 10.1006/jmbi.1998.2212. [DOI] [PubMed] [Google Scholar]
- 32.Hupp T R, Meek D M, Midgley C A, Lane D P. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 33.Bustamante C, Keller D, Guoliang Y. Curr Opin Struct Biol. 1993;3:363–372. [Google Scholar]
- 34.Sarkaria J N, Tibbetts R S, Busby E C, Kennedy A P, Hill D E, Abraham R T. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 35.Izzard R J, Jackson S P, Smith G C M. Cancer Res. 1999;59:2581–2586. [PubMed] [Google Scholar]
- 36.Suwa A, Hirakata M, Takeda Y, Jesch S A, Mimori T, Hardin J A. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blier P R, Griffin A J, Craft J, Hardin J A. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 38.Gately D P, Hittle J C, Chan G K T, Yen T J. Mol Biol Cell. 1998;9:2361–2374. doi: 10.1091/mbc.9.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]