Abstract
The spliceosome is a dynamic ribonucleoprotein complex responsible for the removal of intron sequences from pre-messenger RNA. The highly conserved 5′ end of the U2 small nuclear RNA (snRNA) makes key base-pairing interactions with the intron branch point sequence and U6 snRNA. U2 stem I, a stem–loop located in the 5′ region of U2, has been implicated in spliceosome assembly and may modulate the folding of the U2 and U6 snRNAs in the spliceosome active site. Here we present the NMR structures of U2 stem I from human and Saccharomyces cerevisiae. These sequences represent the two major classes of U2 stem I, distinguished by the identity of tandem wobble pairs (UU/UU in yeast and CA/GU in human) and the presence of post-transcriptional modifications (four 2′-O-methyl groups and two pseudouracils in human). The structures reveal that the UU/UU and CA/GU tandem wobble pairs are nearly isosteric. The tandem wobble pairs separate two thermodynamically distinct regions of Watson–Crick base pairs, with the modified nucleotides in human stem I conferring a significant increase in stability. We hypothesize that the separate thermodynamic stabilities of U2 stem I exist to allow the structure to transition through different folded conformations during spliceosome assembly and catalysis.
Keywords: NMR, RNA structure, U2 snRNA, spliceosome
INTRODUCTION
Pre-messenger RNA splicing refers to the process catalyzed by the spliceosome, in which noncoding intron sequences are excised and coding exons are ligated together through a two-step reaction to form mature mRNA. In the first of two nucleophilic reactions, the 2′-OH of an intronic “branch point” adenosine attacks the phosphodiester backbone at the 5′-splice site, creating a branched lariat intermediate and a free 3′-OH on the 5′ exon. This hydroxyl then attacks the 3′-splice site, ligating the exons and releasing the intron in the lariat form. The chemistry of these two transesterification reactions is relatively simple, and is the exact mechanism catalyzed by group II intron ribozymes. However, pre-mRNA splicing requires exquisite accuracy and precise regulation, and is therefore catalyzed by the spliceosome, one of the largest and most complex molecular machines in the cell.
The spliceosome consists of five small nuclear RNAs (snRNAs)—U1, U2, U4, U5, and U6—that assemble with numerous protein factors to form small nuclear ribonucleoproteins (snRNPs). Accurate splice site selection and catalysis require a highly ordered network of dynamic RNA–RNA and RNA–protein interactions between the spliceosomal snRNPs and the pre-mRNA (for review, see Staley and Guthrie 1998; Brow 2002). The U2 snRNP is a pivotal component of several of these interactions during the splicing cycle. In the early stages of spliceosome assembly, U2 snRNP recognizes the branch point adenosine, and a short intermolecular helix forms between U2 snRNA and the consensus sequence within the intron. This U2–intron duplex serves to position the branch point adenosine for its role as the nucleophile during the first catalytic step. Prior to splicing activation, U2 also base-pairs with U6 snRNA, forming the active site of the spliceosome. The U2–U6 complex acts as a framework for juxtaposing the branch site helix with the 5′-splice site, and may be responsible for catalysis (Valadkhan and Manley 2001, 2003).
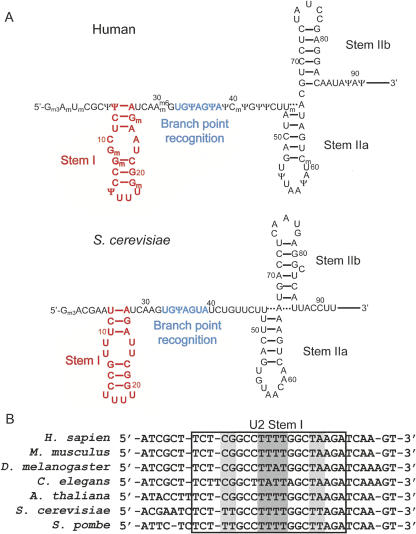
The highly conserved 5′ end of U2 snRNA is made up of several stem–loops and single-stranded regions that interact with U6 snRNA, the intron, and the snRNP proteins. Stem I, just upstream of the branch point sequence, is made up of two short Watson–Crick helices separated by two mismatch pairs, with a tetraloop containing three or four uracil residues capping the stem (Fig. 1; Guthrie and Patterson 1988; Ares and Igel 1990). Sequence alignments of the 5′ end of U2 snRNA reveal significant conservation of stem I (Fig. 1B; Guthrie and Patterson 1988). Previous studies of the U2 snRNA 5′ terminus have revealed potential functions for stem I in initial steps of spliceosome assembly. Post-transcriptional modifications in the 5′ end of metazoan U2, including several in stem I, are required for formation of one of the earliest spliceosomal complexes (Yu et al. 1998; Donmez et al. 2004). It is unknown exactly how these modifications contribute to complex formation, but they may facilitate an interaction between the RNA and a protein. A recent UV cross-linking and RNA cleavage analysis of the U2 snRNP suggests that this protein may be the essential splicing factor SF3a60, which was shown to contact stem I (Dybkov et al. 2006). This discovery further implicates stem I in spliceosome assembly, as SF3a60 is a component of the SF3a complex, which is required for recruitment of U2 snRNP to the nascent spliceosome (Brosi et al. 1993).
FIGURE 1.
Secondary structures of free U2 snRNA and sequence alignment for the 5′ end of U2 snRNA. (A) The 5′ end of U2 snRNA contains the branch point recognition sequence (blue) and three stem–loops, including stem I (red). (B) Watson–Crick pairing is universally conserved and wobble base pairs at the center of the helix (light-gray background) are either tandem U-U pairs in yeast or G-U and C-A pairs in higher eukaryotes. The U-rich tetraloop (dark-gray background) is highly conserved as well.
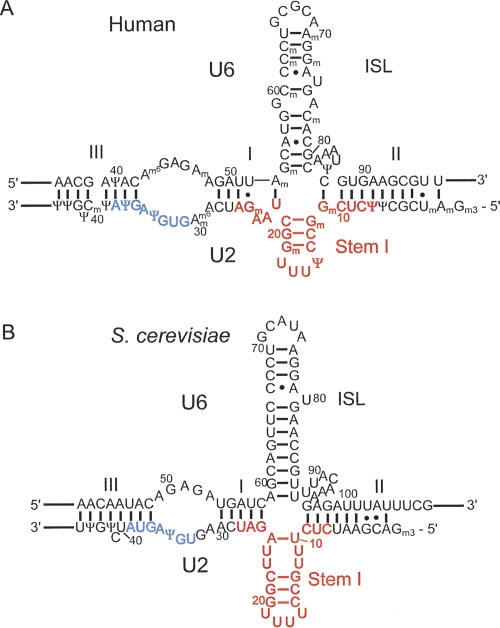
In addition to anchoring essential proteins to U2 snRNA during assembly steps, stem I may also play a role during splicing catalysis. The regions flanking the branch site recognition sequence and U2 stem I are complementary to U6 snRNA, allowing U2 to form two short intermolecular helices (I and II) with U6. These intermolecular helices, along with U2 stem I and the U6 intramolecular stem–loop (ISL), can form a four-way helical junction in all U2-dependent spliceosomes (Fig. 2). Structural studies of the U2–U6 complex indicate that U2 stem I is present within the complex as a stable domain (Fig. 2B; Sashital et al. 2004). The theoretical free energy of folding for the U2–U6 complex also indicates that the U2 stem I-containing four-helix junction is the most stable conformation for the complex (Cao and Chen 2006). Mutational analysis of U2 snRNA suggests that the tetraloop proximal G-C pairs within stem I are very important for splicing activity (McPheeters and Abelson 1992; Sun and Manley 1995). These combined data suggest a potential role for U2 stem I in the active site of the spliceosome.
FIGURE 2.
Secondary structure for U2–U6 complexes from S. cerevisiae and human. (A) The proposed secondary structure of the human U2–U6 complex (Sun and Manley 1995). (B) The NMR-observed secondary structure of the yeast U2–U6 complex (Sashital et al. 2004). Colors correspond to those in Figure 1.
Stem I competes structurally with several potential U2–U6 interactions, indicating that it may be unwound at some point during splicing catalysis. For example, in the yeast spliceosome, U2 stem I must be fully melted prior to the second step of splicing to allow for the formation of the essential intermolecular helix Ib, a short Watson–Crick stem that forms between U2 residues 21-GCU-23 and U6 residues 59-AGC-61 (Madhani and Guthrie 1992; Hilliker and Staley 2004). To that end, the stability of stem I is of significant importance, as has been demonstrated through mutagenesis studies of both the human and yeast sequences. Stabilizing the wobble base pairs at the center of stem I by introducing mutations that form Watson–Crick pairs disrupts splicing, likely through inhibition of the helix I and II interactions between U2 and U6 (Wu and Manley 1992). Mutations that destabilize stem I suppress phenotypic mutations that disrupt helix Ib, suggesting that stem I may regulate formation of helix Ib for the second step of splicing (Hilliker and Staley 2004). These results suggest that a delicate thermodynamic balance exists between different conformations of the U2–U6 complex, and that the stability of U2 stem I must be fine tuned in order to account for this balance.
We have carried out a detailed analysis of U2 stem I in order to gain more insight into the structural and thermodynamic characteristics of the stem–loop. Two stem I sequences, from human and Saccharomyces cerevisiae, were chosen because of the considerable amount of genetic and biochemical information available for both species. The sequences are also representative of two classes of stem I that we have identified based on the identity of two tandem wobble base pairs at the center of the helix (Fig. 1B). Here we report the NMR structures of U2 stem I from S. cerevisiae and the fully modified U2 stem I from human. The solution structures reveal a great degree of similarity between the overall folds of the stem–loops, despite the divergent tandem wobble pairs and presence of modified nucleotides. In contrast to the structural similarity, the stabilities of human and yeast U2 stem I differ greatly. The modifications in human serve to increase the thermodynamic stability of stem I by 18°C. When framed in the context of existing genetic data, we find that the observed thermodynamics of U2 stem I correlate well with the importance of U2 stem I in splicing.
RESULTS
Human and yeast U2 stem I RNA sequences
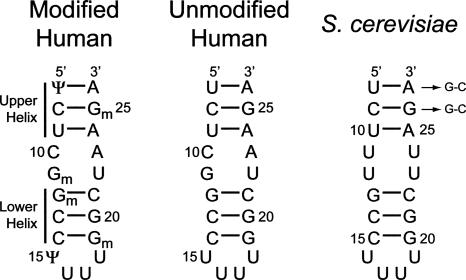
The U2 stem I helix is made up of six Watson–Crick base pairs flanking an internal loop made up of two tandem wobble pairs, and capped by an unusual U-rich tetraloop. The two short Watson–Crick helices are henceforth referred to as the upper helix (three terminal base pairs) and lower helix (three tetraloop–proximal base pairs) (Fig. 3). Interestingly, the identity of the wobble pairs is conserved within yeast strains as U-U wobbles, while all higher eukaryotes contain a tandem C-A and G-U sequence (Fig. 1B). Yeast and human U2 snRNAs also differ greatly in the degree to which they are post-transcriptionally modified. Mammalian U2 snRNA is extensively modified, including four 2′-O-methyl guanosines and two pseudouracils within stem I (Massenet et al. 1998). In contrast, yeast U2 stem I contains no modifications.
FIGURE 3.
Sequences of U2 stem I used for structural and thermodynamic studies. The human sequences represent U2 nucleotides 7–26, and the S. cerevisiae sequence represents nucleotides 8–27. All RNAs were produced via solid-phase synthetic chemistry (Dharmacon, Inc.), except for one S. cerevisiae sequence, in which two G-C pairs were substituted at the helical terminus, as indicated by arrows, to facilitate in vitro transcription using T7 RNA polymerase. The nomenclature used for the short Watson–Crick helices is shown on the left.
In order to investigate the effects of wobble pair identity and presence of modified nucleotides on the structure and stability of U2 stem I, we studied several RNAs corresponding to residues 7–26 and 8–27 of human and S. cerevisiae U2 snRNAs, respectively (Fig. 3). Isotopically labeled yeast stem I RNA for NMR studies was synthesized by substituting the first two base pairs with G-C pairs to facilitate in vitro transcription using T7 RNA polymerase. Changing the first two base pairs to G-C in the yeast stem I sequence has no effect on the overall structure of the molecule, as determined by comparison of the NMR data from the wild-type sequence (data not shown).
NMR analysis
The Watson–Crick base pairs for the human and yeast stem I RNAs were confirmed by two-dimensional nuclear Overhauser effect spectroscopy (2D NOESY) spectra. For the human stem I RNA, a G-U wobble was also observed in the 2D NOESY spectrum. The adenine in the C-A wobble is readily protonated, as evidenced by a pH-dependent upfield shift for the C2 carbon (Legault and Pardi 1997). At pH ≤ 6.4, the A23 C2 chemical shift is shifted upfield to 146 ppm, indicating that the N3 atom is protonated under these conditions (data not shown). At pH > 6.4, the C2 resonance is broadened, which can be attributed to proton exchange (Cai and Tinoco 1996; Flinders and Dieckmann 2004). Thus, it is likely that a protonated A+-C wobble forms at pH lower than 6.4 in human U2 stem I.
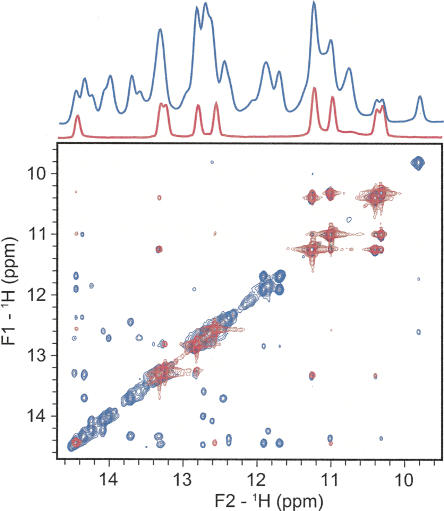
The formation of the tandem U-U wobble pairs in yeast stem I was observed by 2D NOESY. The NOE pattern for the U-U wobbles, as well as other exchangeable protons in the isolated stem I, are an almost perfect subspectrum of the same region of the NOESY for a 73-nucleotide (nt) U2–U6 complex (Fig. 4). We have previously demonstrated that the 73-nt U2–U6 RNA construct is well folded and adopts the same four-helix junction conformation as a 110-nt U2–U6 complex that contains the nearly full-length U2–U6 secondary structure (Sashital et al. 2004). The excellent agreement in the overlay of the NMR data indicates that U2 stem I retains the same structure in isolation as it does in the context of much larger U2–U6 complexes.
FIGURE 4.
Overlay of imino region of 1D 1H and 2D 1H–1H NOESY spectra (150-msec mixing time) for yeast U2 stem I (red) and the 73-nt U2–U6 complex (blue). The 73-nt U2–U6 complex has been investigated and described previously (Sashital et al. 2004).
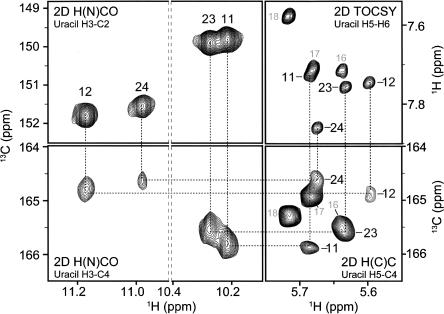
Due to the spatial proximity of imino protons within a U-U wobble, assignments for these protons cannot be distinguished from 2D NOESY spectra alone. The imino protons for the U-U wobbles were assigned using through-bond correlations to the base C4 atom [2D 1H–13C H(N)CO] (Ikura et al. 1990), which was also correlated to the H5 atom [2D 1H–13C H(C)C] (Fig. 5; Fiala et al. 2004). This method allowed for unambiguous assignment of the four uracil imino protons in the tandem U-U wobbles; moreover, chemical shifts for the C4 and C2 atoms obtained in the 2D H(N)CO spectrum provided insight into the identity of the hydrogen-bond acceptors. As has been noted previously for a variety of uracil-containing base pairs, chemical shifts for hydrogen-bonded carbonyls are shifted downfield relative to those of non-hydrogen-bonded carbonyls (Furtig et al. 2003). In the case of U2 stem I, U11 and U23 have similar chemical shift patterns, with the C2 signal shifted downfield and the C4 signal shifted upfield. The opposite is true for U12 and U24, in which C2 is relatively upfield and C4 is downfield. These observations indicate that the two U-U wobbles adopt different conformations, in which uracil residues on the same side of the helix contribute different hydrogen-bond acceptors. The observed downfield carbon chemical shifts strongly suggest that the C2 carbonyls are the hydrogen-bond acceptors in U11 and U23, and the C4 carbonyls are the hydrogen-bond acceptors in U12 and U24. This hydrogen-bonding pattern is consistent with other studies of RNAs containing tandem U-U wobbles (SantaLucia et al. 1991; Lietzke et al. 1996; Theimer et al. 2003).
FIGURE 5.
Determination of tandem U-U wobble conformations in yeast U2 stem I. Imino protons (H3) were assigned by correlation to C4 using 2D H(N)CO (bottom left panel), and correlation of C4 to H5 using 2D HCC (bottom right panel). The 2D HCC H5 assignments were confirmed by 2D 1H–1H TOCSY (upper right panel). The C2 and C4 chemical shifts in the 2D H(N)CO (left panels) identify the hydrogen-bonded carbonyls from their upfield carbon chemical shifts.
Residual dipolar couplings (RDCs) were measured for the 13C-labeled yeast U2 stem I and unlabeled human stem I RNAs. Homonuclear-decoupled TROSY and TROSY-ECOSY methods (Boisbouvier et al. 2004) were used to measure 76 one-bond and two-bond C–H and C–C couplings for the uracil bases within the yeast RNA (Table 1). These experiments provided orientational information for the tandem U-U wobbles, which helped to confirm the conformation of the asymmetric base pairs.
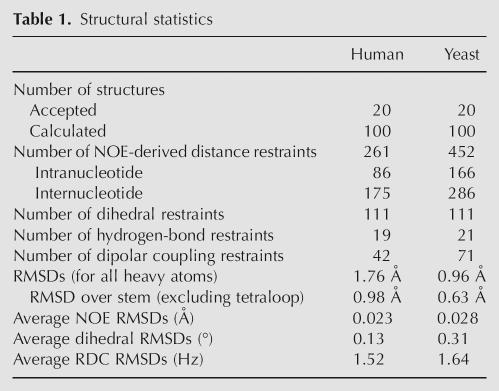
Table 1.
Structural statistics
U2 stem I structures
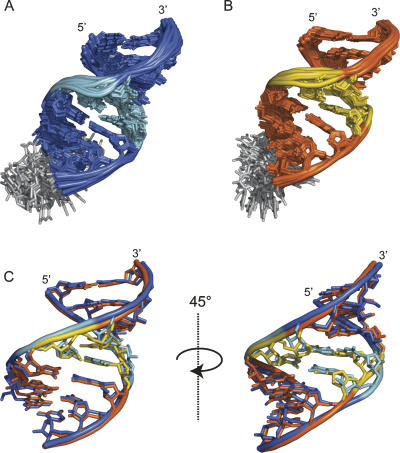
U2 stem I structures were calculated using 42 RDC and 261 NOE-derived distance restraints for the human and 71 RDC and 452 distance restraints for the yeast sequence (Table 1). Despite the differences in sequence and presence of modified nucleotides, the human and yeast U2 stem I structures are quite similar (Fig. 6). The helical backbone atoms for the mean structures overlay with an RMSD of 1.34 Å. However, the tandem wobble regions of the two structures deviate significantly. The U-U base pairs in the yeast structure cause a narrowing of the helix, to compensate for the smaller size of the pyrimidine bases and allow for the formation of hydrogen bonds. For the yeast mean structure, the C1′–C1′ distance across the helix within the U-U wobbles is 8.9 Å, ∼1.8 Å shorter than the same distance in an A-form helix. In the human structure, the minor groove width increases significantly to 10.7 Å at the Gm-U wobble and 11.2 Å at the C-A wobble. The slightly enlarged width of the C-A pair results from the wobble conformation, which places the bulky adenine amino group at the hydrogen-bonding interface, causing a minor protrusion of the backbone. These structural differences result in a pronounced backbone perturbation on the 5′ side of the internal loop (Fig. 6C).
FIGURE 6.
Solution structure of (A) human and (B) yeast U2 stem I. (A,B) The 10 lowest-energy structures superimposed over the helical region. Tandem wobble pairs at the center of the helices are (A) light blue and (B) yellow. The tetraloop is gray. (C) Helices of mean human and yeast structures superimposed over heavy atoms.
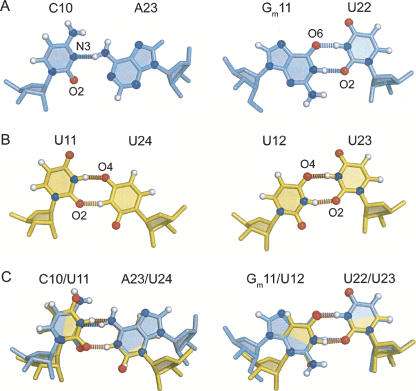
The divergence in backbone width has little effect on the overall similarity of the two structures, likely due to analogous hydrogen-bonding patterns within the internal loops (Fig. 7). Hydrogen bonding within the Gm11-U22 wobble in the human structure is identical to the corresponding U12-U23 wobble in yeast, each containing two imino-carbonyl hydrogen bonds (Fig. 7A,B). The human C10-A23 and yeast U11-U24 wobbles differ in the functional groups that contribute to hydrogen bonding; however, the overall geometry of the base pairs is retained in the two structures. Direct comparison of the wobble pair conformations by superimposition of the pyrimidine rings highlights these similarities (Fig. 7C).
FIGURE 7.
The tandem wobble pairs. (A,B) Hydrogen-bond acceptors are labeled for each base pair. (A) Human C-A and Gm-U pairs. (B) Yeast U-U wobbles. (C) Tandem wobble pairs superimposed over pyrimidine ring heavy atoms.
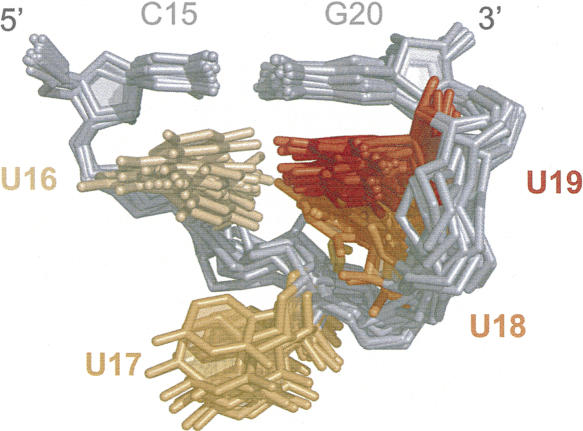
The yeast UUUU tetraloop adopts a preferred structure stabilized by base stacking (Fig. 8). The U16 and U19 bases stack under the closing C15-G20 base pair. U18 is flipped into the minor groove and stacks under U19, while U17 is unstacked and bulged into solution. The tetraloop may also be stabilized by hydrogen bonding. In the NMR ensemble of structures, the O2 of U16 is oriented toward the imino proton of either U18 or U19, and is within the hydrogen-bonding distance of one of these protons in most structures. The human tetraloop structure contains a larger degree of conformational heterogeneity than yeast; however, this is likely due to a smaller number of NOE restraints available for the structure, which was solved using the 1% natural abundance of the 13C isotope.
FIGURE 8.
Structure of yeast UUUU tetraloop. NMR ensemble of residues 15–20 from yeast structure, superimposed over heavy atoms of residues 15, 16, and 20.
Dynamic motions throughout the yeast and human U2 stem I molecules were investigated by measuring the intensities of 1H–13C correlations in HSQC spectra (Zhang et al. 2006). For the yeast structure, all of the correlations have identical resonance intensities, with the exception of the second, third, and fourth uracil residues in the tetraloop (U17–U19), which have intensities that are two- to threefold higher than all other nucleotides. These data indicate that the nucleotides on the 3′ side of the tetraloop experience internal motions that are faster than the overall tumbling of the molecule. In the human structure, only the third and fourth uracil residues of the tetraloop (U17–U18) exhibit similar increased intensities, suggesting that the second uracil (U16) is more ordered due to the presence of Ψ15. Since helical RNA structures of this size have correlation times of ∼4 nsec (Blad et al. 2005), the dynamic motions observed for the 3′-uracil residues of the tetraloops are likely to be on the subnanosecond timescale. The dynamic motions are in agreement with the structure of the yeast tetraloop, which shows the U17 nucleotide to be exposed and the U18–U19 nucleotides to be loosely stacked (Fig. 8). Conversely, there is no evidence for such fast dynamic motions in the first position of the tetraloop either, which stacks upon C15 in all the structures.
Thermodynamic analysis of U2 stem I by UV and NMR monitored melting
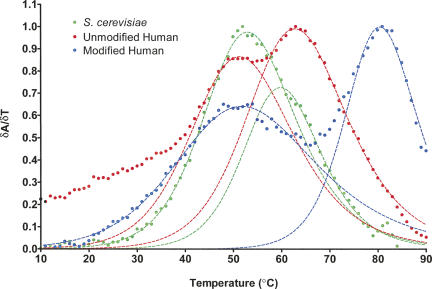
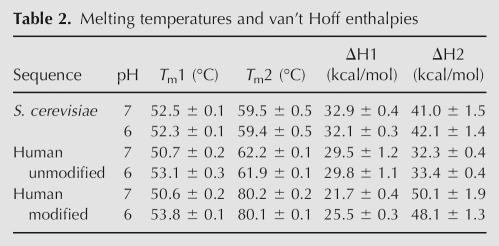
The differences in sequence and presence of covalent modifications in U2 stem I may have a significant effect on the thermal stability of the RNAs. In order to investigate the relative stabilities of the stem–loop based on these variations, we monitored UV absorbance at 260 nm as a function of temperature for the yeast and unmodified and modified human sequences. The first derivative plot for the UV melt profiles at pH 7 is shown in Figure 9. The melting temperatures and van't Hoff enthalpies for the RNAs are reported in Table 2.
FIGURE 9.
First-derivative UV melt profiles for modified and unmodified human and yeast U2 stem I at pH 7. Data points are circles, and the nonlinear least-square fits to the data are dashed lines. (Green) S. cerevisiae; (blue) modified human; (red) unmodified human.
Table 2.
Melting temperatures and van't Hoff enthalpies
Interestingly, two melting transitions are observed for all three RNAs: a lower-temperature melting transition between 50°C and 52°C (T m1); and a higher-temperature transition that varies between 59°C and 80°C (T m2), depending on the sequence and presence of modified nucleotides. The disparate melting transitions reflect a differential stability between various regions of the stem–loop. Based on the relatively similar T m1 values for the three RNAs, a portion of stem I likely retains comparable thermal stability regardless of sequence and modification variations. In contrast, the large difference in T m2 between the unmodified and modified RNAs indicates that the stability of the remainder of the stem–loop is greatly affected by the presence of the modified nucleotides.
Due to the presence of a protonatable C-A pair in human stem I, we reasoned that the stability of the two human RNAs would increase at lower pH. To test this hypothesis, the UV melting experiments were also performed at pH 6 (Table 2). As expected, the T m1 values for both human sequences increase by >2°C relative to pH 7 values. In contrast, the yeast T m1 and all T m2 values remain constant, independent of pH. These data suggest that the C-A wobble pair melts during the first transition, and is therefore within the less-stable region of the stem–loop. Furthermore, the stabilities of the two regions appear to be independent of one another, as the human T m2 values are unaffected by pH and the protonation state of the C-A pair.
In order to gain further insight into the two thermodynamically variable regions, imino signals for the RNAs were monitored as a function of temperature using NMR (data not shown). The spectra indicate that the first three base pairs (upper helix) melt during the first transition, while the second transition corresponds to the melting of the G-C-rich helix that closes the tetraloop (lower helix).
DISCUSSION
We have investigated the structure and stability of two classes of U2 stem I found in yeast and human U2 snRNA. Our results indicate that the sequence divergence has a small effect on the structure of the stem–loop, while the modified nucleotides greatly increase the stability. Below we discuss these results in the context of existing genetic and structural data.
Structural comparison of human and yeast U2 stem I
The electrostatic surface potential for the yeast structure reveals a deeper and more electronegative minor groove pocket compared to the human structure (data not shown). The increased electronegativity at the center of the yeast stem is created by the presence of the U12 O2 and U24 O2 in the minor groove, which are replaced by the Gm11 amino and A23 H2 in the human structure. These structural variations are created by the two classes of tandem wobbles and may present different metal- or protein-binding sites.
The human ΨUUU tetraloop structure is less well defined than the yeast structure, likely because it was solved using the natural abundance of 13C, resulting in fewer distance restraints for this region of the molecule. However, Ψ15 is well stacked under the C14-Gm19 base pair, which closes the human tetraloop in a manner very similar to the yeast tetraloop. Furthermore, the two uracil residues on the 3′ side of the tetraloop have nearly identical 1H and 13C chemical shifts, suggesting that the human loop adopts a similar conformation to that of yeast. The two imino protons for Ψ15 are observable in the exchangeable proton spectra, further indicating that the human tetraloop is stabilized by stacking or hydrogen-bonding, as is observed for the yeast structure.
Previously, a combined molecular dynamics and NMR study of the cUUUUg tetraloop indicated that the loop is highly dynamic and does not adopt a single conformation (Koplin et al. 2005). The MD simulation resulted in four conformational states containing stacking patterns for the last three uracil residues of the loop, which differ greatly from the stacking that occurs in the yeast structure. In contrast to the MD simulation structures, we observe that the structure of the yeast cUUUUg tetraloop is nearly homogeneous in conformation, with one preferred stacking arrangement (Fig. 8). Since our structure determination represents the time-averaged ensemble of the major conformation, it is possible that minor or transient conformers corresponding to those in the MD simulations may form within the population of structures. Previous thermodynamic studies revealed that cUUUUg loops stabilize helices by −3 kcal/mol, whereas the highly stable cUUCG tetraloop stabilizes helices by −5.7 kcal/mol (Antao et al. 1991). These thermodynamic data are also consistent with the observed stacking of the cUUUUg tetraloop.
Stability of U2 stem I and its biological implications
The modifications within the human sequence confer an 18°C increase in melting temperature to the lower helix of stem I. This heightened stability is likely due to the presence of three 2′-O-methyl guanosine residues in the lower helix, and perhaps also increased stacking of Ψ15 on C14 in the modified tetraloop. The 2′-O-methyl modifications stabilize the C3′-endo conformation of the ribose because of steric clashing that occurs between the methyl and base when the ribose adopts the 2′-endo conformation (Lee and Tinoco 1977; Kawai et al. 1992). Surprisingly, the Ψ7 and Gm25 modifications in the upper helix seem to have no effect on stability, as the T m1 values for the unmodified and modified sequences are nearly identical. This unexpected result may be due to the proximity of the two modified residues to the terminus of the stem–loop. In the context of the full U2 snRNP, the melting of the upper stem may also be influenced by the presence of flanking RNA sequences and/or protein contacts. Recent evidence suggests that the upper helix may be melted at a very early stage in assembly, supporting the observed instability of the upper helix (Dybkov et al. 2006).
Melting of the yeast stem occurs more cooperatively than for the human stem, as evidenced by the overlap of the transitions in the UV melt data (Fig. 9), and the uniformity of temperature-dependent imino peak broadening in the NMR data (data not shown). In contrast, the human UV melting transitions are well defined, even for the unmodified sequence, which has similar melting temperatures to yeast. This discrepancy may be caused by the G-U wobble in the human sequence, which is relatively stable compared to the U-U wobbles based on the temperature dependence of the NMR spectra.
The observed stability of the lower helix from the unmodified human U2 stem I is somewhat surprising. This helix is more stable by 2°C than the corresponding stem in yeast, which is of identical sequence. Therefore, the difference in stability may be attributed to the adjacent tandem mismatch pairs. Previous studies of tandem mismatch pairs indicate that C-A wobbles are generally destabilizing (Xia et al. 1997), while tandem U-U wobbles are relatively stable (Wu et al. 1995). However, the sequence of the flanking base pairs is a major factor in the stability of tandem mismatches. To our knowledge, this work represents the first study of the thermodynamics of tandem mismatches with nonsymmetric (U-A/G-C) flanking base pairs.
The importance of the lower helix of stem I for splicing activity has been demonstrated in both yeast and human (McPheeters and Abelson 1992; Sun and Manley 1995). Our thermodynamic study suggests that the nucleotide modifications in U2 snRNA may have evolved in order to increase the stability of the critical lower helix. In human stem I, two of the 2′-O-methyl modifications that contribute to the stability of the lower helix are required for spliceosome assembly (Donmez et al. 2004), further correlating stability with function. The tandem wobble pairs and modifications delineate the human U2 stem I into two thermodynamically distinct helical regions, providing a potential RNA-mediated mechanism for retaining the lower helix upon melting of the upper helix.
The relative uniformity of yeast stem I melting is of interest, given the importance of the competing helix Ib structure for the second step of splicing in yeast (Madhani and Guthrie 1992; Hilliker and Staley 2004). Helix Ib may not be essential in human (Sun and Manley 1995); therefore, the lower stem I helix may never melt during the human splicing cycle. In contrast, stem I must melt completely in yeast to accommodate the formation of helix Ib. The relatively low stability and cooperative melting of yeast stem I would facilitate this transition.
The potential function of U2 stem I as a regulatory element for the formation of helix Ib is intriguing in light of a recent competition-based model for conformational changes within the spliceosome active site (Query and Konarska 2004). The active site of the spliceosome must undergo a conformational change between the two steps of splicing in order to accommodate the different substrates that are involved in each step (Moore and Sharp 1993; Steitz and Steitz 1993; Sontheimer et al. 1997). Konarksa and Query proposed that the two active-site conformations compete kinetically and that destabilization of one conformation drives the complex and catalysis toward the other state (Query and Konarska 2004). The competition between U2 stem I and helix Ib may be an illustration of this kinetic model. Because helix Ib must form during the second step of splicing, it is possible that the competing U2 stem I structure forms during the first step, within the context of a U2–U6 four-way helical junction (Sashital et al. 2004). The conformational change from a four- to three-helix junction could be driven by the melting of U2 stem I. In this way, U2 stem I could modulate formation of the second-step active site in the spliceosome.
MATERIALS AND METHODS
Preparation of RNA samples
Wild-type yeast and both human U2 stem I RNA sequences were obtained from Dharmacon, Inc., and deprotected as recommended. Yeast U2 stem I was also synthesized through in vitro transcription using purified His6-tagged T7 RNA polymerase, synthetic DNA oligonucleotide templates (Integrated DNA Technologies), and commercially available unlabeled (Sigma-Aldrich) or 13C/15N-labeled (Cambridge Isotope Labs) NTPs. The first two base pairs of the yeast sequence were substituted with G-C pairs to optimize transcription yields. All RNAs were purified on denaturing 17% polyacrylamide gels, DEAE anion exchange, and a G-15 Sephadex desalt column as previously described. RNAs were lyophilized and resuspended in water, and pH was adjusted to 7.0 using 1 M NaOH. Partial alignment of modified human and 13C/15N-labeled yeast RNAs for residual dipolar coupling measurements was achieved by addition of 17 mg/mL Pf1 filamentous bacteriophage (ASLA Biotech, Ltd.) in deuterium oxide.
NMR spectroscopy
All spectra were collected on a 750 MHz Bruker Avance DMX or 900 MHz Varian Inova spectrometer at the National Magnetic Resonance Facility (University of Wisconsin–Madison), equipped with a single z-axis gradient HCN cryoprobe and conventional room-temperature probe, respectively. Exchangeable proton resonances were assigned by reference to 2D NOESY spectra, and collected with a 150-msec mixing time at 283 K for RNA at pH 7.0 in 90% H2O/10% D2O using a 1–1 spin-echo water suppression scheme. Ambiguous imino proton assignments for uracils involved in U-U wobbles in yeast U2 stem I were confirmed using a combination of 2D H1-C4 H(N)CO (298 K at pH 7.0, 90% H2O/10% D2O, water suppression using a water flip-back pulse) (Muhandiram and Kay 1994) and 2D H5-C4 H(C)C (298 K at pH 7.0, 99.99% D2O) (Fiala et al. 2004) spectra. Nonexchangeable proton resonances were assigned by reference to 2D NOESY (mixing times: 25, 50, 100, 150, 250, and 350 msec) and 2D (1H–1H) TOCSY for all RNA constructs in 99.99% D2O at pH 7.0 and 303 K. Assignments were confirmed for modified human U2 stem I RNA by reference to 2D (13C–1H) HSQC spectra collected at natural 13C abundance, and for yeast U2 stem I RNA by reference to 2D (13C–1H) HSQC, 3D (1H–13C–1H) TOCSY, and 3D (1H–13C–1H) COSY spectra collected for a fully 13C/15N-labeled sample.
Residual dipolar couplings for the modified human U2 stem I RNA were measured using natural abundance (13C–1H) HSQC spectra with coupling in the proton dimension. RDCs for fully 13C/15N-labeled yeast U2 stem I RNA were measured using J-modulated (13C–1H) CT-HSQC (20 planes) (Ottiger et al. 1998) and resolution-optimized homonuclear 1H–1H decoupled-TROSY and TROSY-ECOSY spectra (Boisbouvier et al. 2004).
Melting of purified wild-type S. cerevisiae, unmodified human, and modified human U2 stem I RNAs (0.4–0.6 mM RNA, 90% H2O/10% D2O at pH 7) was monitored by collecting a series of 1D 1H spectra (1–1 echo water suppression) at increasing temperature. Data were collected on a Bruker 500 MHz spectrometer equipped with a single z-axis gradient HCN conventional probe. The temperature was increase by 5°C from 5°C to 80°C, or until imino peaks were no longer visible. All spectra were collected with 16 scans.
Structure calculations
NOE distance restraints were qualitatively categorized as strong (1.8–3.0 Å), medium (2.0–4.5 Å), or weak (3.0–5.5 Å) based on NOESY spectra with mixing times of 50, 150, 250, and 350 msec. Torsion angle restraints were set to standard A-form values in all Watson–Crick regions (±15°). No A-form torsion angle restraints were used for the wobble pairs or tetraloops. The use of A-form torsion angle restraints in the Watson–Crick base-paired regions of the molecules was validated through structure calculations in the complete absence of A-form restraints, with the exception of the experimentally validated glycosidic bond angle (χ) and sugar pucker (ν1 and ν2). The structures without A-form torsion angle restraints are essentially identical to those solved with the A-form restraints, with only slightly higher RMSD values. The 20 lowest-energy structures without A-form restraints converged to RMSD values of 1.34 Å and 0.90 Å (superimposed over all heavy atoms, excluding the tetraloop) for human and yeast, respectively. The corresponding RMSD values obtained with the A-form restraints are 0.98 Å and 0.63 Å (Table 1). Importantly, the structures with and without A-form restraints overlay with an RMSD of 1.24 Å and 0.85 Å for human and yeast, respectively. These results indicated that the geometries of the wobble pairs were not influenced by the A-form restraints for the Watson–Crick regions of the molecule. The potential impact of including nonexperimental A-form restraints on the Watson–Crick pairs was analyzed for the tetraloops in an analogous manner. It was found that the tetraloop structures are essentially identical with and without the A-form dihedral restraints on the adjacent Watson–Crick base pairs. The RMSD values for the 20 lowest-energy tetraloop structures (including the adjacent Watson–Crick base pair) calculated in the presence and absence of A-form restraints were 2.06 Å and 1.38 Å for human and yeast, respectively, which is quite close to the values obtained for the individual ensembles calculated with A-form restraints (2.08 Å and 1.03 Å for human and yeast, respectively) and without A-form restraints (1.88 Å and 1.43 Å for human and yeast, respectively). Therefore, we conclude that the structure of the tetraloop was not influenced by the A-form restraints on the adjacent helical nucleotides. The internal loop and tetraloop backbone dihedral angles were unrestrained in all structure calculations, with the exception of the experimentally validated glycosidic bond angles and sugar puckers. The sugar pucker angles (ν1, ν2) for the last three uracil residues in the tetraloop were initially unrestrained. Based on strong H1′–H2′ signals in the 2D 1H–1H TOCSY spectra (40 msec mixing time) and ribose conformations in converged initial structures, the last three uracil residues in the tetraloop were restrained as C2′-endo (±30°) in subsequent calculations. All other residues were restrained as C3′-endo.
Hydrogen-bond restraints were used for all Watson–Crick pairs and the human Gm-U wobble. Initial structures were calculated without hydrogen-bond restraints for the C-A wobble in human and the U-U wobbles in yeast in order to confirm the conformation of the base pairs based solely on NOE and RDC information. For the C-A wobble, a hydrogen bond between the adenine amino and cytosine N3 was confirmed in the converged initial structures. For the U-U wobbles, the hydrogen-bonding pattern detected by correlation experiments (C2 carbonyl hydrogen-bond acceptors for U11 and U23; C4 carbonyl hydrogen-bond acceptors for U12 and U24) was confirmed in the converged initial structures. These hydrogen bonds were restrained in all subsequent structure calculations. Very weak planarity restraints (1 kcal mol−1 Å−2) were used for all Watson–Crick pairs, but not for the wobble pairs.
Angle and bond parameters as well as atomic charges for modified nucleotides were calculated with Gaussian version 98 (http://www.gaussian.com) at Hartree–Fock levels using a 6-31G* basis set for previously described nucleotide geometries (Cornell et al. 1995). An extended structure generated in CNS 1.1 (Brunger et al. 1998) was used to calculate 100 starting structures using NOE, dihedral, hydrogen bonding, and planarity restraints. Structures were subjected to 60 psec (15-fsec time steps) of restrained molecular dynamics in torsion angle space, 90 psec of slow cooling, and 30 psec (5-fsec time steps) of restrained molecular dynamics in Cartesian coordinate space. The 100 structures were then refined with additional RDC restraints using XPLOR-NIH (Schwieters et al. 2003). Structures were heated to 3000 K and cooled to 100 K in 58 cycles for 28 psec of restrained molecular dynamics in Cartesian coordinate space, followed by 500 steps of energy minimization using the Powell algorithm and simulated annealing.
Structures were selected based on criteria of lowest overall energy and lack of NOE violations >0.2 Å and dihedral violations >5°. Mean structures were calculated in MOLMOL (Koradi et al. 1996) and energy minimized in Amber 9 (http://amber.scripps.edu/). Electrostatic surface potentials for mean structures were created using QNIFFT (Chin et al. 1999) and visualized using the APBS tool (www-personal.umich.edu/∼mlerner/PyMOL/index.html) in PyMol (www.pymol.org). Figures were created using PyMol.
Optical spectroscopy
Thermal stability of purified wild-type S. cerevisiae, unmodified human, and modified human U2 stem I RNAs was monitored using a Cary Model 100 Bio UV-visible spectrophotometer equipped with a Peltier heating accessory and temperature probe. Samples containing 10 mM potassium phosphate buffer (pH 6.0 or 7.0), 100 mM KCl, and 4 μM RNA were heated to 95°C and cooled to 5°C at a rate of 1°C/min. Absorbance at 260 nm was detected in 1°C increments. Scans were run on three samples for each RNA. The data were derivatized, smoothed (∼3°C), normalized (with primary peak set to 1), and fit using nonlinear least-square fit to solve for T m and ΔH as previously described (John and Weeks 2000). Each transition was fit separately in order to deconvolute the individual melting transitions. Data analysis was performed using Prism 4.0 (GraphPad).
Coordinates
Coordinates for the human and yeast U2 stem I have been deposited into the Protein Data Bank (accession codes 2O32 for human and 2O33 for yeast). NMR chemical shift assignments for both RNAs have been deposited into the BioMagResBank (accession code 15081 for human and 15080 for yeast).
ACKNOWLEDGMENTS
We thank Frits Abildgaard, Marco Tonelli, and the National Magnetic Resonance Facility at Madison (NMRFAM) staff for technical support. We also thank Jared Davis, Ryan Marcheschi, Nicholas Reiter, David Staple, and Kirk Vander Meulen for helpful discussions. All NMR studies were performed at the NMRFAM (www.nmrfam.wisc.edu) with support from U.S. National Institutes of Health (NIH) grants P41RR02301 (Biomedical Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment funding for the facility was provided by the University of Wisconsin, the National Institutes of Health (P41GM66326, P41RR02301, RR02781, RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture. D.G.S. was supported by NIH Predoctoral training grant T32 GM007215, a Sullivan Wisconsin Distinguished Graduate Fellowship, and a Wharton Predoctoral Fellowship. This work was supported by NIH grant GM65166 to S.E.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.418407.
REFERENCES
- Antao, V.P., Lai, S.Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M., Jr, Igel, A.H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes & Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Blad, H., Reiter, N.J., Abildgaard, F., Markley, J.L., Butcher, S.E. Dynamics and metal ion binding in the u6 RNA intramolecular stem–loop as analyzed by NMR. J. Mol. Biol. 2005;353:540–555. doi: 10.1016/j.jmb.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Boisbouvier, J., Bryce, D.L., O'Neil-Cabello, E., Nikonowicz, E.P., Bax, A. Resolution-optimized NMR measurement of 1DCH, 1DCC and 2DCH residual dipolar couplings in nucleic acid bases. J. Biomol. NMR. 2004;30:287–301. doi: 10.1007/s10858-005-1846-5. [DOI] [PubMed] [Google Scholar]
- Brosi, R., Groning, K., Behrens, S.E., Luhrmann, R., Kramer, A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grossekunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system—A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cai, Z., Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- Cao, S., Chen, S.J. Free-energy landscapes of RNA/RNA complexes: With applications to snRNA complexes in spliceosomes. J. Mol. Biol. 2006;357:292–312. doi: 10.1016/j.jmb.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, K., Sharp, K.A., Honig, B., Pyle, A.M. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., Kollman, P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Donmez, G., Hartmuth, K., Luhrmann, R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybkov, O., Will, C.L., Deckert, J., Behzadnia, N., Hartmuth, K., Luhrmann, R. U2 snRNA–protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol. Cell. Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala, R., Munzarova, M.L., Sklenar, V. Experiments for correlating quaternary carbons in RNA bases. J. Biomol. NMR. 2004;29:477–490. doi: 10.1023/B:JNMR.0000034358.12599.d1. [DOI] [PubMed] [Google Scholar]
- Flinders, J., Dieckmann, T. The solution structure of the VS ribozyme active site loop reveals a dynamic “hot-spot.”. J. Mol. Biol. 2004;341:935–949. doi: 10.1016/j.jmb.2004.06.084. [DOI] [PubMed] [Google Scholar]
- Furtig, B., Richter, C., Wohnert, J., Schwalbe, H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., Patterson, B. Spliceosomal snRNAs. Annu. Rev. Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hilliker, A.K., Staley, J.P. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura, M., Kay, L.E., Bax, A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: Heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- John, D.M., Weeks, K.M. van't Hoff enthalpies without baselines. Protein Sci. 2000;9:1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, G., Yamamoto, Y., Kamimura, T., Masegi, T., Sekine, M., Hata, T., Iimori, T., Watanabe, T., Miyazawa, T., Yokoyama, S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- Koplin, J., Mu, Y., Richter, C., Schwalbe, H., Stock, G. Structure and dynamics of an RNA tetraloop: A joint molecular dynamics and NMR study. Structure. 2005;13:1255–1267. doi: 10.1016/j.str.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lee, C.H., Tinoco I., Jr Studies of the conformation of modified dinucleoside phosphates containing 1,N6-ethenoadenosine and 2′-O-methylcytidine by 360-MHz 1H nuclear magnetic resonance spectroscopy. Investigation of the solution conformations of dinucleoside phosphates. Biochemistry. 1977;16:5403–5414. doi: 10.1021/bi00644a001. [DOI] [PubMed] [Google Scholar]
- Legault, P., Pardi, A. Unusual dynamics and pK a shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 1997;119:6621–6628. [Google Scholar]
- Lietzke, S.E., Barnes, C.L., Berglund, J.A., Kundrot, C.E. The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure. 1996;4:917–930. doi: 10.1016/s0969-2126(96)00099-8. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Guthrie, C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Massenet, S., Mougin, A., Branlant, C. Post-transcriptional modifications in the U small nuclear RNAs. In: Grosjean H., Benne R., editors. Modifications and editing of RNA. ASM Press; Washington, DC: 1998. pp. 201–228. [Google Scholar]
- McPheeters, D.S., Abelson, J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992;71:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- Moore, M.J., Sharp, P.A. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Muhandiram, D.R., Kay, L.E. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B. 1994;103:203–216. [Google Scholar]
- Ottiger, M., Delaglio, F., Marquardt, J.L., Tjandra, N., Bax, A. Measurement of dipolar couplings for methylene and methyl sites in weakly oriented macromolecules and their use in structure determination. J. Magn. Reson. 1998;134:365–369. doi: 10.1006/jmre.1998.1546. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Konarska, M.M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek, R., Turner, D.H. Stabilities of consecutive A·C, C·C, G·G, U·C, and U·U mismatches in RNA internal loops: Evidence for stable hydrogen-bonded U·U and C·C·+ pairs. Biochemistry. 1991;30:8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- Sashital, D.G., Cornilescu, G., McManus, C.J., Brow, D.A., Butcher, S.E. U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., Marius Clore, G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Sun, S., Piccirilli, J.A. Metal ion catalysis during splicing of premessenger RNA. Nature. 1997;388:801–805. doi: 10.1038/42068. [DOI] [PubMed] [Google Scholar]
- Staley, J.P., Guthrie, C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Steitz, T.A., Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J.S., Manley, J.L. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., Trantirek, L., Feigon, J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Manley, J.L. Multiple functional domains of human U2 small nuclear RNA: Strengthening conserved stem I can block splicing. Mol. Cell. Biol. 1992;12:5464–5473. doi: 10.1128/mcb.12.12.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., McDowell, J.A., Turner, D.H. A periodic table of symmetric tandem mismatches in RNA. Biochemistry. 1995;34:3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- Xia, T., McDowell, J.A., Turner, D.H. Thermodynamics of nonsymmetric tandem mismatches adjacent to G·C base pairs in RNA. Biochemistry. 1997;36:12486–12497. doi: 10.1021/bi971069v. [DOI] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., Steitz, J.A. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Sun, X., Watt, E.D., Al-Hashimi, H.M. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]