Abstract
Barramundi (Lates calcarifer) is an important farmed marine food fish species. Its compact genome (∼700 Mb) is among the smallest genomes of food fish species. We established a first-generation genetic linkage map of Barramundi with a mapping panel containing three parents (two males and one female) and 93 progeny. A total of 240 microsatellite markers were mapped into 24 linkage groups. Among these markers, 10 were located in ESTs and known genes. The total lengths of the female and male maps were 873.8 and 414.5 cM with an average marker spacing of 6.20 and 4.70 cM, respectively. Comparing the flanking sequences of the 240 Barramundi microsatellites with the assembled whole-genome sequences of Tetraodon nigrovidiris revealed 55 homologous sequences located in 19 of the 21 chromosomes of T. nigrovidiris. The map will not only enable the mapping of quantitative trait loci, but also provide new resources for understanding the evolution of fish genomes.
BARRAMUNDI (Lates calcarifer), also called Asian sea bass, is one of the nine Lates species of the family Latidae and is widely distributed in the coastal and freshwater of the tropical Indo-west Pacific from the Persian Gulf to India to northern Australia (Nelson 1994; Berra 2001). Barramundi reach sexual maturity at the age of 2–3 years and are protandrous; that is, the fish mature first as males and become females when they grow older and larger. At each spawning, females produce >1 million eggs. The genome size of Barramundi is rather small with a haploid C-value of 0.7 pg, according to the Animal Genome Size Database (http://www.genomesize.com/fish.htm). The genome of Barramundi contains 24 chromosome pairs (Carrey and Mather 1999). Barramundi can be raised easily under captive conditions and in controlled aquatic laboratories. This fish is becoming an important farmed marine food fish species with global annual production of nearly 400,000 metric tons according to Food and Agriculture Organization of the United Nations statistics. Although Barramundi has been cultured for >20 years, no detailed selective breeding program has been reported. With the strong support of the Singapore government, we have conducted a selective breeding program of Barramundi since April 2003. Such a selective breeding program requires the development of large number of DNA markers for the management of stocks, detection of quantitative trait loci (QTL), and genetic improvement through marker-assisted selection.
Genetic linkage maps are essential in the localization of QTL for marker-assisted selection (Dekkers and Hospital 2002). Linkage maps have been constructed for almost all economically important farm animals (Andersson and Georges 2004) and model vertebrate species. Although fish (∼25,000 species) compose half of all known vertebrate species (Nelson 1994), linkage maps are available for only a few fish species, including zebrafish (Gates et al. 1999), medaka (Kimura et al. 2005), tiger pufferfish (Kai et al. 2005), European sea bass (Chistiakov et al. 2005), tilapia (Kocher et al. 1998; Lee et al. 2005), rainbow trout (Sakamoto et al. 2000), and salmon (Gilbey et al. 2004).
Microsatellites are short (1–6 bp) repetitive DNA sequences, which are highly abundant and almost evenly distributed in genomes (Weber and May 1989). Because of their high abundance and polymorphism as well as the ease of scoring by PCR, microsatellites have been extensively used in the construction of linkage maps (Murray et al. 1994; Kappes et al. 1997; Lindgren et al. 1998). Linkage maps based on microsatellites in fish are relatively scarce (Gates et al. 1999; Chistiakov et al. 2005; Lee et al. 2005). In this article, we report a first-generation microsatellite-based linkage map in Barramundi that will provide an indispensable tool for selective breeding, for mapping economically important QTL for this species, and for comparative genomic studies.
MATERIALS AND METHODS
Microsatellites:
Seventy-six microsatellites selected from published data (Yue et al. 2002; Sim and Othman 2005), together with 18 microsatellites derived from known genes (Yue et al. 2001) and 4800 ESTs sequences (Xu et al. 2006), were included in this study. Additional microsatellites were isolated from microsatellite-enriched libraries. Briefly, three partial genomic DNA libraries enriched for CA, GA, and CAA repeats were constructed according to the protocol of Fischer and Bachmann (1998) with some modifications (Yue et al. 2000). Repeat-enriched DNA fragments of 400–1200 bp in length were cloned into pGEM-T vector (Promega, San Luis Obispo, CA), and transformed into XL-10 blue supercompetent cells (Stratagene, La Jolla, CA). The libraries were arrayed into 96-well plates for bidirectional sequencing on an ABI3730xl DNA sequencer (ABI, Foster City, CA) using the BigDye V3.0 kit and M13 and M13 reverse primers (Zhu et al. 2005). Redundant and overlapping sequences were grouped using Sequencher (Gene Codes, Ann Arbor, MI). Unique sequences were compared to known microsatellite sequences of Barramundi prior to primer design to remove redundancy. Sequences containing CA >7, GA >7, and CAA >5 were subjected to primer design using PrimerSelect (DNASTAR, Brighton, MA), targeting a product size between 100 and 400 bp.
Mapping panel:
A whole broodstock containing 94 brooders, including 48 males and 46 females collected from the wild in Southeast Asia 4 years ago, were genotyped with nine polymorphic microsatellites as described previously (Zhu et al. 2006). One female and two male brooders were selected for constructing a mapping panel because of their high allelic diversity and genetic differences. By crossing the female and two male brooders, millions of eggs were produced. A total of 47 and 46 full-sib progeny were randomly collected from the two full-sib families, respectively. Fin clips of the three parents were collected and kept in absolute ethanol, whereas the whole body of each offspring at the age of 90 days posthatch was cut into small pieces, soaked in absolute ethanol, and kept in a −80° freezer. DNA was isolated from tissues using a new method that we developed (Yue and Orban 2005). The quality of DNA was checked on 1% agarose gel, and the quantity was measured using Nanodrop (NanoDrop, Wilmington, DE). The DNA concentration of each fish was adjusted to 2.5 ng/μl and was arrayed into 96-well PCR plates for later use. The 93 F1 individuals and their three parents were genotyped for linkage analysis.
Genotyping:
Primers were designed for each unique sequence using PrimerSelect (DNASTAR, Madison, WI). One primer of each pair was labeled with FAM or HEX fluorescent dyes at the 5′-end. The PCR program for microsatellite amplifications on PTC-100 PCR machines (MJ Research, Santa Cruz, CA) consisted of the following steps: 94° for 2 min followed by 37 cycles of 94° for 30 sec, 55° for 30 sec, and 72° for 45 sec, with a final step of 72° for 5 min. Each PCR reaction consisted of 1× PCR buffer (Finnzymes, Espoo, Finland) with 1.5 mm MgCl2, 200 nm of each PCR primer, 50 μm of each dNTP, 10 ng genomic DNA and 1 unit of DNA–polymerase (Finnzymes, Espoo, Finland). Products were analyzed using the DNA sequencer ABI3730xl, and genotyping was carried out to determine fragment size against the size standard GS-ROX-500 (Applied Biosystems, Foster City, CA) with software GeneMapper V3.5 (Applied Biosystems) (Zhu et al. 2006).
Linkage mapping:
Linkage analyses were conducted through a series of pairwise comparisons between loci using LINKMFEX version 1.5 (available at http://www.uoguelph.ca/∼rdanzman/software/LINKMFEX). The analyses were executed using segregation data for all 250 microsatellite markers in 93 progeny. Pairwise recombination estimates obtained with module LINKMFEX were used as input into module MAPORD to determine linear assignments of markers within a linkage group (LG). All linkage maps reported here have been constructed using sex-specific data (i.e., data generated from the female or the male parent) and a minimum LOD score of 3.0 to assign markers to linkage groups. Map distances were calculated using the Kosambi function with module MAPDIS. The linkage maps were constructed with the two full-sib families, respectively. Using the module MERGE, new linkage group orders were built for preexisting linkage group orders that were obtained from the two full-sib families. Map construction was not averaged across the sexes due to the large differences in recombination rate detected between sexes (Johnson et al. 1987); instead, sex-specific linkage maps were generated. Map graphics were drawn with MapMaker software (Lander et al. 1987).
Comparative mapping:
The fish species Tetraodon nigrovidis was used for comparative mapping, because its whole genome has been sequenced, and DNA sequences were assembled for each chromosome (Jaillon et al. 2004). Furthermore, the phylogenetic tree including >30 fish species, constructed using 12 mitochondrial genes, showed that Barramundi is more closely related to T. nigrovidis than to zebrafish and medaka (Lin et al. 2006). Comparison of flanking sequences of each Barramundi microsatellite with the assembled genomic DNA sequences of the T. nigrovidis (Jaillon et al. 2004) was conducted using BLASTn (http://www.genoscope.cns.fr/blat-server/cgi-bin/tetraodon/webBlat).
RESULTS AND DISCUSSION
Microsatellite markers:
From the three partial genomic DNA libraries enriched for CA, GA, and CAA repeats, 4800 clones were sequenced in both directions. A total of 2361 clones contained repeat DNA sequences of CA >7 or GA >7 or AAC >5, yielding 1754 unique sequences with microsatellites. Among the 1754 sequences, 1208 had enough flanking regions for primer design. Primers were designed for 306 microsatellites. Together with 18 microsatellites from genes (Yue et al. 2001) and ESTs (Xu et al. 2006) and 76 microsatellites published previously (Yue et al. 2002; Sim and Othman 2005; Zhu et al. 2006), a total of 400 primer pairs were tested on the three parents of the mapping panel containing two full-sib families to check the informativeness of markers for mapping. In one full-sib family, 194 markers were informative, while in another full-sib family, 184 markers were informative. A total of 250 microsatellite loci were informative in the mapping panel. Of the 250 markers, 10 were located in known genes (Lca001 in IGF-II, Lca002 in IGF-II, and Lca003 in GH, PVALB1, and TH-I) and ESTs (LcaE01, LcaE02, LcaE10, LcaE19, and LcaE20), while 240 were isolated from genomic DNA libraries. Details about primer sequences, GenBank accession number, annealing temperature for PCR, PCR product size, and locations of the 250 markers are summarized in supplemental Table 1 at http://www.genetics.org/supplemental/. The large collection of microsatellite markers is not only useful for mapping, but also critical for studies on genetic diversity, population structure, and brood stock management.
Linkage map:
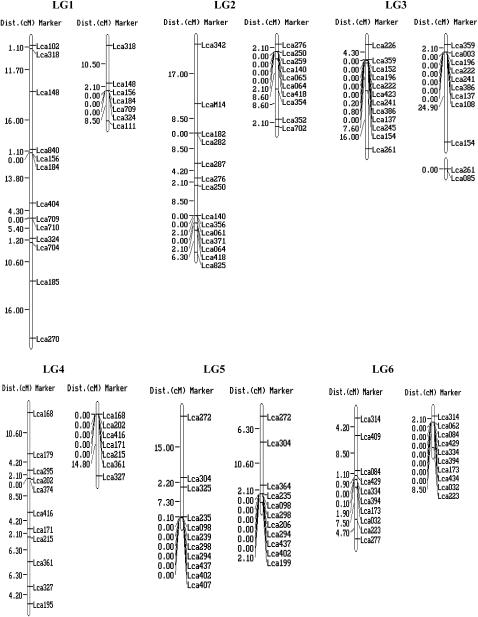
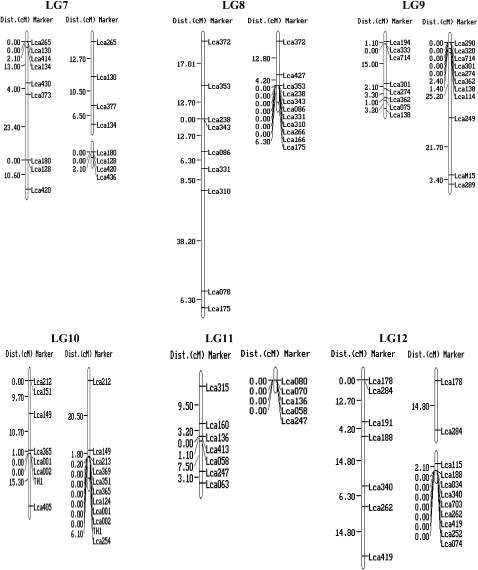
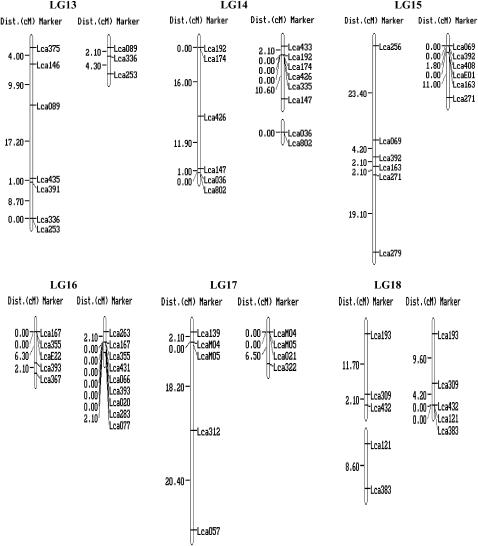
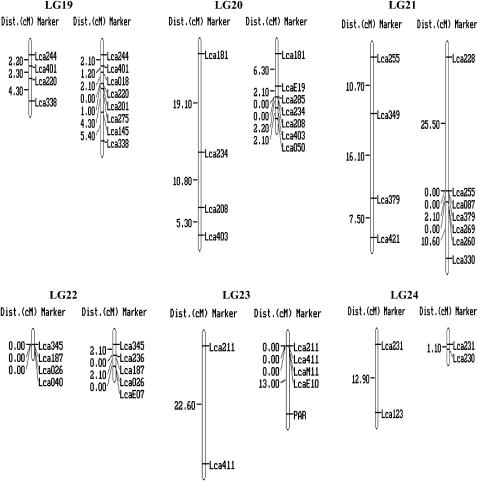
For each of the 250 markers, genotypes were obtained for all 93 offspring. Two microsatellites (Lca122 and Lca127) generated four alleles instead of two. As it is impossible to score them correctly, these two markers were excluded from the linkage analysis. Genotype data of the remaining 248 markers were passed forward into linkage analysis. As a result, 240 microsatellites, including the 10 gene-based markers, were assembled into 24 linkage groups (Figure 1), ranging in length from 1.1 to 101.7 cM (Table 1). The remaining eight microsatellites markers were unlinked to the other markers tested (supplemental Table 1 at http://www.genetics.org/supplemental/). Most of the markers showed normal Mendelian segregation. Segregation ratios that departed from the Mendelian expectation at P < 0.05 were detected at 23 markers in the female map and 16 in the male map as indicated by asterisks in supplemental Table 1 at http://www.genetics.org/supplemental/. Interestingly, most of the segregation distortion markers were clustered on LG5, -13, and -15 in the female map, and on LG5 and -12 in the male map. It can be deduced that the suggested segregation distortion loci at these marker regions might link to deleterious alleles. If these loci in the regions are removed from the analysis, <5% of the remaining markers show significant distortion, as expected by chance. Similar phenomena were reported in the linkage maps of Tilapia (Lee et al. 2005) and rainbow trout (Sakamoto et al. 2000).
Figure 1.—
A genetic linkage map of Barramundi based on microsatellite markers (left, the female map; right, the male map). Estimates of map distances between markers are indicated in Kosambi centimorgans.
TABLE 1.
Number of markers and genetic length for each linkage group
| No. of markers | Female
|
Male
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| LG | LG | No. of markers | Length (cM) | cM/marker | LG | No. of markers | Length (cM) | cM/marker | |
| LG1 | 14 | LG1 | 13 | 81.2 | 6.25 | LG1 | 7 | 21.1 | 3.01 |
| LG2 | 19 | LG2 | 14 | 59.3 | 4.24 | LG2 | 9 | 23.5 | 2.35 |
| LG3 | 15 | LG3 | 12 | 28.9 | 2.41 | LG3-M1 | 9 | 27 | 3 |
| LG3-M2 | 2 | 0 | 0 | ||||||
| LG4 | 11 | LG4 | 11 | 48.5 | 4.41 | LG4 | 7 | 14.8 | 2.11 |
| LG5 | 14 | LG5 | 11 | 24.6 | 2.24 | LG5 | 11 | 21.1 | 1.92 |
| LG6 | 12 | LG6 | 10 | 28.9 | 2.89 | LG6 | 10 | 10.6 | 1.06 |
| LG7 | 11 | LG7 | 9 | 53.1 | 5.90 | LG7-M1 | 4 | 29.7 | 7.43 |
| LG7-M2 | 4 | 2.1 | 0.53 | ||||||
| LG8 | 12 | LG8 | 9 | 101.7 | 11.30 | LG8 | 10 | 23.3 | 2.33 |
| LG9 | 14 | LG9 | 8 | 25.7 | 3.21 | LG9 | 11 | 54.1 | 4.92 |
| LG10 | 13 | LG10 | 8 | 36.7 | 4.59 | LG10 | 11 | 28.6 | 2.6 |
| LG11 | 9 | LG11 | 7 | 26.5 | 3.31 | LG11 | 5 | 0 | 0 |
| LG12 | 12 | LG12 | 7 | 52.8 | 7.54 | LG12 | 11 | 16.9 | 1.54 |
| LG13 | 7 | LG13 | 7 | 40.8 | 5.83 | LG13 | 3 | 6.4 | 2.13 |
| LG14 | 8 | LG14 | 6 | 28.9 | 4.82 | LG14-M1 | 6 | 12.7 | 2.12 |
| LG14-M2 | 2 | 0 | 0 | ||||||
| LG15 | 8 | LG15 | 6 | 50.9 | 8.48 | LG15 | 6 | 12.8 | 2.13 |
| LG16 | 11 | LG16 | 5 | 8.4 | 1.68 | LG16 | 9 | 4.2 | 0.47 |
| LG17 | 7 | LG17 | 5 | 40.7 | 8.14 | LG17 | 4 | 6.5 | 1.63 |
| LG18 | 5 | LG18-F1 | 3 | 13.8 | 4.60 | LG18 | 5 | 13.8 | 2.76 |
| LG18-F2 | 2 | 8.6 | 4.30 | ||||||
| LG19 | 8 | LG19 | 4 | 8.8 | 2.20 | LG19 | 8 | 16.1 | 2.01 |
| LG20 | 7 | LG20 | 4 | 35.2 | 8.80 | LG20 | 7 | 12.7 | 1.81 |
| LG21 | 9 | LG21 | 4 | 34.3 | 8.58 | LG21 | 11 | 38.2 | 3.47 |
| LG22 | 6 | LG22 | 4 | 0 | 0.00 | LG22 | 5 | 4.2 | 0.84 |
| LG23 | 5 | LG23 | 2 | 22.6 | 11.30 | LG23 | 5 | 13 | 2.6 |
| LG24 | 3 | LG24 | 2 | 12.9 | 6.45 | LG24 | 2 | 1.1 | 0.55 |
| Total | 240 | 174/144a | 873.8 | 5.0/6.20b | 184/88a | 414.5 | 2.3/4.70b | ||
M1, linkage group for male 1; M2, linkage group for male 2.
Data are shown as number of markers mapped/unique locations.
Data are shown as centimorgans/marker and centimorgans/unique marker location.
The female map consisted of 25 linkage groups with 174 markers located in 144 unique positions (Figure 1 and Table 1). The maps spanned 873.8 cM, with an average distance between two markers of 6.2 cM. The sizes of linkage groups ranged from 0 to 101.7 cM (mean 36.4 cM). The number of markers per linkage group varied from 2 to 14, with an average of 7.3 markers per group.
The male map consisted of 27 linkage groups with 184 markers located in 88 unique positions (Figure 1 and Table 1). The map spanned 414.5 cM and the average spacing between two markers is 4.7 cM. The sizes of linkage groups ranged from 0 to 54.1 cM (mean 17.3 cM). The number of markers per linkage group varied from 2 to 11, with an average of 7.7 markers/group. A large portion of markers (96) were mapped at the same locations in different linkage groups, which might be due to the relatively small number (93) of F1 individuals used for mapping. Similar phenomenon occurred on the linkage map of the European sea bass Dicentrarchus labrax L., which was constructed with 50 F1 offspring (Chistiakov et al. 2005). This problem could be solved by genotyping more F1 individuals. However, genotyping of more F1 individuals increases the cost substantially.
By using markers that were polymorphic among the three parents, homologous pairs of linkage groups of the male and female parents were identified, and then these maps were arranged in 24 linkage groups, designated LG1–LG24 (Figure 1 and Table 1). Ultimately, a total of the 240 markers were employed to successfully condense the current maps into 24 linkage groups corresponding to the number of chromosome pairs in Barramundi (Carrey and Mather 1999). Among the 240 mapped DNA markers, 10 were located in genes and ESTs. More type I markers are required to be developed and mapped into linkage groups of Barramundi. SNPs and microsatellites in ESTs and genes would be the best choice for such a purpose.
The total length of the 24 linkage groups is smaller than that of tilapia (1311 cM) (Lee et al. 2005), rainbow trout (∼2627.5 cM) (Young et al. 1998), common carp (4500 cM) (Sun and Liang 2004), catfish (∼1958–1993 cM) (Waldbieser et al. 2001; Liu et al. 2003), medaka (1401.5 cM) (Naruse et al. 2004), yellowtails (∼901.7–1715.3 cM) (Ohara et al. 2005), salmon (99 and 39 M for female and male) (Woram et al. 2004), and zebrafish (3011 cM) (Kelly et al. 2000), but is somewhat similar to that of European sea bass (905.5 and 564.7 cM in the female and male maps) (Chistiakov et al. 2005) and Japanese tiger pufferfish (1213.5 and 697.1 cM in the female and male maps) (Kai et al. 2005). The short length of the Barramundi linkage maps is in agreement with its small haploid C-value (0.7 pg), indicating that Barramundi has the smallest genome among the economically important food fish species. For QTL analysis, the required intermarker distance is generally <20 cM (Dekkers and Hospital 2002). The overall 6.2 and 4.7 cM average marker distances on female and male maps offer sufficient marker density for genetic dissection of quantitative traits. Male-specific reduction in the recombination ratio found in Barramundi can be used to facilitate efficient experimental design in genomewide linkage analysis (Lander and Schork 1994; Glazier et al. 2002; Singer et al. 2002). A decreased rate of recombination in males is an advantage for mapping genetic traits in preliminary low-resolution analysis, especially when analyzing QTL with minor effect (Glazier et al. 2002). On the other hand, using a higher frequency of recombination in females would be a better choice for fine mapping of these loci.
Differences in recombination between sexes:
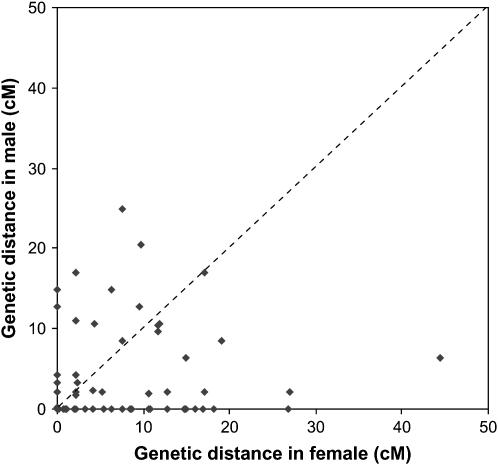
The sexes showed substantial differences in recombination rate for the same pairs of linked markers; individual pairwise female:male recombination differences varied from infinity to 0.0. Figure 2 shows a comparison of male and female recombination ratios between the common intervals flanked by paired markers from all the linkage groups. The number of intervals that showed a higher recombination ratio in females was larger than those in males. Summing up the length of the common interval for each linkage group on both the male and female maps gave a total length of 260.9 and 536.5 cM, respectively. Thus, the recombination rates in the female were estimated to be 2.06 times higher than in the male. Different recombination rates between the sexes have been reported in mammals (Dib et al. 1996) and other fish species (Woram et al. 2004) with female map distances usually greater than those in male maps (Dib et al. 1996; Sakamoto et al. 2000). There are, however, exceptions where in LG9, -19, and -21 of Barramundi, the recombination rates in some marker intervals are higher in males than in females. Similar phenomena were observed in trout and zebrafish (Sakamoto et al. 2000; Singer et al. 2002).
Figure 2.—
Differences in recombination ratio between males and females among adjacently paired markers on all 24 linkage groups.
Comparative mapping:
Flanking sequences of 55 microsatellites showed high similarity (e-values >10−8) to known genomic DNA sequences of T. nigroviridis. The distribution of the 55 hits on 19 of the 21 chromosomes of T. nigroviridis is shown in Table 2. Unique correspondences were detected in six chromosome pairs (LG3–CH21, LG4–CH15, LG5–CH18, LG9–CH4, LG10–CH13, and LG12-CH11), while the markers located on LG17 of Barramundi did not hit any T. nigroviridis sequences. Although genetic linkage maps have been constructed for some fish species, including zebrafish (Kelly et al. 2000), medaka (Naruse et al. 2004), fugu (Kai et al. 2005), rainbow trout (Young et al. 1998), Atlantic salmon (Woram et al. 2004), catfish (Waldbieser et al. 2001; Liu et al. 2003), tilapia (Lee et al. 2005), common carp (Sun and Liang 2004), and European sea bass (Chistiakov et al. 2005), comparative mapping among them was difficult as some of these maps were constructed using nonsequenced markers (e.g., AFLP and RAPD markers) and a few type I markers. Whole-genome sequencing has been started with four fish species: zebrafish, Fugu, T. nigroviridis, and medaka. The developed linkage map of Barramundi, coupled with the whole-genome sequences of the four model fish species, would provide new insights into the evolution of fish chromosomes. However, in the current linkage map of Barramundi, there are only 10 type I markers; more type I markers (i.e., SNP and microsatellites in genes) are needed for comparative mapping to gain new insights into the evolution of fish chromosome. We are in the process in developing more type I markers, which will be mapped to the linkage map in the near future.
TABLE 2.
Oxford plot comparing the linkage maps of L. calcarifer and T. nigroviridis
|
T. nigroviridis chromosome
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. calcarifer | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| LG1 | 2 | ||||||||||||||||||||
| LG2 | 5 | ||||||||||||||||||||
| LG3 | 1 | 1 | 1 | 1 | |||||||||||||||||
| LG4 | 3 | ||||||||||||||||||||
| LG5 | 3 | ||||||||||||||||||||
| LG6 | 2 | ||||||||||||||||||||
| LG7 | 2 | 1 | |||||||||||||||||||
| LG8 | 1 | 1 | 1 | ||||||||||||||||||
| LG9 | 1 | ||||||||||||||||||||
| LG10 | 6 | ||||||||||||||||||||
| LG11 | 2 | 1 | |||||||||||||||||||
| LG12 | 4 | ||||||||||||||||||||
| LG13 | 1 | ||||||||||||||||||||
| LG14 | 1 | ||||||||||||||||||||
| LG15 | 2 | 1 | |||||||||||||||||||
| LG16 | 3 | ||||||||||||||||||||
| LG17 | |||||||||||||||||||||
| LG18 | 1 | ||||||||||||||||||||
| LG19 | 1 | ||||||||||||||||||||
| LG20 | 1 | ||||||||||||||||||||
| LG21 | 1 | ||||||||||||||||||||
| LG22 | 1 | ||||||||||||||||||||
| LG23 | 1 | 1 | |||||||||||||||||||
| LG24 | 1 | ||||||||||||||||||||
Numbers indicate the number of L. calcarifer markers with BLASTn hits to particular Tetraodon chromosomes. Italic numbers indicate linkage groups with unique correspondences.
Barramundi as a model for studying complex traits in aquatic environments:
Fishes are a diverse group of >25,000 species placed in 42 orders, 431 families, and 4075 genera (Nelson 1994). Because of the enormous species diversity involved, the ancient origin that goes back ∼400 million years ago, and the wide-ranging variations not only in morphology but also in behavior, ecology, and physiology, fish offer unique systems for genomic studies. Despite high levels of conservation in both genomes and biological functions, the extent to which fish can adapt to various environments is beyond imagination for any higher vertebrates such as mammals. Therefore, genomic studies using a fish species provide unique scientific information concerning genetic mechanisms governing performance traits in aquatic environments in relation to genome evolution. Teleost fishes such as cichlid and stickleback have been suggested as model vertebrate species for identifying the genetic variation responsible for phenotypic differences among related species (Cresko et al. 2004; Kocher 2004). As a teleost, the Barramundi could be an excellent model organism for genomic studies due to its compact genome (∼700 Mb), particularly for studying the mechanisms underlying the complex traits in marine aquatic environments. Its compact genome makes the genomewide scan for QTL affecting complex traits much easier than that in other fish species with a bigger genome. Moreover, Barramundi's extremely high fecundity allows for the production of a large number of offspring under controlled laboratory conditions. However, one principle drawback of Barramundi as a model organism is the lack of whole-genome sequences, a common problem of all food fish species. Nevertheless, the Barramundi may be a good candidate for future whole-genome sequencing due to its compact genome.
In conclusion, the first generation of the linkage map of Barramundi containing 240 microsatellites has been constructed. The map will not only facilitate selective breeding and mapping of QTL, but also provide new data for comparative genomic studies.
Acknowledgments
The authors thank L. Orban, R. Chou, H. S. Lim, J. Tan, H. Y. Pang, and H. M. Liu for their support in the project. We are grateful to the members of the Animal Genome Community for their suggestions and information about software for linkage analyses. This study is part of the project “Molecular Breeding of Marine Food Fish” funded by Agri-Food and Veterinary Authority of Singapore.
References
- Andersson, L., and M. Georges, 2004. Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet. 5: 202–212. [DOI] [PubMed] [Google Scholar]
- Berra, T. M., 2001. Freshwater Fish Distribution. Academic Press, San Diego.
- Carrey, G., and P. Mather, 1999. Karyotypes of four species: Melanotaenia duboulayi, Bidyanus bidyanus, Macquaria novemaculeata and Lates calcarifer. Cytobios 100: 137–146. [Google Scholar]
- Chistiakov, D. A., B. Hellemans, C. S. Haley, A. S. Law, C. S. Tsigenopoulos et al., 2005. A microsatellite linkage map of the European sea bass Dicentrarchus labrax L. Genetics 170: 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko, W. A., A. Amores, C. Wilson, J. Murphy, M. Currey et al., 2004. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA 101: 6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers, J. C. M., and F. Hospital, 2002. Multifactoral genetics: the use of molecular genetics in the improvement of agricultural populations. Nat. Rev. Genet. 3: 22–32. [DOI] [PubMed] [Google Scholar]
- Dib, C., S. Faure, C. Fizames, D. Samson, N. Drouot et al., 1996. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380: 152–154. [DOI] [PubMed] [Google Scholar]
- Fischer, D., and K. Bachmann, 1998. Microsatellite enrichment in organisms with large genomes (Allium cepa L.). Biotechniques 24: 796–802. [DOI] [PubMed] [Google Scholar]
- Gates, M. A., L. Kim, E. S. Egan, T. Cardozo, H. I. Sirotkin et al., 1999. A genetic linkage map for zebrafish: comparative analysis and localization of genes and expressed sequences. Genome Res. 9: 334–347. [PubMed] [Google Scholar]
- Gilbey, J., E. Verspoor, A. McLay and D. Houlihan, 2004. A microsatellite linkage map for Atlantic salmon (Salmo salar). Anim. Genet. 35: 98–105. [DOI] [PubMed] [Google Scholar]
- Glazier, A. M., J. H. Nadeau and T. J. Aitman, 2002. Finding genes that underlie complex traits. Science 298: 2345–2349. [DOI] [PubMed] [Google Scholar]
- Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann et al., 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957. [DOI] [PubMed] [Google Scholar]
- Johnson, K. R., J. E. Wright and B. May, 1987. Linkage relationships reflecting ancestral tetraploidy in salmonid fish. Genetics 116: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, W., K. Kikuchi, M. Fujita, H. Suetake, A. Fujiwara et al., 2005. A genetic linkage map for the tiger pufferfish, Takifugu rubripes. Genetics 171: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes, S. M., J. W. Keele, R. T. Stone, T. S. Sonstegard, T. P. L. Smith et al., 1997. A second-generation linkage map of the bovine genome. Genome Res. 7: 235–249. [DOI] [PubMed] [Google Scholar]
- Kelly, P. D., F. Chu, I. G. Woods, P. Ngo-Hazelett, T. Cardozo et al., 2000. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 10: 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, T., K. Yoshida, A. Shimada, T. Jindo, M. Sakaizumi et al., 2005. Genetic linkage map of medaka with polymerase chain reaction length polymorphisms. Gene 363: 24–31. [DOI] [PubMed] [Google Scholar]
- Kocher, T. D., 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 5: 288–298. [DOI] [PubMed] [Google Scholar]
- Kocher, T. D., W. J. Lee, H. Sobolewska, D. Penman and B. McAndrew, 1998. A genetic linkage map of a cichlid fish, the tilapia (Oreochromis niloticus). Genetics 148: 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and N. J. Schork, 1994. Genetic dissection of complex traits. Science 265: 2037–2048. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lee, B. Y., W. J. Lee, J. T. Streelman, K. L. Carleton, A. E. Howe et al., 2005. A second-generation genetic linkage map of tilapia (Oreochromis spp.). Genetics 170: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, G., L. C. Lo, Z. Y. Zhu, F. Feng, R. Chou et al., 2006. The complete mitochondrial genome sequence and characterization of single-nucleotide polymorphisms in the control region of the Asian seabass (Lates calcarifer). Mar. Biotechnol. 8: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren, G., K. Sandberg, H. Persson, S. Marklund, M. Breen et al., 1998. A primary male autosomal linkage map of the horse genome. Genome Res. 8: 951–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., A. Karsi, P. Li, D. Cao and R. Dunham, 2003. An AFLP-based genetic linkage map of channel catfish (Ictalurus punctatus) constructed by using an interspecific hybrid resource family. Genetics 165: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J. C., K. H. Buetow, J. L. Weber, S. Ludwigsen, T. Scherpbier-Heddema et al., 1994. A comprehensive human linkage map with centimorgan density. Science 265: 2049–2054. [DOI] [PubMed] [Google Scholar]
- Naruse, K., M. Tanaka, K. Mita, A. Shima, J. Postlethwait et al., 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J., 1994. Fishes of the World. John Wiley & Sons, New York.
- O'Hara, E., T. Nishimura, Y. Nagakura, T. Sakamoto, K. Mushiake et al., 2005. Genetic linkage maps of two yellowtails (Seriola quinqueradiata and Seriola lalandi). Aquaculture 244: 41–48. [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, M. P., and A. S. Othman, 2005. Isolation and characterization of microsatellite DNA loci in sea bass, Lates calcarifer Bloch. Mol. Ecol. Notes 4: 873–875. [Google Scholar]
- Singer, A., H. Perlman, Y. Yan, C. Walker, G. Corley-Smith et al., 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. W., and L. Q. Liang, 2004. A genetic linkage map of common carp (Cyprinus carpio L.) and mapping of a locus associated with cold tolerance. Aquaculture 238: 165–172. [Google Scholar]
- Waldbieser, G. C., B. G. Bosworth, D. J. Nonneman and W. R. Wolters, 2001. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics 158: 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, J. L., and P. E. May, 1989. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain-reaction. Am. J. Hum. Genet. 44: 388–396. [PMC free article] [PubMed] [Google Scholar]
- Woram, R. A., C. McGowan, J. A. Stout, K. Gharbi, M. M. Ferguson et al., 2004. A genetic linkage map for Arctic char (Salvelinus alpinus): evidence for higher recombination rates and segregation distortion in hybrid versus pure strain mapping parents. Genome 47: 304–315. [DOI] [PubMed] [Google Scholar]
- Xu, Y. X., Z. Y. Zhu, L. C. Lo, C. M. Wang, G. Lin et al., 2006. Characterization of two parvalbumin genes and their association with growth traits in Asian seabass (Lates calcarifer). Anim. Genet. 37: 266–268. [DOI] [PubMed] [Google Scholar]
- Young, W. P., P. A. Wheeler, V. H. Coryell, P. Keim and G. H. Thorgaard, 1998. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, G. H., and L. Orban, 2005. A simple and affordable method for high throughput DNA extraction from animal tissues for PCR. Electrophoresis 26: 3081–3083. [DOI] [PubMed] [Google Scholar]
- Yue, G. H., F. Chen and L. Orban, 2000. Rapid isolation and characterization of microsatellites from the genome of Asian arowana (Scleropages formosus, Osteoglossidae, Pisces). Mol. Ecol. 9: 1007–1009. [DOI] [PubMed] [Google Scholar]
- Yue, G., Y. Li and L. Orban, 2001. Characterization of microsatellites in the IGF-2 and GH genes of Asian seabass (Lates calcarifer). Mar. Biotechnol. 3: 1–3. [DOI] [PubMed] [Google Scholar]
- Yue, G. H., Y. Li, T. M. Chao, R. Chou and L. Orban, 2002. Novel microsatellites from Asian sea bass (Lates calcarifer) and their application to broodstock analysis. Mar. Biotechnol. 4: 503–511. [DOI] [PubMed] [Google Scholar]
- Zhu, Z. Y., L. C. Lo, G. Lin, Y. X. Xu and G. H. Yue, 2005. Isolation and characterization of polymorphic microsatellites from red coral grouper (Plectropomus maculatus). Mol. Ecol. Notes 3: 579–581. [Google Scholar]
- Zhu, Z. Y., G. Lin, L. C. Lo, Y. X. Xu, C. Renee et al., 2006. Genetic analyses of Asian seabass stocks using novel polymorphic microsatellites. Aquaculture 256: 167–173. [Google Scholar]