Abstract
A previous polymorphism survey of the type 2 diabetes gene CAPN10 identified a segment showing an excess of polymorphism levels in all population samples, coinciding with localized breakdown of linkage disequilibrium (LD) in a sample of Hausa from Cameroon, but not in non-African samples. This raised the possibility that a recombination hotspot is present in all populations and we had insufficient power to detect it in the non-African data. To test this possibility, we estimated the crossover rate by sperm typing in five non-African men; these estimates were consistent with the LD decay in the non-African, but not in the Hausa data. Moreover, resequencing the orthologous region in a sample of Western chimpanzees did not show either an excess of polymorphism level or rapid LD decay, suggesting that the processes underlying the patterns observed in humans operated only on the human lineage. These results suggest that a hotspot of recombination has recently arisen in humans and has reached higher frequency in the Hausa than in non-Africans, or that there is no elevation in crossover rate in any human population, and the observed variation results from long-standing balancing selection.
LINKAGE disequilibrium (LD), the nonrandom association of linked alleles, has attracted great attention in human genetics because of the hope that LD-based association studies may help dissect the genetic basis of common diseases (Risch and Merikangas 1996). Local LD patterns are influenced by a broad range of different factors, which include variation in recombination and mutation rate, natural selection, and demography (Pritchard and Przeworski 2001). Indeed, fine-scale variation in LD decay with distance between sites is well documented in the human genome, and this variation was shown to exceed what is expected if mutation and recombination occurred at a uniform rate and all variation were evolutionarily neutral (Crawford et al. 2004; McVean et al. 2004; Altshuler et al. 2005; Myers et al. 2005). Moreover, recent analyses of LD using population-genetic approaches have suggested that there is extensive heterogeneity in recombination rates over the scale of kilobases (Crawford et al. 2004; McVean et al. 2004; Altshuler et al. 2005; Myers et al. 2005).
The notion that variation in LD decay is due largely to variation in recombination rates was bolstered by the analysis of recombinants in sperm samples (Hubert et al. 1994; Arnheim et al. 2003). Sperm-typing studies have demonstrated that crossover events cluster in narrow intervals of 1–2 kb corresponding to regions of localized breakdown of LD (Han et al. 2000; Jeffreys et al. 2001). Moreover, they showed that substantial interindividual variability in recombination rate exists, raising the possibility that hotspots may be short-lived features of the human genome (Yu et al. 1996; Jeffreys and Neumann 2002). Recent cross-species comparisons of LD patterns have further shown that there is little overlap between inferred hotspots in humans and closely related nonhuman primates (Wall et al. 2003; Ptak et al. 2004a, 2005; Winckler et al. 2005). These findings suggest that hotspots evolve quickly, at least relative to the divergence time of human and chimpanzee. However, even though recombination rates on a fine scale appear to have changed substantially over evolutionary time, on a larger scale recombination rates are relatively similar across populations and possibly across species (Myers et al. 2005; Ptak et al. 2005; Serre et al. 2005).
Despite the general concordance between LD-based estimates of recombination rate and empirical estimates based on genetic maps or sperm typing, interesting deviations have also been detected. LD and sperm-typing analysis of a region of 206 kb on chromosome 1 revealed several hotspots in areas of strong LD (in addition to hotspots in regions of rapid LD decay) (Jeffreys et al. 2005). Because LD analyses estimate average recombination rates over both sexes and over long-term evolutionary time, while sperm-based estimates are specific to a sample of contemporary males, the observed discrepancies may reflect the presence of hotspots that are too young to have left a signature in LD patterns. The opposite deviation, i.e., low sperm-typing estimates of recombination rate in regions of rapid LD decay, has also been observed (Jeffreys et al. 2005; Kauppi et al. 2005). Proposed explanations for these findings include unusually large differences in male- vs. female-specific recombination rate, a polymorphic hotspot that is absent in the men sampled for sperm analysis, or an extinct hotspot that was active in the relatively recent past, such that patterns of LD are still shaped by it. While these explanations are plausible, the possible role of natural selection in leading to apparent variation in recombination rate has received little attention (but see Reed and Tishkoff 2006 and Stephan et al. 2006).
A previous resequencing study of CAPN10, a susceptibility gene for type 2 diabetes, detected a local excess of polymorphism levels in all populations examined; the same segment also showed localized breakdown of LD in one of the surveyed populations (Hausa of Cameroon) (Vander Molen et al. 2005). This pattern was shown to be inconsistent with a coalescent model with uniform recombination rate and neutral evolution; specifically, a sliding-window analysis of an estimate of the population crossover rate parameter showed that the ratio of the maximum to the median window value was significantly larger than expected by chance in a random-mating population of constant size (Vander Molen et al. 2005). We previously showed that a model of long-standing balancing selection maintaining multiple alleles fit the CAPN10 Hausa data, for up to nine selected alleles (Takahata 1990; Vander Molen et al. 2005). A selection scenario is particularly relevant because the genetic susceptibility to type 2 diabetes is widely thought to be the result of “thrifty” alleles that conferred a selective advantage by increasing the efficiency of energy use and storage. Diabetes risk variants may therefore be metabolic adaptations to an ancient lifestyle characterized by feast and famine (Neel 1962). Thus, it is plausible that balancing selection resulted in a local elevation of polymorphism levels and rapid LD decay in an ancestral human population prior to the dispersal out of Africa, with a signature in LD decay retained only in the African population.
An alternative scenario is that the pattern of LD at CAPN10 is due to the presence of a recombination hotspot present in all populations, but detected only in one. Testing this neutral explanation is the major focus of the work presented here. To this end, we used a combination of empirical and statistical approaches to evaluate whether the LD patterns could be due to a recombination hotspot. More specifically, we measured the crossover rate in sperm samples and analyzed the LD data to explicitly test for the presence of a hotspot; we show that the crossover rate estimates based on sperm typing are consistent with LD patterns in the non-African samples, but not in the Hausa data. In addition, by resequencing the orthologous region in a population sample of Western chimpanzees, we show that the excess of polymorphism levels and LD decay is human specific.
MATERIALS AND METHODS
DNA samples:
DNA from 35 semen samples was extracted using the Puregene DNA isolation kit (Gentra Systems, Research Triangle Park, NC); these samples were from anonymous donors of non-African ancestry. DNA concentrations were determined spectrophotometrically. However, the number of amplifiable genomes per mass unit of DNA may vary from sample to sample due to a variety of reasons (including fragmentation) and, in turn, affect the number of effective recombinants detected in sperm-typing assays. Hence, we used kinetic (kt) PCR with primers upstream and downstream of the region of study to compare the number of amplifiable genomes of each sperm sample to a high-quality genomic DNA control (BD Biosciences). The number of genomes was then adjusted to the measured number of amplifiable genomes (see supplemental data at http://www.genetics.org/supplemental/). The use of sperm samples from anonymous donors was approved by the Institutional Review Boards of the University of Southern California and of the University of Chicago.
Whole blood samples (5–10 ml) were taken from 15 chimpanzees during routine veterinary examinations, and DNA was isolated using a standard phenol/chloroform-based extraction protocol. The chimpanzees in this sample are wild born and were exported from Africa prior to the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) enacted in 1975. The geographic origins of the individuals in the sample are unknown, except that two individuals were probably captured in Sierra Leone. Subspecies status was determined through mitochondrial DNA and Y chromosome sequences (Stone et al. 2002 and our unpublished data). All of the chimpanzees in this sample were identified as Western chimpanzees (Pan troglodytes verus) and were sequenced at intron 13 of CAPN10 to examine the patterns of polymorphism and LD in a close outgroup species. These results were compared to previously published resequencing data for a segment of 33,465 bp in 48 humans (Vander Molen et al. 2005), comprising equal numbers of Hausa of Cameroon, Han Chinese, and Italians. All nucleotide positions in this article refer to reference sequence AF158748.
Sequencing and haplotyping of sperm samples:
Heterozygous individuals for two pairs of SNPs flanking each side of the region of study were identified by sequencing. The four SNPs selected were at positions 30952 (C/T) and 31449 (C/G) 5′ of the target region and 35020 (C/A) and 35309 (G/C) 3′. Of the 35 sperm samples, we identified five heterozygotes at all four SNPs (test individuals) as well as two nonrecombinant homozygotes; the latter were used as negative controls.
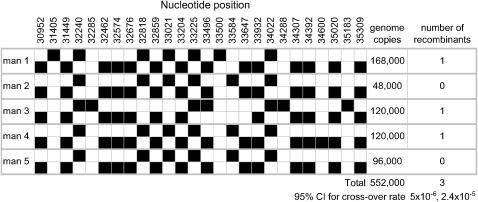
By directly sequencing the first allele-specific (AS)–PCR product (see below), it was possible to establish the haplotype phase for the internal SNPs at nt positions (pos.) 31,449 and 35,020, as well as all other SNPs present in these individuals (Figure 1). All test individuals have the same nonrecombinant haplotype configuration at the four flanking SNPs (C-C-C-G and T-G-A-C).
Figure 1.—
Visual haplotypes of the five informative individuals assayed by sperm typing. The ancestral and derived alleles, inferred on the basis of a sequence alignment of nine primate species (Clark et al. 2005), are shown as open and solid cells, respectively. Nucleotide positions of each polymorphic site are shown across the top. Haplotype phase was assigned by sequencing the AS–PCR product for each individual.
Recombination detection in sperm DNA:
We measured the recombination fraction of a 4357-bp region in intron 13 of CAPN10 located on chromosome 2q37 (centered on nt pos. 33,100 in AF158748, i.e., nt pos. chr2:241261505 in hg17) by counting the number of recombinant molecules in pools of 3000 sperm genomes each using long-range AS–PCR (Cheng et al. 1994; Han et al. 2000; Jeffreys et al. 2001; Arnheim et al. 2003). The recombinant molecules were selectively amplified in two rounds of PCR using AS primers (Operon Biotechnologies, Alameda, CA) (see supplemental data at http://www.genetics.org/supplemental/). AS primers were designed so that the 3′ end of the primer forms a mismatch with the abundant nonrecombinant molecules and a perfect match with one of the recombinant alleles. This design allows for the preferential amplification of a single recombinant copy over the nonrecombinant genomes, at the appropriate dilution. To increase the specificity of the PCR the last four phosphate bonds in the primer were phosphorothiolated and a mismatch substitution was added 2 or 3 bases from the 3′ end of each second-round primer. However, because we were unable to achieve complete allele specificity for haplotype T-G-C-G, the crossover rate could not be measured for this recombination product. Throughout, we assume that both products of a crossover are produced at the same frequency as observed in other sperm-typing studies in which both crossover products were evaluated (Jeffreys and Neumann 2002).
AS–PCR was performed on the informative individuals using the Expand Long Template PCR system (Roche, Indianapolis) (see supplemental data at http://www.genetics.org/supplemental/). For each of the five test sperm samples, we amplified in the same experiment multiple independent aliquots of each test sample, an equal number of negative controls (containing both nonrecombinant haplotypes), and a set of positive controls consisting of 13 aliquots with 1 recombinant molecule on average in 3000 nonrecombinants (1:3000 dilution controls) and 1 aliquot containing 30 recombinant molecules in 3000 nonrecombinants (1:10 dilution control). For each sample, the total amount of sperm DNA used was adjusted to account for variation in the number of amplifiable genomes (as described above). In all experiments, the first round and the nested second round of PCR were set up in different biological safety cabinets to minimize cross-contamination.
Two methods were used to detect recombinant molecules: visualization of PCR products on an agarose gel or evaluation by kt–PCR. For the gel detection method, an experiment was considered successful if the 1:3000 dilution controls produced the Poisson-expected number of positives (50–65% positives) and there was little or no evidence of non-AS–PCR in the negative or nonrecombinant controls (see supplemental Figure 1 at http://www.genetics.org/supplemental/). kt–PCR was performed on an ABI7700 or an ABI7900 real-time PCR machine (Applied Biosystems, Foster City, CA). Amplification plots were evaluated using Sequence Detection Systems (Applied Biosystems) software (v. 1.9 or 2.1) to determine the cycle threshold (Ct) (i.e., the cycle at which the PCR yield achieves a user-specified threshold) for each PCR reaction. A kt–PCR experiment was considered successful if there was a distinct difference between positive and negative PCR reactions (i.e., the latest positives reached the specified Ct value at least three cycles before the earliest negatives) and the expected proportions of 1:3000 dilution reactions were positive (see supplemental Figure 2 at http://www.genetics.org/supplemental/).
Resequencing survey in Western chimpanzees:
A segment of 10,268 bp, which aligns to the human reference sequence AF158748 between nt pos. 26,079 and 36,346, was amplified and sequenced as previously described (Vander Molen et al. 2005). Where necessary, additional primers were designed on a chimpanzee reference sequence to fill in gaps. All sequences were aligned and visualized using the Phred/Phrap/Consed system and scored using Polyphred (Nickerson et al. 1997); sequence chromatograms were visually inspected to confirm the putative polymorphisms and all genotype calls. The total length of scored sequence was 10,171 bp.
Statistical analyses:
Summary statistics of polymorphism were calculated using the online program SLIDER (http://genapps.uchicago.edu/slider/index.html). The population crossover rate parameter (ρ = 4Nec, where Ne is the effective population size and c the crossover rate) was estimated for the segment of ∼10 kb centered on the peak of 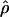 (Vander Molen et al. 2005), using the online program MAXDIP (http://home.uchicago.edu/∼rhudson1/source.html), which implements the composite-likelihood estimator described in Hudson (2001). Following Frisse et al. (2001), we assumed that recombination events resolve either as gene conversion or as crossover and that the gene conversion tract lengths are exponentially distributed with mean length L bp. The model therefore has three parameters: the population crossover rate per base pair, ρ = 4Nec; the ratio of gene conversion to crossover events, f; and L. Since simulations suggested that there is not enough information in polymorphism data from a single locus to coestimate the three parameters (Frisse et al. 2001; Ptak et al. 2004b; Wall 2004), we considered different models of gene conversion and estimated ρ under each. Specifically, we set f = 0 (i.e., no gene conversion), f = 2 and L = 500 bp, or f = 5 and L = 60 bp (Jeffreys and May 2004; Ptak et al. 2004b). In each case, we estimated ρ on a grid from 0 to 0.01 (with an increment of 10−5). We ran MAXDIP using the ancestral allele information as inferred from outgroup sequences and we excluded the two microsatellite loci in humans. The estimates of ρ obtained in this way are denoted ρLD. To derive an estimate of the crossover rate, we divided ρLD by 4Ne* for each population, where Ne* is an estimate of the effective population size (11,990 for Hausa, 9028 for Italians, 8323 for Chinese, and 8019 for Western chimpanzees). This estimate of Ne* was obtained from diversity (the number of segregating sites) and sequence divergence levels (using the orangutan) for the surveyed segment. We assumed 6 MYA to the common ancestor of human and chimpanzee and 25 and 15 years per generation for the two lineages, respectively.
(Vander Molen et al. 2005), using the online program MAXDIP (http://home.uchicago.edu/∼rhudson1/source.html), which implements the composite-likelihood estimator described in Hudson (2001). Following Frisse et al. (2001), we assumed that recombination events resolve either as gene conversion or as crossover and that the gene conversion tract lengths are exponentially distributed with mean length L bp. The model therefore has three parameters: the population crossover rate per base pair, ρ = 4Nec; the ratio of gene conversion to crossover events, f; and L. Since simulations suggested that there is not enough information in polymorphism data from a single locus to coestimate the three parameters (Frisse et al. 2001; Ptak et al. 2004b; Wall 2004), we considered different models of gene conversion and estimated ρ under each. Specifically, we set f = 0 (i.e., no gene conversion), f = 2 and L = 500 bp, or f = 5 and L = 60 bp (Jeffreys and May 2004; Ptak et al. 2004b). In each case, we estimated ρ on a grid from 0 to 0.01 (with an increment of 10−5). We ran MAXDIP using the ancestral allele information as inferred from outgroup sequences and we excluded the two microsatellite loci in humans. The estimates of ρ obtained in this way are denoted ρLD. To derive an estimate of the crossover rate, we divided ρLD by 4Ne* for each population, where Ne* is an estimate of the effective population size (11,990 for Hausa, 9028 for Italians, 8323 for Chinese, and 8019 for Western chimpanzees). This estimate of Ne* was obtained from diversity (the number of segregating sites) and sequence divergence levels (using the orangutan) for the surveyed segment. We assumed 6 MYA to the common ancestor of human and chimpanzee and 25 and 15 years per generation for the two lineages, respectively.
The method of McVean et. al. (2004) was used to examine rate variation across the ∼33.5-kb segment surveyed in humans (Vander Molen et al. 2005) and the ∼10-kb subsegment surveyed in Western chimpanzees. As in McVean et. al. (2004), a smoothing parameter of 20 was used in the analysis. A range of smoothing parameters between 15 and 50 was investigated, and similar results were observed (results not shown).
We also assessed the support in polymorphism data for a recombination hotspot using the method of Crawford et al. (2004), as implemented in PHASE 2.2.1. In humans, we used variation in the entire ∼33.5-kb segment to estimate recombination rate parameters for the ∼10-kb subsegment. For this analysis, we assumed there is at most one hotspot, of unknown location, and then estimated the intensity of the hotspot (λ) and its location, as well as the total recombination rate. We assessed the support for the presence of a hotspot by estimating the posterior probability that λ > 5 and λ > 10, in effect defining a hotspot as a short segment that experiences a uniform rate of genetic exchange that is greater than five times the background rate; the prior distributions were as in Crawford et al. (2004). To compare the crossover rates estimated by PHASE to those obtained from sperm typing, we divided the estimate of ρ for the ∼10-kb segment by 4Ne*.
It is important to note that all three methods for estimating recombination parameters rely on a simple model of a random-mating population of constant size, the method of McVean et al. (2004) and MAXDIP explicitly and PHASE implicitly (Hudson 2001; Li and Stephens 2003). Simulations suggest that the estimates of ρ tend to decrease under a bottleneck model (Li and Stephens 2003; Smith and Fearnhead 2005). Simulations of a two-island model gave similar results, whether sampling from each island was equal or not (Li and Stephens 2003); it was also shown that estimates of ρ tend to decrease under a four-island model with equal sampling from all islands (Smith and Fearnhead 2005). However, the behavior of the estimators of ρ under more complex models of population structure that may apply to humans has not been investigated (Wakeley 1999; Wakeley et al. 2001).
To assess whether the estimates of crossover rate obtained from LD data and sperm typing are consistent, we generated 1000 simulated polymorphism data sets under a model of uniform recombination, with the rate obtained from sperm-typing experiments. We then estimated ρ using MAXDIP and tabulated the proportion of simulated data sets for which the estimate was as high or higher than that observed for the actual polymorphism data. Simulations were run using the software MS (Hudson 2002); the program produces a list of haplotypes, which were randomly paired to form genotypes. In the simulations, we matched the actual number of individuals sequenced (16 for each of three human populations and 15 for Western chimpanzees), the length of the surveyed segment (10,268 and 10,171 bp in humans and chimpanzees, respectively), and the fraction of missing genotype calls (1%) or missing ancestral information (one site in humans). Moreover, the value of θ = 4N0μ (where N0 is the effective population size at present and μ is the mutation rate) was chosen to yield the same average number of segregating sites as that observed. We considered the same three models of gene conversion as for the actual data and set ρ = 4N0c*, where c* is the upper 97.5th percentile estimate of crossover obtained from sperm typing. The point estimates ρLD were obtained under the same gene conversion models as those under which the simulated data were generated.
We ran two sets of simulations: (1) under the standard neutral model of a random-mating population of constant size and (2) under random-mating models of population size change shown to provide a good fit to a 50-locus data set collected in the same populations (Voight et al. 2005). The latter allowed us to examine the effect that population size changes might have on our estimate of ρLD. Specifically, we assumed a model of a constant population size followed by exponential growth to the present for the Hausa (Marjoram and Donnelly 1994; Pluzhnikov et al. 2002; Voight et al. 2005), while for the Chinese and Italians we used a simple bottleneck model where a population size decreases and then recovers to the same size (Fay and Wu 1999). Parameters were as follows: 2.1-fold growth starting 1000 generations ago for the Hausa, a 60% reduction in population size from 3200 to 200 generations ago for the Chinese, and a 50% reduction from 3200 to 100 generations ago for the Italians (cf. Voight et al. 2005). We also tried other bottleneck parameters and the qualitative conclusions were unchanged (results not shown). It should be noted that we did not consider models of population structure. It was previously shown that, under a simplified model of population structure, estimates of absolute ρ tend to be lower than those under random mating models, but that there is a slightly higher probability of incorrectly inferring a hotspot (Li and Stephens 2003); it remains possible that the latter effect is greater under more complex models of population structure (Wakeley 1999).
RESULTS
Estimation of crossing-over rate based on sperm typing:
We used the strategy for “recombination detection using total sperm DNA” described in (Arnheim et al. 2003) to measure the crossover rate in five informative sperm samples in a segment of 4357 bp centered on nt pos. 33,100 of the reference sequence. Sequencing of the AS–PCR products of the informative samples reveals a total of six different haplotypes (Figure 1). These haplotypes fall within three of the five major groups of haplotypes previously described in this segment of the CAPN10 gene and include the deepest lineages in the gene genealogy (Clark et al. 2005).
The AS–PCR experiments in the sperm samples yielded a total of three recombinants in 552,000 genomes (meioses), comprised of 184 pools of an estimated 3000 amplifiable genomes each. The distribution of the recombinants detected and the number of genomes queried in each individual are detailed in Figure 1. Since our AS–PCR assay recovers only one of the two possible recombination products, our results effectively correspond to finding six recombinants in 552,000 meioses. Therefore, the estimated crossover rate for the entire segment of 4357 bp is 1.09 × 10−5. Since the number of recombinants is expected to be binomially distributed, the 95% confidence interval for the crossover rate is ∼(5 × 10−6, 2.4 × 10−5) for the entire segment or (1.2 × 10−9, 5.4 × 10−9)/bp. Note that effectively this represents an estimate of the crossing-over rate, c, as gene conversion is unlikely to coconvert both SNPs targeted by the two rounds of AS–PCR.
Patterns of sequence variation in Western chimpanzees and humans:
A sliding-window analysis of resequencing data for the ∼33.5 kb spanning the CAPN10 gene identified a peak of polymorphism levels and LD decay in the Hausa population in intron 13 (nt pos. 32,000–33,000 of reference sequence AF158748); a peak of polymorphism levels was observed also in the other three population samples at approximately the same position (Vander Molen et al. 2005). Coalescent simulations showed that these peaks of polymorphism and LD decay are not consistent with a neutral model with uniform mutation and recombination rate (Vander Molen et al. 2005). Here, we have collected resequencing data for an orthologous subsegment of 10,268 bp, centered on the peak, in 15 Western chimpanzees (P. troglodytes verus). This subspecies was chosen because a previous analysis of noncoding regions showed that patterns of variation are in rough accordance with the expectation of a standard neutral model (Fischer et al. 2004). Summary statistics of the polymorphism data in each sample are shown in Table 1. To compare polymorphism levels across samples, we use the estimator θW of the population mutation rate parameter θ = 4Neμ that is based on the number of polymorphic sites and sample size (Watterson 1975), and for allele frequencies we use a summary of the folded frequency spectrum, Tajima's D (Tajima 1989a). The chimpanzees show two- to threefold lower polymorphism levels compared to humans and a slight skew toward rare variants. Both polymorphism levels and the frequency spectrum are within the range of previous polymorphism surveys in this chimpanzee subspecies (Yu et al. 2003; Fischer et al. 2004). Moreover, a sliding-window analysis of θW in the chimpanzee and human population samples shows distinct patterns of variation, with no prominent peak of polymorphism in the chimpanzees (data not shown).
TABLE 1.
Summary statistics of polymorphism data in the subsegment of ∼10 kb
| Estimates of c (×10−8) based on ρLDe
|
|||||||
|---|---|---|---|---|---|---|---|
| Sa | πb | θWc | Dd | f = 0 | f = 2, L = 500 | f = 5, L = 60 | |
| Western chimpanzees | 31 | 6.62 | 7.83 | −0.56 | 0.90 | 0.62 | 0.75 |
| Hausa | 85 | 15.05 | 21.11 | −1.08 | 13.99 | 5.92 | 8.69 |
| Italians | 64 | 17.20 | 15.89 | 0.31 | 2.88 | 1.55 | 2.22 |
| Chinese | 59 | 15.98 | 14.65 | 0.34 | 1.65 | 0.99 | 1.35 |
Number of polymorphic sites.
Nucleotide diversity (×10−4) (Tajima 1989b).
Watterson's estimator of the population mutation rate parameter θ = 4Neμ (×10−4) (Watterson 1975).
Tajima's D (Tajima 1989b).
Estimate of the crossover rate (×10−8) based on the composite-likelihood estimate of ρLD calculated using MAXDIP (see materials and methods).
Estimation of crossing-over rate based on LD data:
To characterize levels of LD in the surveyed segment, we estimated the population crossover rate parameter, ρ = 4Nec, under different models of gene conversion using the program MAXDIP (Hudson 2001). To obtain estimates of the crossing-over rate (c) (given in Table 1), each estimate of ρLD was divided by 4Ne* (where Ne* is an estimate of the effective population size based on levels of interspecies divergence and on the polymorphism levels observed in each population sample). Consistent with previous analyses, the point estimates of c for the Hausa are higher than those for both non-African populations (for all rates of gene conversion). The point estimates for the Western chimpanzees are lower than those obtained for all human populations. The consistency between estimates of c based on ρLD and those based on sperm typing is assessed in the next section.
To investigate the fine-scale variation in LD decay across the surveyed segment, we use the method of McVean et al. (2004), which extends the composite-likelihood estimator implemented in MAXDIP to estimate variation in ρ along the sequence. The method produces a smoothed map of the population crossover rate, with the smoothing parameter reflecting the prior assumption about the rate heterogeneity in the region. Consistent with previous analyses, this method detects a peak in estimates of ρ in the Hausa data approximately at nt pos. 32,500 of the reference sequence (Figure 2). No substantial peak is observed in the other human population samples (Figure 2) and there is no evidence of rate variation in the Western chimpanzees (data not shown). Interestingly, the same statistical methodology applied to genomewide SNP genotyping data in a pool of ethnically diverse samples (Myers et al. 2005) did not find evidence for a hotspot in CAPN10.
Figure 2.—
Estimates of the population crossing-over rate parameter (ρ) per base pair across the CAPN10 gene in Hausa, Italians, and Chinese. Estimates were obtained using the method of McVean et al. (2004).
To investigate explicitly the hypothesis that the pattern of LD decay in the Hausa is due to the presence of a recombination hotspot, we used the method of Crawford et al. (2004) to estimate the total crossover rate, the intensity of the hotspot (λ), and its location. An advantage of this method is that it provides an estimate of the uncertainty associated with estimates of ρ, therefore allowing for hypothesis testing. Table 2 reports the estimated 95% credible intervals for the total crossover rate (c), obtained by dividing the estimate of the total population recombination rate in the segment by 4Ne* (see materials and methods). Consistent with the results presented in Table 1, the credible interval for c obtained under the hotspot model is higher in the Hausa than in the other samples and does not overlap with estimates from the other three populations. The credible intervals of the Italians, Chinese, and Western chimpanzees, however, do show substantial overlap. Hence, consistent with previous reports (Hudson 2001; Li and Stephens 2003), the uncertainty around the point estimates of ρLD can be large and could significantly affect the interpretation of statistical tests if not properly taken into account. In agreement with other analyses suggesting unusual heterogeneity of recombination rate (sliding-window analyses in Vander Molen et al. 2005 and Figure 2), the results of PHASE provide moderate and very strong support, respectively, for a hotspot with λ > 10 and λ > 5 in the Hausa. In the non-African and Western chimpanzee data, in contrast, there is little support for a hotspot.
TABLE 2.
Estimated crossing-over rate (c) per base pair (×108) based on a hotspot model as implemented in the program PHASE
| Western chimpanzees | Hausa | Italians | Chinese | |
|---|---|---|---|---|
| Total rate, 2.5th percentile | 0 | 4.16 | 0.05 | 0 |
| Total rate, 97.5th percentile | 3.03 | 11.82 | 1.01 | 0.23 |
| Pr (λ > 5) | 0.06 | 0.98 | 0.22 | 0.24 |
| Pr (λ > 10) | 0.03 | 0.65 | 0.19 | 0.16 |
The prior probability for a hotspot was set to 0.5.
Comparison of crossing-over rates estimated on the basis of sperm-typing and LD data:
As shown in Table 1, the point estimates of c based on ρLD fall above the 95% confidence interval (i.e., 1.2 × 10−9, 5.4 × 10−9/bp) estimated by sperm-typing data for all population samples. However, when we consider an explicit hotspot model and the uncertainty of the ρLD estimates is taken into account (see Table 2), a discrepancy between estimates of c is observed only in the Hausa LD data. Although estimates of c based on ρLD depend on an estimate of Ne, they remain inconsistent with those based on sperm typing unless we assume a Ne* of ∼92,000, a value over seven times higher than that computed from diversity estimates and much higher than that estimated for other regions (Frisse et al. 2001; Voight et al. 2005). Hence, the difference between estimates of c is unlikely to reflect error in Ne* alone.
In non-African populations, the estimate of c from sperm typing falls within the 95% credible intervals estimated from LD, using the hotspot model (Table 2). However, the point estimates obtained under the method of Hudson (2001) are almost an order of magnitude higher than expected (Table 1). To examine whether this reflects error in the estimator, we simulated data sets of sequence variation under a null model of uniform recombination rate and neutral evolution, assuming that crossover occurs at the highest rate compatible with our sperm-typing estimates (i.e., the upper boundary of the 95% confidence interval), and used MAXDIP to estimate ρ in each replicate. The simulations were performed under a standard neutral model of constant population size and random mating as well as under other demographic models shown to provide a reasonable fit to noncoding sequence variation data for each of the non-African population samples (Voight et al. 2005). We then calculated the proportion of simulated data sets in which ρLD is as high or higher than that observed. The results show that, under demographic models appropriate for the non-African samples, a point estimate of ρLD as high as that reported in Table 1 for Italians and Chinese is not unusual (Table 3); this suggests that the difference between point estimates of ρLD and the sperm-typing estimates is due to chance. Moreover, the data in the Western chimpanzees are compatible with the estimates of c obtained by sperm typing in humans (Table 3).
TABLE 3.
The proportion of 1000 simulated data sets in which the estimate of ρLD was as high or higher than that observed
| Western chimpanzees | Hausa | Italians | Chinese | |
|---|---|---|---|---|
| Standard neutral | ||||
| f = 0 | 0.530 | 0.000 | 0.068 | 0.261 |
| f = 2, L = 500 | 0.592 | 0.000 | 0.138 | 0.364 |
| f = 5, L = 60 | 0.575 | 0.000 | 0.115 | 0.315 |
| Other demographya | ||||
| f = 0 | — | 0.000 | 0.290 | 0.326 |
| f = 2, L = 500 | — | 0.000 | 0.325 | 0.390 |
| f = 5, L = 60 | — | 0.000 | 0.278 | 0.339 |
Includes a model of constant population size followed by exponential growth for the Hausa and a bottleneck model for the Italians and the Chinese (see materials and methods).
DISCUSSION
Our previous analysis of sequence variation in the CAPN10 gene detected an excess of polymorphism levels in all population samples examined, a pattern shown to be consistent with the action of long-standing balancing selection (Vander Molen et al. 2005). Localized LD breakdown, also consistent with long-standing balancing selection in a randomly mating population, was observed only in the Hausa population sample. Here, we showed that the LD breakdown in the Hausa (but not in the non-African samples) is also consistent with the presence of a hotspot of recombination. Moreover, when we directly measured the crossover rate in five sperm samples of non-African descent, we obtained estimates that are too low to explain the rapid LD decay in the Hausa, but are consistent with LD data in non-Africans. Finally, resequencing data in a sample of Western chimpanzees showed that there is neither an excess of polymorphism levels nor localized LD breakdown, suggesting that the processes underlying the patterns observed in humans operated only on the human lineage. These results may reflect the presence of a newly evolved hotspot that occurs at sufficiently different frequencies in Hausa vs. non-African populations to result in population-specific patterns of LD decay, with all variation being neutral. Alternatively, the pattern of LD in the Hausa may be due to the action of long-standing balancing selection maintaining multiple alleles. Finally, although we considered demographic models that provide a good fit to patterns of noncoding variation observed in the same population samples (Voight et al. 2005), it is possible that additional demographic models, such as complex models of population structure (Wakeley 1999), could generate spurious evidence for a hotspot in the Hausa. These explanations are discussed in greater detail below.
Despite the close correlation between LD patterns and estimates of ρ across human populations, marked differences in regions of rapid LD decay between African and non-African samples have previously been documented. For example, two large surveys of SNP variation in ethnically diverse populations showed that, at a global level, estimates of ρ are strongly correlated across populations, but regions of localized LD breakdown are not always shared (Clark et al. 2003; Evans and Cardon 2005). Moreover, when Crawford et al. (2004) specifically tested for the presence of recombination hotspots in samples of African Americans and European Americans, they found that 16 of 35 hotspots show substantive evidence in one population, but not in the other. However, additional power analyses suggested that these hotspots may actually be present in both populations, but with insufficient information in the LD data to be detected in both populations (Crawford et al. 2004). The LD pattern we observe at CAPN10 is similar to those of Crawford et al. (2004) in that we find statistical support for recombination rate heterogeneity only in the Hausa, but not in the non-African populations. In our case, however, we also have direct estimates of crossover rate obtained by sperm typing in samples of non-African ancestry. These allow us to rule out the possibility that a hotspot with similar frequency and/or intensity is present in all human populations and we simply lack the power to detect it in our non-African samples. Thus, if there is a hotspot in the Hausa, it is truly population specific, at least relative to the populations we surveyed. More generally, our results suggest that typing of sperm samples from ethnically diverse individuals may provide important insights into the evolution of hotspots and will help with the interpretation of LD patterns in human populations.
The possibility of a hotspot with sufficiently different frequency and/or intensity across populations to generate disparate LD patterns has important implications for the evolution of recombination. Previous work revealed that hotspots may vary not only between species but also across individuals. In particular, sperm-typing analysis showed that the DNA2 hotspot in the HLA region experienced different rates of recombination in individuals depending on the genotype at a SNP located near the center of the hotspot (Jeffreys and Neumann 2002). In light of these results, it is particularly interesting to note that SNP63, one of the CAPN10 variants implicated in diabetes risk, exhibits unusually large differences in allele frequencies between Africans and non-Africans (FST = 0.605) (Fullerton et al. 2002). The parallel between the divergence of SNP63 allele frequency and the marked differences in LD patterns between Africans and non-Africans raises the possibility that the allele found at high frequency in the Hausa might be associated with increased crossover rate. However, the only sperm sample heterozygous for this allele, man 3, has similar crossover rate estimates to the other samples. Thus, although we have low power to detect subtle changes in the rate of crossover in man 3, it seems unlikely that his genotype at SNP63 results in any substantial rate increase.
A caveat to the above analyses is that the evidence for a hotspot was assessed under models of random mating. Simulations of a simple two-island model showed that the estimates of ρ tend to be lower compared to those obtained under random mating (Li and Stephens 2003); this might imply that our conclusion about the presence of a hotspot in the Hausa is robust to violation of the random-mating assumption. However, the same simulations showed that the type I error under this model is 0.07 (see Table 3 in Li and Stephens 2003). If more complex models of population structure (Wakeley 1999) result in a higher false-positive rate and such models fit the Hausa polymorphism data, the pattern observed at CAPN10 may be consistent with uniform recombination rate.
The alternative interpretation, i.e., long-standing balancing selection maintaining multiple alleles, is strengthened by considering the LD data together with polymorphism levels. Indeed, while we find a substantial elevation of polymorphism coinciding with rapid LD decay in Hausa, an analysis of resequencing data from 74 genes did not find that this is a common feature of recombination hotspots (Crawford et al. 2004). Likewise, a resequencing study of the β-globin hotspot, a hotspot verified by sperm typing (Schneider 1999), also did not reveal an elevation in polymorphism levels (Wall et al. 2003). In contrast, we previously tested an explicit model of long-standing balancing selection maintaining multiple alleles by coalescent simulations. We showed that this model can explain not only the observed LD breakdown in the Hausa, but also other features of the Hausa data, including the excess polymorphism levels and the spectrum of allele frequency, without requiring changes in the underlying mutation and recombination rates (Takahata 1990; Vander Molen et al. 2005). As described by Takahata (1990), this model is specified by two parameters: M, the number of alleles at the selected site maintained at equal frequency by selection, and Q, the rate at which alleles at the selected site are lost and replaced (“turnover”). In our previous simulation analyses, we determined that the Hausa data at CAPN10 are consistent with the above selection model for high turnover rates (Q = 1) and low number of alleles (M = 5–6); because Q and M depend on the selection coefficient, varying their values implicitly varies the strength of selection (Takahata 1990; Vander Molen et al. 2005). As shown by Takahata (1990), this model of long-standing balancing selection results in a local deepening of an effectively neutral gene genealogy, hence allowing more mutation and recombination events than at neutrally evolving regions. Consistent with balancing selection maintaining multiple alleles, the gene genealogy for intron 13 of CAPN10 was shown to have five deep haplotype lineages (Clark et al. 2005). Moreover, phylogenetic shadowing of the same region revealed several moderately to highly conserved sequence segments immediately 5′ to the apex of the peak of polymorphism and LD decay (Clark et al. 2005). Because of the estimated time depth for this segment of intron 13 (∼2–3 MYA) (Vander Molen et al. 2005), the signal in the data would reflect the onset of new selective pressures in an ancestral human population living in sub-Saharan Africa and possibly acting in all populations since the dispersal out of Africa. Subsequent events during the history of non-African populations may have eroded the signature of selection on patterns of LD decay but not on polymorphism levels, thereby generating the observed excess of polymorphism in all populations and retaining the rapid LD decay only in the Hausa. A greater difference between Africans and non-Africans in LD levels compared to polymorphism levels is observed also at other genomic regions, supporting the notion that the pattern observed at CAPN10 may be due to differences in demographic history rather than in selective pressures (Frisse et al. 2001; Voight et al. 2005). In particular, bottleneck models predict a greater effect on LD levels than on polymorphism levels (Ardlie et al. 2002; Wall et al. 2002).
A model of positive selection is particularly appropriate for CAPN10 because of its role in type 2 diabetes risk. A positional cloning study identified CAPN10 as a susceptibility gene for type 2 diabetes in Mexican Americans and proposed a complex model whereby heterozygosity for two haplotypes, defined by three diagnostic variants (SNP43-INDEL19-SNP63), increased risk to disease (Horikawa et al. 2000). In that respect, it is worth noting that SNP63 (nt pos. 34,288) is located immediately 3′ to the region of excess polymorphism and LD decay; in addition, SNP30, just 1267 bp upstream of SNP63 (nt pos. 33,021), is in perfect LD (r2 > 0.8) with INDEL19 in all populations tested including Mexican American cases and controls (Fullerton et al. 2002; Clark et al. 2005). Thus, our population and sperm-typing data may be compatible with long-standing balancing selection acting on thrifty variants in an ancestral human population.
Whether our results reflect a hotspot generating population-specific patterns of LD decay or the action of long-standing balancing selection, they have interesting implications for using LD as a means of dissecting the genetic bases of common diseases and of type 2 diabetes in particular. If our results are due to a hotspot present in one population but at low frequency or absent in others, this implies that significant interethnic variation may be seen also in some fraction of the 25,000–50,000 hotspots estimated to occur in the human genome (although those estimates are based on LD data, not direct measurement of the recombination rate) (McVean et al. 2004; Myers et al. 2005). This may affect the association signals between markers and disease causative variants observed in different populations and the ability to replicate a reported association in study populations with varying ethnic composition. Likewise, if multiple SNPs at a gene influence disease risk, the repertoire of haplotypes carrying those SNPs will also vary across populations. Although there is no shortage of unreplicated association signals in the literature, additional work is necessary to determine whether any of them result from variation in hotspot frequency across populations.
If instead our results are due to natural selection, this implies that crucial changes in selective pressures acting on the biological processes underlying diabetes risk occurred early in human evolution, possibly reflecting climate and diet changes associated with the onset of the Ice Ages (Miller and Colagiuri 1994; Colagiuri and Brand Miller 2002; Vander Molen et al. 2005), and raises the possibility that a similar signature is present in other susceptibility genes for type 2 diabetes.
Acknowledgments
We thank the Riverside Zoo, the Sunset Zoo, the Detroit Zoo, the Primate Foundation of Arizona, and the New Iberia Primate Center for the Western chimpanzee samples. The chimpanzee sample collection and subspecies assignment were supported by grants from the Wenner–Gren Foundation for Anthropological Research (grant 6266) and the National Science Foundation (BCS-0073871) to A.C.S. This work was supported by National Institutes of Health (NIH) grant R01 DK56670 to A.D. G.C. was supported by NIH grant R01 HG02772. M.P. is supported by an Alfred P. Sloan fellowship in Computational Molecular Biology. V.J.C. is supported by a National Research Service Award postdoctoral fellowship (DK66974).
References
- Altshuler, D., L. D. Brooks, A. Chakravarti, F. S. Collins, M. J. Daly et al., 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie, K. G., L. Kruglyak and M. Seielstad, 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3: 299–309. [DOI] [PubMed] [Google Scholar]
- Arnheim, N., P. Calabrese and M. Nordborg, 2003. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am. J. Hum. Genet. 73: 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S., C. Fockler, W. M. Barnes and R. Higuchi, 1994. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc. Natl. Acad. Sci. USA 91: 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., R. Nielsen, J. Signorovitch, T. C. Matise, S. Glanowski et al., 2003. Linkage disequilibrium and inference of ancestral recombination in 538 single-nucleotide polymorphism clusters across the human genome. Am. J. Hum. Genet. 73: 285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, V. J., N. J. Cox, M. Hammond, C. L. Hanis and A. Di Rienzo, 2005. Haplotype structure and phylogenetic shadowing of a hypervariable region in the CAPN10 gene. Hum. Genet. 117: 258–266. [DOI] [PubMed] [Google Scholar]
- Colagiuri, S., and J. Brand Miller, 2002. The ‘carnivore connection’—evolutionary aspects of insulin resistance. Eur. J. Clin. Nutr. 56(Suppl. 1): S30–S35. [DOI] [PubMed] [Google Scholar]
- Crawford, D. C., T. Bhangale, N. Li, G. Hellenthal, M. J. Rieder et al., 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700–706. [DOI] [PubMed] [Google Scholar]
- Evans, D. M., and L. R. Cardon, 2005. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am. J. Hum. Genet. 76: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 1999. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 16: 1003–1005. [DOI] [PubMed] [Google Scholar]
- Fischer, A., V. Wiebe, S. Paabo and M. Przeworski, 2004. Evidence for a complex demographic history of chimpanzees. Mol. Biol. Evol. 21: 799–808. [DOI] [PubMed] [Google Scholar]
- Frisse, L., R. R. Hudson, A. Bartoszewicz, J. D. Wall, J. Donfack et al., 2001. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton, S. M., A. Bartoszewicz, G. Ybazeta, Y. Horikawa, G. I. Bell et al., 2002. Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. Am. J. Hum. Genet. 70: 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, L. L., M. P. Keller, W. Navidi, P. F. Chance and N. Arnheim, 2000. Unequal exchange at the Charcot-Marie-Tooth disease type 1A recombination hot-spot is not elevated above the genome average rate. Hum. Mol. Genet. 9: 1881–1889. [DOI] [PubMed] [Google Scholar]
- Horikawa, Y., N. Oda, N. J. Cox, X. Li, M. Orho-Melander et al., 2000. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26: 163–175. [DOI] [PubMed] [Google Scholar]
- Hubert, R., M. MacDonald, J. Gusella and N. Arnheim, 1994. High resolution localization of recombination hot spots using sperm typing. Nat. Genet. 7: 420–424. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., and C. A. May, 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36: 151–156. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., and R. Neumann, 2002. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 31: 267–271. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., L. Kauppi and R. Neumann, 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217–222. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., R. Neumann, M. Panayi, S. Myers and P. Donnelly, 2005. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 37: 601–606. [DOI] [PubMed] [Google Scholar]
- Kauppi, L., M. P. Stumpf and A. J. Jeffreys, 2005. Localized breakdown in linkage disequilibrium does not always predict sperm crossover hot spots in the human MHC class II region. Genomics 86: 13–24. [DOI] [PubMed] [Google Scholar]
- Li, N., and M. Stephens, 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165: 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram, P., and P. Donnelly, 1994. Pairwise comparisons of mitochondrial DNA sequences in subdivided populations and implications for early human evolution. Genetics 136: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean, G. A., S. R. Myers, S. Hunt, P. Deloukas, D. R. Bentley et al., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Miller, J. C., and S. Colagiuri, 1994. The carnivore connection: dietary carbohydrate in the evolution of NIDDM. Diabetologia 37: 1280–1286. [DOI] [PubMed] [Google Scholar]
- Myers, S., L. Bottolo, C. Freeman, G. McVean and P. Donnelly, 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Neel, J. V., 1962. Diabetes Mellitus: A “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 14: 353–362. [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D. A., V. O. Tobe and S. L. Taylor, 1997. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov, A., A. Di Rienzo and R. R. Hudson, 2002. Inferences about human demography based on multilocus analyses of noncoding sequences. Genetics 161: 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., and M. Przeworski, 2001. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S. E., A. D. Roeder, M. Stephens, Y. Gilad, S. Paabo et al., 2004. a Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S. E., K. Voelpel and M. Przeworski, 2004. b Insights into recombination from patterns of linkage disequilibrium in humans. Genetics 167: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S. E., D. A. Hinds, K. Koehler, B. Nickel, N. Patil et al., 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37: 429–434. [DOI] [PubMed] [Google Scholar]
- Reed, F. A., and S. A. Tishkoff, 2006. Positive selection can create false hotspots of recombination. Genetics 172: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch, N., and K. Merikangas, 1996. The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- Schneider, J. A., 1999. Genetic recombination in the human beta-globin gene cluster. Ph.D. Thesis, University of Oxford, Oxford.
- Serre, D., R. Nadon and T. J. Hudson, 2005. Large-scale recombination rate patterns are conserved among human populations. Genome Res. 15: 1547–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N. G., and P. Fearnhead, 2005. A comparison of three estimators of the population-scaled recombination rate: accuracy and robustness. Genetics 171: 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., Y. S. Song and C. H. Langley, 2006. The hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics 172: 2647–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, A. C., R. C. Griffiths, S. L. Zegura and M. F. Hammer, 2002. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc. Natl. Acad. Sci. USA 99: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. a DNA polymorphism in a subdivided population: the expected number of segregating sites in the two-subpopulation model. Genetics 123: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. b Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., 1990. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87: 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Molen, J., L. M. Frisse, S. M. Fullerton, Y. Qian, L. del Bosque-Plata et al., 2005. Population genetics of CAPN10 and GPR35: implications for the evolution of type 2 diabetes variants. Am. J. Hum. Genet. 76: 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., A. M. Adams, L. A. Frisse, Y. Qian, R. R. Hudson et al., 2005. Interrogating multiple aspects of variation in a full re-sequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. USA 102: 18508–18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley, J., 1999. Non-equilibrium migration in human evolution. Genetics 153: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley, J., R. Nielsen, S. N. Liu-Cordero and K. Ardlie, 2001. The discovery of single-nucleotide polymorphisms—and inferences about human demographic history. Am. J. Hum. Genet. 69: 1332–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., 2004. Estimating recombination rates using three-site likelihoods. Genetics 167: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., L. A. Frisse, R. R. Hudson and A. Di Rienzo, 2003. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am. J. Hum. Genet. 73: 1330–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Winckler, W., S. R. Myers, D. J. Richter, R. C. Onofrio, G. J. McDonald et al., 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308: 107–111. [DOI] [PubMed] [Google Scholar]
- Yu, J., L. Lazzeroni, J. Qin, M. M. Huang, W. Navidi et al., 1996. Individual variation in recombination among human males. Am. J. Hum. Genet. 59: 1186–1192. [PMC free article] [PubMed] [Google Scholar]
- Yu, N., M. I. Jensen-Seaman, L. Chemnick, J. R. Kidd, A. S. Deinard et al., 2003. Low nucleotide diversity in chimpanzees and bonobos. Genetics 164: 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]